Abstract
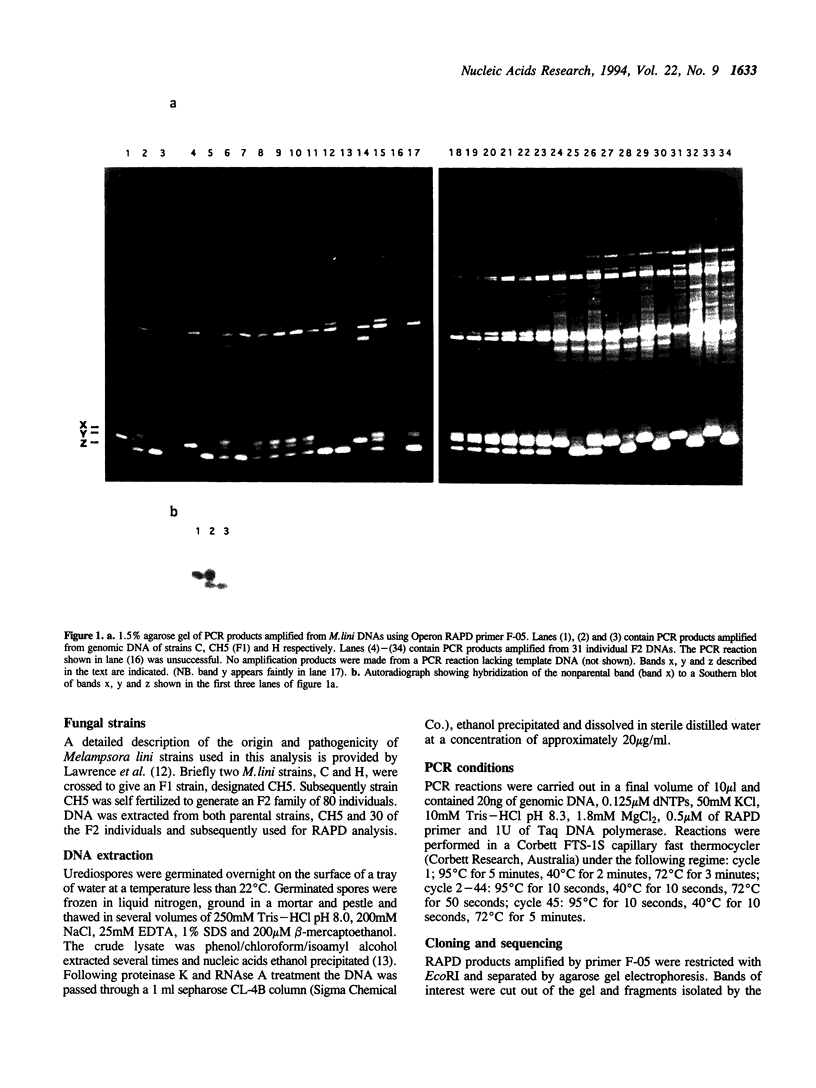
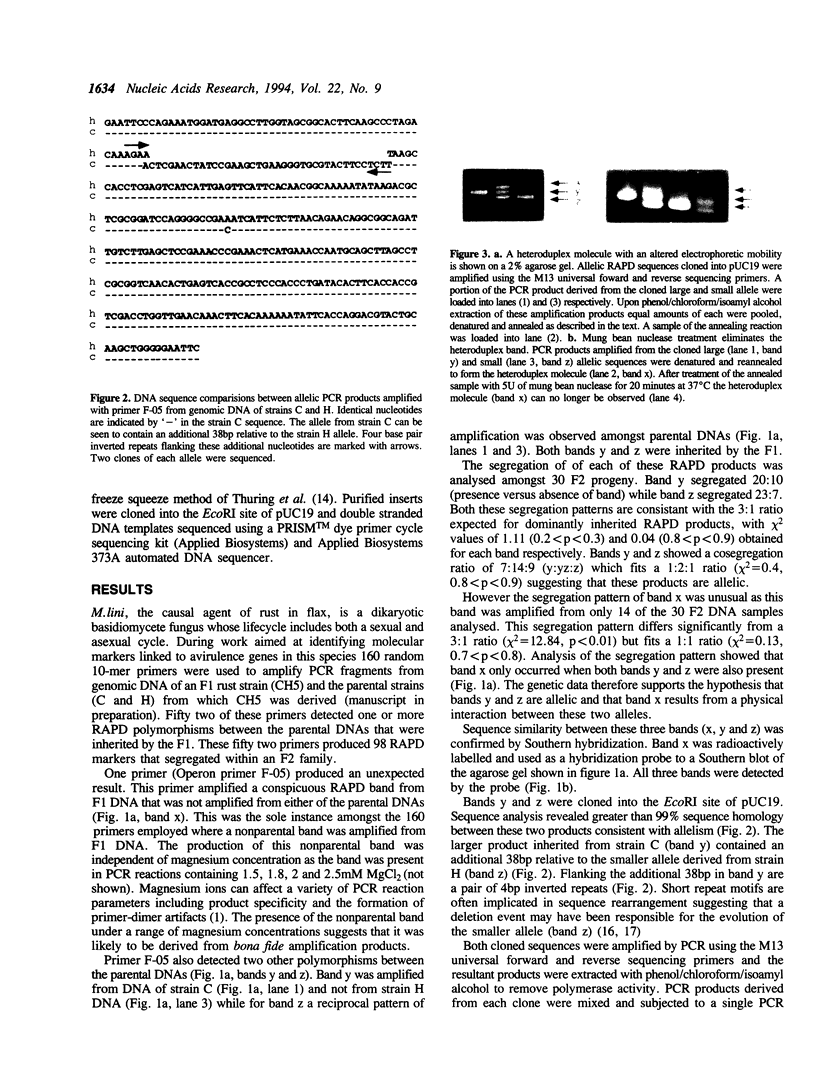
Primers (10-mers) of random sequence were used to amplify RAPD bands from genomic DNA of an F1 strain of flax rust (Melampsora lini) and its two parent strains. One primer out of 160 tested was unusual in that it amplified a product from F1 DNA that was not amplified from either parental DNAs. The same primer also generated two RAPD bands that segregated as codominant alleles amongst F2 progeny. The nonparental band was only generated from DNAs of F2 individuals that were heterozygous for these two allelic sequences. Sequence analysis of the two RAPD alleles demonstrated greater than 99% sequence identity, although the larger allele possessed an additional 38bp relative to the smaller. Mixing of the two allelic sequences followed by denaturation and annealing in the absence of polymerase activity resulted in the formation of the nonparental band. Thus the nonparental band present in some RAPD reactions consisted of a heteroduplex molecule formed between two allelic sequences of different size. These data demonstrate that heteroduplex molecules formed between allelic RAPD products are a potential source of artifactual polymorphism that can arise during RAPD analysis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Echt C. S., Erdahl L. A., McCoy T. J. Genetic segregation of random amplified polymorphic DNA in diploid cultivated alfalfa. Genome. 1992 Feb;35(1):84–87. doi: 10.1139/g92-014. [DOI] [PubMed] [Google Scholar]
- Harrison S. P., Mytton L. R., Skøt L., Dye M., Cresswell A. Characterisation of Rhizobium isolates by amplification of DNA polymorphisms using random primers. Can J Microbiol. 1992 Oct;38(10):1009–1015. doi: 10.1139/m92-166. [DOI] [PubMed] [Google Scholar]
- Nagley P. Coordination of gene expression in the formation of mammalian mitochondria. Trends Genet. 1991 Jan;7(1):1–4. doi: 10.1016/0168-9525(91)90002-8. [DOI] [PubMed] [Google Scholar]
- Reiter R. S., Williams J. G., Feldmann K. A., Rafalski J. A., Tingey S. V., Scolnik P. A. Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1477–1481. doi: 10.1073/pnas.89.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedy M. F., Hamilton W. J., 3rd, Aquadro C. F. Excess of non-parental bands in offspring from known primate pedigrees assayed using RAPD PCR. Nucleic Acids Res. 1992 Feb 25;20(4):918–918. doi: 10.1093/nar/20.4.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Haymes K. M., Williams S. M. Parentage analysis using RAPD PCR. Nucleic Acids Res. 1992 Oct 25;20(20):5493–5493. doi: 10.1093/nar/20.20.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]