Abstract
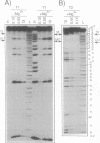
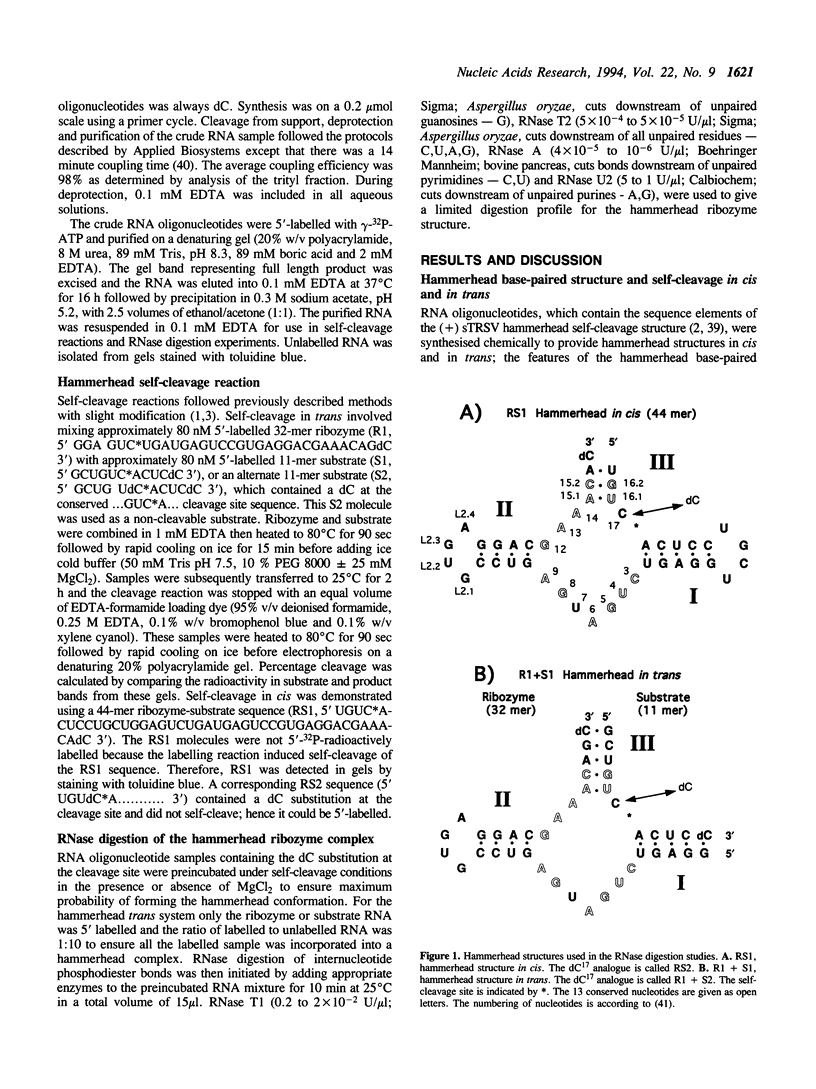
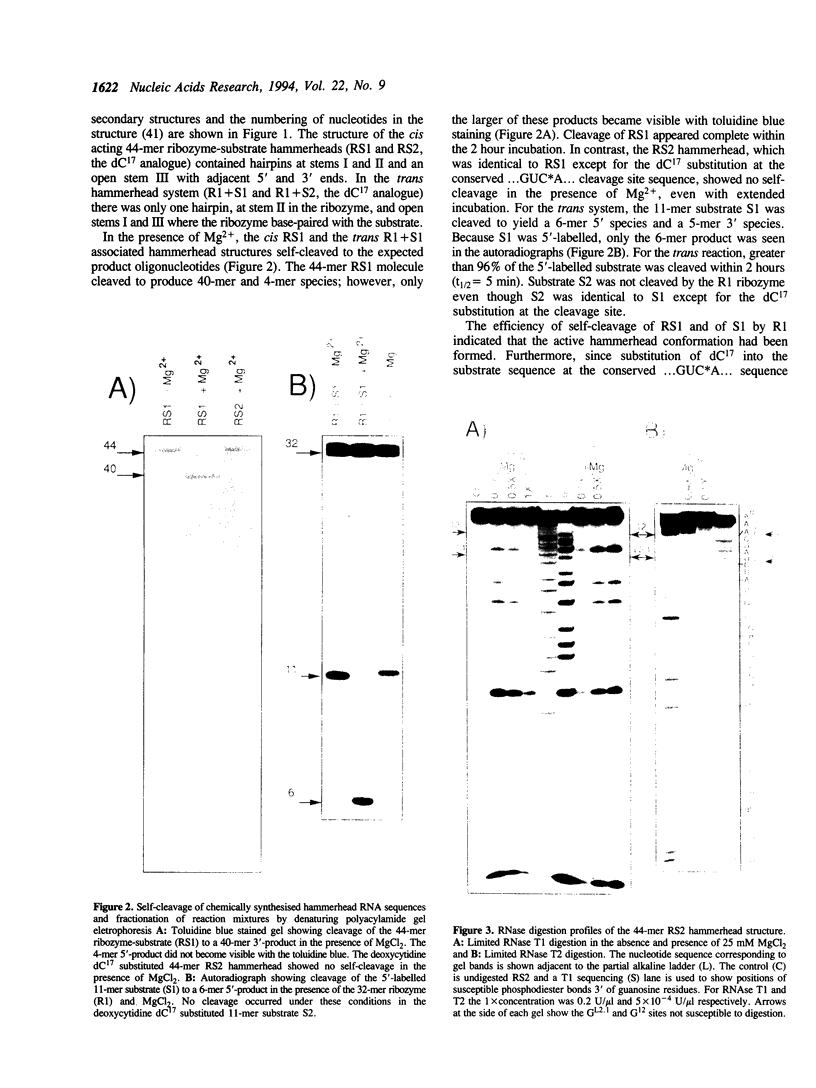
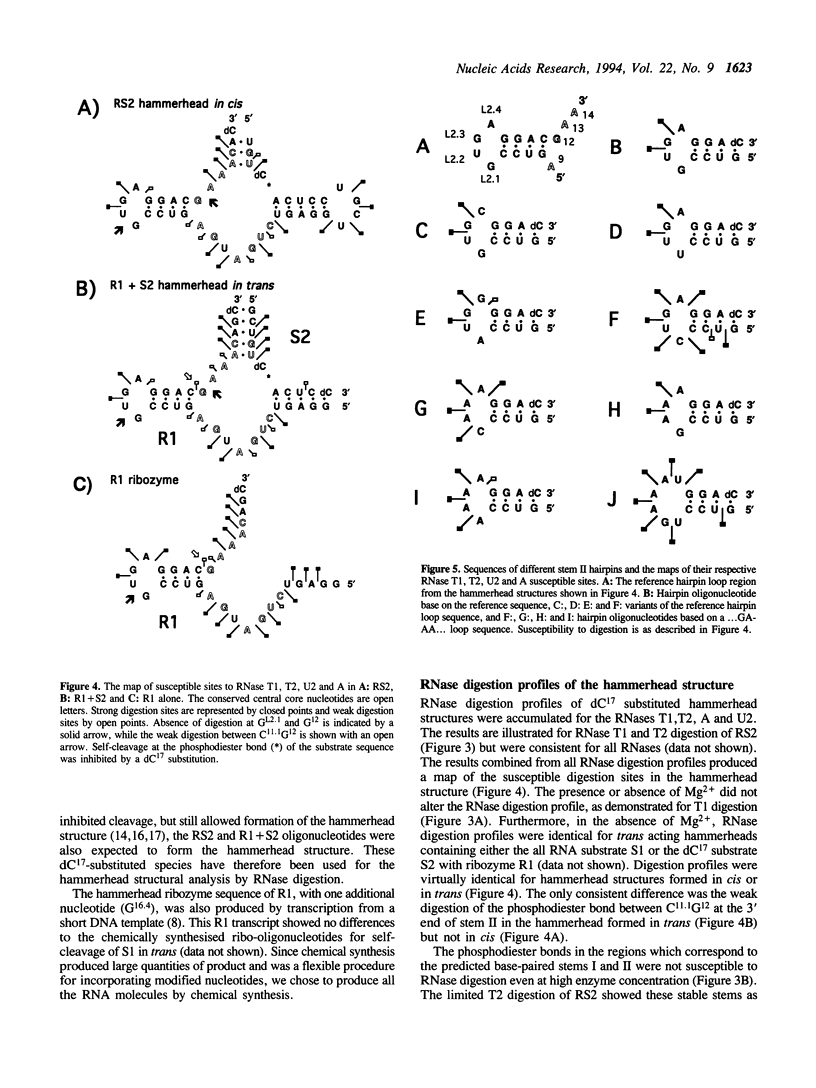
Susceptibility to RNase digestion has been used to probe the conformation of the hammerhead ribozyme structure prepared from chemically synthesised RNAs. Less than about 1.5% of the total sample was digested to obtain a profile of RNase digestion sites. The observed digestion profiles confirmed the predicted base-paired secondary structure for the hammerhead. Digestion profiles of both cis and trans hammerhead structures were nearly identical which indicated that the structural interactions leading to self-cleavage were similar for both systems. Furthermore, the presence or absence of Mg2+ did not affect the RNase digestion profiles, thus indicating that Mg2+ did not modify the hammerhead structure significantly to induce self-cleavage. The base-paired stems I and II in the hammerhead structure were stable whereas stem III, which was susceptible to digestion, appeared to be an unstable region. The single strand domains separating the stems were susceptible to digestion with the exception of sites adjacent to guanosines; GL2.1 in the stem II loop and G12 in the conserved GAAAC sequence, which separates stems II and III. The absence of digestion at GL2.1 in the stem II hairpin loop of the hammerhead complex was maintained in uncomplexed ribozyme and in short oligonucleotides containing only the stem II hairpin region. In contrast, the G12 site became susceptible when the ribozyme was not complexed with its substrate. Overall the results are consistent with the role of Mg2+ in the hammerhead self-cleavage reaction being catalytic and not structural.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm S. C., Uhlenbeck O. C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991 Oct 1;30(39):9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Denman R. B. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques. 1993 Dec;15(6):1090–1095. [PubMed] [Google Scholar]
- Doudna J. A., Grosshans C., Gooding A., Kundrot C. E. Crystallization of ribozymes and small RNA motifs by a sparse matrix approach. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7829–7833. doi: 10.1073/pnas.90.16.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Kinetics of intermolecular cleavage by hammerhead ribozymes. Biochemistry. 1992 Dec 8;31(48):12042–12054. doi: 10.1021/bi00163a012. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Jeffries A. C., Sheldon C. C., Symons R. H. Structural and ionic requirements for self-cleavage of virusoid RNAs and trans self-cleavage of viroid RNA. Cold Spring Harb Symp Quant Biol. 1987;52:249–259. doi: 10.1101/sqb.1987.052.01.030. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Fu D. J., McLaughlin L. W. Importance of specific adenosine N7-nitrogens for efficient cleavage by a hammerhead ribozyme. A model for magnesium binding. Biochemistry. 1992 Nov 17;31(45):10941–10949. doi: 10.1021/bi00160a001. [DOI] [PubMed] [Google Scholar]
- Fu D. J., McLaughlin L. W. Importance of specific purine amino and hydroxyl groups for efficient cleavage by a hammerhead ribozyme. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3985–3989. doi: 10.1073/pnas.89.9.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D. J., Rajur S. B., McLaughlin L. W. Importance of specific guanosine N7-nitrogens and purine amino groups for efficient cleavage by a hammerhead ribozyme. Biochemistry. 1993 Oct 12;32(40):10629–10637. doi: 10.1021/bi00091a013. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Major F., Cedergren R. Computer modeling and display of RNA secondary and tertiary structures. Methods Enzymol. 1990;183:318–330. doi: 10.1016/0076-6879(90)83021-z. [DOI] [PubMed] [Google Scholar]
- Grasby J. A., Jonathan P., Butler G., Gait M. J. The synthesis of oligoribonucleotides containing O6-methylguanosine: the role of conserved guanosine residues in hammerhead ribozyme cleavage. Nucleic Acids Res. 1993 Sep 25;21(19):4444–4450. doi: 10.1093/nar/21.19.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Nuclear magnetic resonance studies of the hammerhead ribozyme domain. Secondary structure formation and magnesium ion dependence. J Mol Biol. 1991 Jan 5;217(1):113–124. doi: 10.1016/0022-2836(91)90615-d. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Uhlenbeck O. C., Pardi A. Sequence-dependent structural variations of hammerhead RNA enzymes. Nucleic Acids Res. 1990 Mar 11;18(5):1103–1108. doi: 10.1093/nar/18.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- Li Y., Zon G., Wilson W. D. NMR and molecular modeling evidence for a G.A mismatch base pair in a purine-rich DNA duplex. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):26–30. doi: 10.1073/pnas.88.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall M. J., Hendry P., Jennings P. A. Minimal sequence requirements for ribozyme activity. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5710–5714. doi: 10.1073/pnas.89.13.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H. Y., Kaaret T. W., Bruice T. C. A computational approach to the mechanism of self-cleavage of hammerhead RNA. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9727–9731. doi: 10.1073/pnas.86.24.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odai O., Kodama H., Hiroaki H., Sakata T., Tanaka T., Uesugi S. Synthesis and NMR study of ribo-oligonucleotides forming a hammerhead-type RNA enzyme system. Nucleic Acids Res. 1990 Oct 25;18(20):5955–5960. doi: 10.1093/nar/18.20.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen D. B., Benseler F., Aurup H., Pieken W. A., Eckstein F. Study of a hammerhead ribozyme containing 2'-modified adenosine residues. Biochemistry. 1991 Oct 8;30(40):9735–9741. doi: 10.1021/bi00104a024. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Labuda D., Usman N., Yang J. H., Cedergren R. Relationship between 2'-hydroxyls and magnesium binding in the hammerhead RNA domain: a model for ribozyme catalysis. Biochemistry. 1991 Apr 23;30(16):4020–4025. doi: 10.1021/bi00230a029. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Wu T. F., Cousineau B., Ogilvie K. K., Cedergren R. Mixed deoxyribo- and ribo-oligonucleotides with catalytic activity. Nature. 1990 Apr 5;344(6266):565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- Pieken W. A., Olsen D. B., Benseler F., Aurup H., Eckstein F. Kinetic characterization of ribonuclease-resistant 2'-modified hammerhead ribozymes. Science. 1991 Jul 19;253(5017):314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- Pley H. W., Lindes D. S., DeLuca-Flaherty C., McKay D. B. Crystals of a hammerhead ribozyme. J Biol Chem. 1993 Sep 15;268(26):19656–19658. [PubMed] [Google Scholar]
- Serra M. J., Lyttle M. H., Axenson T. J., Schadt C. A., Turner D. H. RNA hairpin loop stability depends on closing base pair. Nucleic Acids Res. 1993 Aug 11;21(16):3845–3849. doi: 10.1093/nar/21.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Symons R. H. Mutagenesis analysis of a self-cleaving RNA. Nucleic Acids Res. 1989 Jul 25;17(14):5679–5685. doi: 10.1093/nar/17.14.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimayama T., Nishikawa F., Nishikawa S., Taira K. Nuclease-resistant chimeric ribozymes containing deoxyribonucleotides and phosphorothioate linkages. Nucleic Acids Res. 1993 Jun 11;21(11):2605–2611. doi: 10.1093/nar/21.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slim G., Gait M. J. Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res. 1991 Mar 25;19(6):1183–1188. doi: 10.1093/nar/19.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slim G., Gait M. J. The role of the exocyclic amino groups of conserved purines in hammerhead ribozyme cleavage. Biochem Biophys Res Commun. 1992 Mar 16;183(2):605–609. doi: 10.1016/0006-291x(92)90525-p. [DOI] [PubMed] [Google Scholar]
- Symons R. H. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Taira K., Uebayasi M., Maeda H., Furukawa K. Energetics of RNA cleavage: implications for the mechanism of action of ribozymes. Protein Eng. 1990 Aug;3(8):691–701. doi: 10.1093/protein/3.8.691. [DOI] [PubMed] [Google Scholar]
- Thomson J. B., Tuschl T., Eckstein F. Activity of hammerhead ribozymes containing non-nucleotidic linkers. Nucleic Acids Res. 1993 Dec 11;21(24):5600–5603. doi: 10.1093/nar/21.24.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T., Eckstein F. Hammerhead ribozymes: importance of stem-loop II for activity. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):6991–6994. doi: 10.1073/pnas.90.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T., Ng M. M., Pieken W., Benseler F., Eckstein F. Importance of exocyclic base functional groups of central core guanosines for hammerhead ribozyme activity. Biochemistry. 1993 Nov 2;32(43):11658–11668. doi: 10.1021/bi00094a023. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Perreault J. P., Labuda D., Usman N., Cedergren R. Mixed DNA/RNA polymers are cleaved by the hammerhead ribozyme. Biochemistry. 1990 Dec 25;29(51):11156–11160. doi: 10.1021/bi00503a002. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Usman N., Chartrand P., Cedergren R. Minimum ribonucleotide requirement for catalysis by the RNA hammerhead domain. Biochemistry. 1992 Jun 2;31(21):5005–5009. doi: 10.1021/bi00136a013. [DOI] [PubMed] [Google Scholar]
- van Tol H., Buzayan J. M., Feldstein P. A., Eckstein F., Bruening G. Two autolytic processing reactions of a satellite RNA proceed with inversion of configuration. Nucleic Acids Res. 1990 Apr 25;18(8):1971–1975. doi: 10.1093/nar/18.8.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]