Abstract
In this issue, Meyer-Ficca et al. demonstrate that PARP1 and PARG regulate TOP2B mediated DNA double-strand breaks during spermiogenesis.
Cell Programmed Sperm DNA Breakage by Topoisomerase (DNA) II Beta
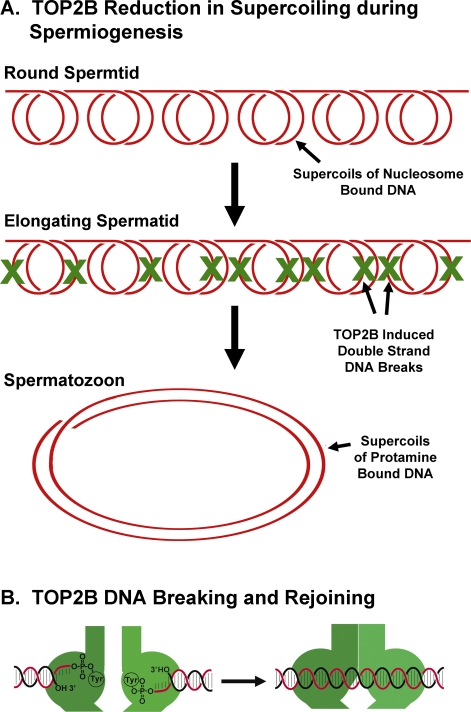
During the chromatin condensation that accompanies spermiogenesis, the entire paternal genome suffers approximately 5 to 10 million double-strand DNA breaks as a matter of necessity (Fig. 1A). These double-strand DNA breaks are caused by topoisomerase (DNA) II beta (TOP2B) [1–3], one of two variants of topoisomerase II (TOP2), which unwinds and untangles DNA by creating a transient DNA double-strand break and passing one DNA strand through the break (see Fig. 1B) [4, 5]. Spermiogenesis has a unique requirement for decreased supercoiling as histones are removed and protamines are deposited in their place. This is because protamine-bound DNA is less supercoiled than histone-bound DNA; protamines induce wider supercoils that are more efficient for packing DNA into a smaller space than histones [6, 7] (Fig. 1A). TOP2B relieves the supercoils, facilitating the displacement of histones during spermiogenesis.
FIG. 1.
Release of supercoils by TOP2B during spermiogenesis. A) DNA is bound to histones in round spermatids as it is in most other cells. Each 200 bp of DNA is coiled twice around a histone octamer to form a nucleosome. Here, the supercoils for six nucleosomes are shown. To relieve these supercoils, TOP2B makes up to 10 double-strand DNA breaks in six nucleosomes (the actual breaks probably occur sequentially and are distal to the nucleosome; the X's are shown only as a way to illustrate the number of breaks necessary). The resulting DNA forms two larger coils that are part of the protamine toroid in spermiogenesis (see [23] for a detailed explanation of this calculation). B) To unwind DNA, TOP2 forms a double-strand break through which a DNA strand is passed. The ends of the cleaved DNA are covalently attached to the TOP2B homodimers. After strand passage, the DNA is resealed.
Given the fact that the main function of the spermatozoon is to deliver a pristine copy of the paternal genome to the oocyte that will then be copied a trillion times during the life of the newly formed embryo, the discovery of this process understandably lead to concerns about how it is regulated to ensure that every break is correctly repaired. It has been proposed, for example, that one cause of male infertility is residual DNA double-strand breaks in mature spermatozoa resulting from incomplete DNA strand passage by TOP2B during spermiogenesis [1, 8]. In this issue, Meyer-Ficca et al. [9] provide the first evidence for the regulation of this DNA breakage during spermiogenesis. They show that poly(ADP-ribose) polymerase 1 (PARP1) and poly(ADP-ribose) glycohydrolase (PARG) cycling is required for each TOP2B-induced DNA double-strand break. Because one of the two TOP2 variants is ubiquitous in all cell types, their results also have wider implications for chromatin structure.
PARP/PARG Regulation of TOP2B Connects DNA Breakage with Histone Removal
PARP1 is the founding member of a family of nuclear proteins that regulate chromatin structure [10]. Recent studies suggested that PARP1 might play a role in TOP2 activity, but Meyer-Ficca et al. [9] were able to demonstrate that PARP1 can inhibit TOP2B activity directly in vitro using recombinant enzymes and that PARG prevents this inhibition. They also provide evidence that a PARP1 inhibitor increases the number of DNA breaks induced by TOP2B in vivo using two strains of mice that lack Parp1 or Parg. The authors propose an intriguing model in which each cycle of TOP2B DNA breakage and rejoining is accompanied by a cycling of PARP and PARG activity. In this model, PARP1 binds TOP2B as it cleaves DNA, then undergoes autopoly(ADP-ribosyl)ation. This causes TOP2B to release the DNA while completing its DNA strand passage. PARG then digests the poly(ADP-ribose) from PARP1, which, in turn, separates it from TOP2B, allowing the cycle to repeat itself. This model suggests an elaborate biochemical regulation of each TOP2B DNA unwinding cycle. The identification of PARP1 as the TOP2B regulator during histone removal and protamine deposition is particularly interesting because of PARP1's well-established roles in histone modifications.
TOP2 at the Crossroads Between DNA Replication (Life) and DNA Degradation (Death)
TOP2 is ubiquitously involved in functions that require manipulation of DNA folding. In addition to its role in histone displacement during spermiogenesis, TOP2 is important for DNA replication [11, 12], chromosome segregation [5, 13], and apoptosis [14, 15]. Its role in cell death has been the most enigmatic because, in this case, the strand passage reaction appears to stop midway at the point where the DNA is cleaved (Fig. 1B). Nucleases then complete the apoptotic digestion of the chromatin [14–16]. It is possible that TOP2 is part of an as yet undefined checkpoint that determines whether the cell progresses along its course of cell replication or differentiation (such as spermiogenesis) or defaults to apoptosis. TOP2B's role in apoptosis has been suggested as another possible source of double-strand DNA breaks in mature spermatozoa that result from so-called abortive apoptosis during spermiogenesis [8]. In epididymal spermatozoa, TOP2B can be induced to cleave the entire paternal genome into fragments of 25–50 kb [17, 18], suggesting that this potential regulator remains active in fully condensed sperm chromatin. There are no adequate explanations as to why TOP2 is temporarily frozen at this intermediate stage during apoptosis or why the cell requires this signal before nucleases can digest the DNA. The report by Meyer-Ficca et al. [9] suggests an intriguing solution to the first part of this quandary, that is, PARP1 and PARG may also play a role in TOP2 regulation during apoptosis. Perhaps TOP2 cannot complete its DNA strand passage in apoptosis because PARP1 is not present to induce the last step of the cycle.
Is TOP2B the Biochemical Signal for DNA Damage?
An important aspect of TOP2B transient DNA breaks is that they are not seen by the cell, as such. Each of the cleaved DNA strands is covalently linked by the 5′ OH to a tyrosine residue in one of the homodimers that make up TOP2B (see Fig. 1B) [19]. Based on the crystallographic structure of this complex, the free 3′ ends appear to be sterically concealed by the TOP2 [20]. The double-strand breaks caused by TOP2 are therefore not typical DNA damage motifs. This point was raised by Meyer-Ficca et al. [9] when they stated, “Typically, the enzyme-bridged DSBs transiently introduced by type II topoisomerases do not elicit a DNA damage response.” It is therefore probable that PARG recognizes the conformational change in TOP2B as it transitions from DNA binding to the cleavable complex, rather than the DNA break itself. The authors may have been thinking along these lines because the summary model that they present shows PARP1 binding to one conformation of TOP2B and not the other. In their version, PARP1 seems to stimulate TOP2B to complete its DNA strand passage, thereby resealing the DNA break. If so, this may set a larger precedent for the regulation of TOP2 in its other functions in apoptosis and DNA replication, that is, TOP2B's conformational changes, rather than the DNA breaks it mediates, may initiate the cell's responses to DNA breaks.
Reproductive Biology as a Window to Cell Biology
The unique requirement for DNA unwinding during spermiogenesis helped bring this aspect of TOP2 regulation to light, illustrating once again that reproductive biology provides novel insights to the larger field of cell biology. This is not the first time that the unique chromatin and nuclear models in reproductive biology have led the way toward solving more general biological problems. The most famous example of this is the identification of cyclins controlling the cell cycle; these proteins were originally discovered in Xenopus oocytes [21, 22], but there are others. The work by Meyer-Ficca et al. [9] on the regulation of TOP2B during spermiogenesis helps unravel some of the questions surrounding the functions of the ubiquitous TOP2 chromatin regulator.
Footnotes
Supported by NIH grant 1R01HD060722.
REFERENCES
- Laberge RM, Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol Reprod 2005; 73: 289 296 [DOI] [PubMed] [Google Scholar]
- Chen JL, Longo FJ. Expression and localization of DNA topoisomerase II during rat spermatogenesis. Mol Reprod Dev 1996; 45: 61 71 [DOI] [PubMed] [Google Scholar]
- Roca J, Mezquita C. DNA topoisomerase II activity in nonreplicating, transcriptionally inactive, chicken late spermatids. EMBO J 1989; 8: 1855 1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 2001; 70: 369 413 [DOI] [PubMed] [Google Scholar]
- DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A 1984; 81: 2616 2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risley MS, Einheber S, Bumcrot DA. Changes in DNA topology during spermatogenesis. Chromosoma 1986; 94: 217 227 [DOI] [PubMed] [Google Scholar]
- Hud NV, Allen MJ, Downing KH, Lee J, Balhorn R. Identification of the elemental packing unit of DNA in mammalian sperm cells by atomic force microscopy. Biochem Biophys Res Commun 1993; 193: 1347 1354 [DOI] [PubMed] [Google Scholar]
- Sakkas D, Mariethoz E, Manicardi G, Bizzaro D, Bianchi PG, Bianchi U. Origin of DNA damage in ejaculated human spermatozoa. Rev Reprod 1999; 4: 31 37 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar JD, Ihara M, Meistrich ML, Ausitn CA, Meyer RG. Poly(ADP-ribose) polymerases PARP1 and PARP2 modulate topoisomerase II beta (TOP2B) function during chromatin condensation in mouse spermiogenesis. Biol Reprod 2011; 84: 900 909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 2010; 39: 8 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, Liu LF, Coffey DS. Newly replicated DNA is associated with DNA topoisomerase II in cultured rat prostatic adenocarcinoma cells. Nature 1986; 322: 187 189 [DOI] [PubMed] [Google Scholar]
- McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem 2005; 280: 39337 39345 [DOI] [PubMed] [Google Scholar]
- Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J Cell Biol 2003; 160: 645 655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyan VT, Bezvenyuk ZA, Salminen A, Austin CA, Courtney MJ. The role of topoisomerase II in the excision of DNA loop domains during apoptosis. J Biol Chem 2002; 277: 21458 21467 [DOI] [PubMed] [Google Scholar]
- Li TK, Chen AY, Yu C, Mao Y, Wang H, Liu LF. Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes Dev 1999; 13: 1553 1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem 2005; 94: 1078 1087 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Ward WS. Topoisomerase II mediated breaks in spermatozoa cause the specific degradation of paternal DNA in fertilized oocytes. Biol Reprod 2007; 76: 666 672 [DOI] [PubMed] [Google Scholar]
- Shaman JA, Prisztoka R, Ward WS. Topoisomerase IIB and an extracellular nuclease interact to digest sperm DNA in an apoptotic-like manner. Biol Reprod 2006; 75: 741 748 [DOI] [PubMed] [Google Scholar]
- Morrison A, Cozzarelli NR. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell 1979; 17: 175 184 [DOI] [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature 1996; 379: 225 232 [DOI] [PubMed] [Google Scholar]
- Maller J, Gautier J, Langan TA, Lohka MJ, Shenoy S, Shalloway D, Nurse P. Maturation-promoting factor and the regulation of the cell cycle. J Cell Sci Suppl 1989; 12: 53 63 [DOI] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell 1989; 58: 361 372 [DOI] [PubMed] [Google Scholar]
- Ward WS. Deoxyribonucleic acid loop domain tertiary structure in mammalian spermatozoa. Biol Reprod 1993; 48: 1193 1201 [DOI] [PubMed] [Google Scholar]