Abstract
To achieve the specialized nuclear structure in sperm necessary for fertilization, dramatic chromatin reorganization steps in developing spermatids are required where histones are largely replaced first by transition proteins and then by protamines. This entails the transient formation of DNA strand breaks to allow for, first, DNA relaxation and then chromatin compaction. However, the nature and origin of these breaks are not well understood. We previously reported that these DNA strand breaks trigger the activation of poly(ADP-ribose) (PAR) polymerases PARP1 and PARP2 and that interference with PARP activation causes poor chromatin integrity with abnormal retention of histones in mature sperm and impaired embryonic survival. Here we show that the activity of topoisomerase II beta (TOP2B), an enzyme involved in DNA strand break formation in elongating spermatids, is strongly inhibited by the activity of PARP1 and PARP2 in vitro, and this is in turn counteracted by the PAR-degrading activity of PAR glycohydrolase. Moreover, genetic and pharmacological PARP inhibition both lead to increased TOP2B activity in murine spermatids in vivo as measured by covalent binding of TOP2B to the DNA. In summary, the available data suggest a functional relationship between the DNA strand break-generating activity of TOP2B and the DNA strand break-dependent activation of PARP enzymes that in turn inhibit TOP2B. Because PARP activity also facilitates histone H1 linker removal and local chromatin decondensation, cycles of PAR formation and degradation may be necessary to coordinate TOP2B-dependent DNA relaxation with histone-to-protamine exchange necessary for spermatid chromatin remodeling.
Keywords: chromatin remodeling, condensation, gametogenesis, histone H1 linker, PARG, PARP, poly(ADP-ribose) glycohydrolase, poly(ADP-ribose) polymerase, poly(ADP-ribosyl)ation, spermatid, spermatogenesis, transition protein, topoisomerase II beta, TOP2A, TOP2B
Poly(ADP-ribose)polymerase (PARP) activity regulates topoisomerase II beta (TOP2B) activity in vitro and in spermatids, providing a mechanism for regulate endogenous DNA strand breaks during spermatid chromatin condensation.
INTRODUCTION
Nuclear DNA integrity and the composition of chromatin proteins are important parameters of normal sperm function. The development of postmeiotic male germ cells (spermatids) is characterized by global changes in cell physiology and nuclear organization, such as a dramatic decrease in nuclear size that is associated with a complete reorganization of the haploid genome from histone-bound nucleosomal DNA to a very condensed and mostly protamine-bound form (Fig. 1). During this process, most histones are removed from the DNA and first replaced by the transition proteins TP1 and TP2 and subsequently by protamines P1 and P2 [1]. Only a small, but apparently well-defined, fraction of the sperm genome remains histone associated in mature sperm [2–5]. However, the regulation of spermatid chromatin reorganization leading to the intricate, conserved sperm nuclear structure is not well understood despite the fact that abnormal chromatin composition and the presence of DNA strand breaks in sperm represent important clinically relevant factors of male infertility [6–10].
FIG. 1.
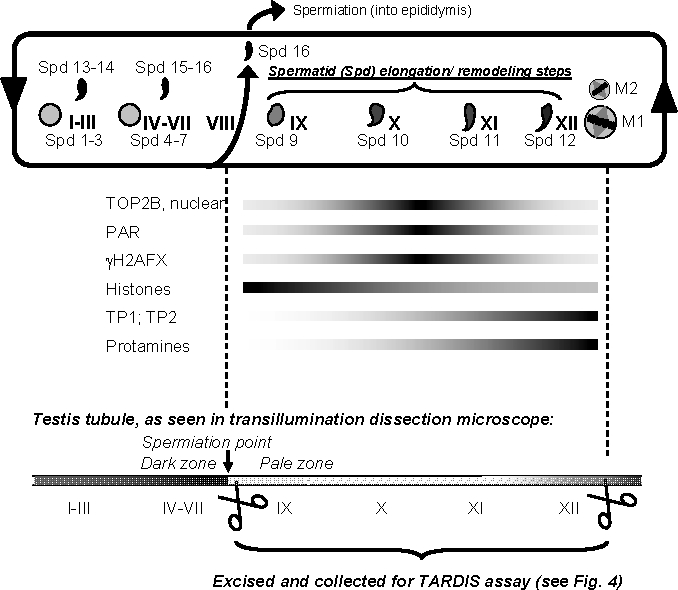
Schematic overview of spermiogenic development and isolation of relevant steps 9–12 spermatids. After meiosis, haploid spermatids (Spd) undergo an intricate differentiation process, called spermiogenesis, that comprises fundamental remodeling of the nucleus. In mice, spermatids are categorized into 16 developmental steps (Spd 1–16) that are found throughout the 12 stages of spermatogenesis (I–XII). Shapes of spermatid nuclei vary as they progress through development, which is shown only very schematically. Round step 1 spermatids (Spd 1) appear after stage XII, the stage where the meiotic divisions (M1 and M2) take place, that is, in stage I. In stage I, condensing spermatids (Spd 13) are also present. Spd 16 are moved into the epididymis during tubule stage VIII (spermiation). Therefore, tubule of stages IX–XII only contain Spd 9–12 that are in varying degrees of elongation. These developmental stages are marked by the formation of DSBs [12, 13] causing H2AFX phosphorylation (γH2AFX) [14, 15], the formation of PAR [14], and the replacement of histones by transition proteins TP1 and TP2 and protamines (reviewed in [67]). While TOP2B expression is detectable in all spermatid steps, TOP2B association with the nucleus is mostly restricted to tubule stages X and XI (see also Supplemental Fig. S2). This schematic shows that elongating spermatids (Spd 9–12) are only present in stages IX–XII where they represent the only haploid cell fraction. This is relevant to this study because these tubule sections were specifically isolated and utilized for in vivo TOP2B DNA binding studies (TARDIS assays). Because condensing spermatids diffract light more efficiently than other cells, transmission illumination was utilized to specifically excise tubule stages that appear darker or clearer using a dissecting scope. Stages IX–XII were readily identified as a pale zone, and such sections were collected and prepared for TARDIS assays (see Fig. 3).
Notably, the exchange of histones, which only support a supercoiled DNA structure, by transition proteins and protamines, which stabilize a relaxed, more linear conformation of the DNA, is accompanied by a large number of physiological DNA strand breaks during midspermiogenesis (Fig. 1) [11–14]. Robust phosphorylation of histone H2AFX (γH2AFX, also known as γH2AX) is also detected during these steps in spermatid development, indicating that these breaks induce DNA damage response signaling [14, 15]. The DNA strand breaks have been attributed to the activity of topoisomerase II beta (TOP2B) although its regulation in this process has not been fully elucidated [15–20].
Like other type II topoisomerases, TOP2B is capable of removing DNA supercoiling by generating a transient protein-bound DNA double-strand break (DSB) through which a second DNA helix may pass before religating the first DNA double strand (for more details, see, for example, [21]). This allows for catenation and decatenation as well as knotting or unknotting reactions of DNA. Typically, the enzyme-bridged DSBs transiently introduced by type II topoisomerases do not elicit a DNA damage response. Therefore, H2AFX phosphorylation as well as TUNEL detection of DNA strand breaks in spermatids may be explained by abortive TOP2B reactions followed by a DNA repair step that involves the creation of a DNA strand break [15]. At the same time, TOP2B has been shown to mediate sperm DNA fragmentation, possibly in association with an unknown nuclease, that occurs when sperm are treated with detergents in the presence of calcium and manganese; this reaction can also trigger fragmentation of paternal DNA in the zygote [22, 23].
In line with these notions, we showed previously that DNA strand breaks in elongating rat spermatids trigger formation of poly(ADP-ribose) (PAR) by PAR polymerases (i.e., PARP1 and PARP2) [14]. Generally, both single and double DNA strand breaks as well as cruciform DNA structures are recognized by PARP1 and PARP2, which triggers their enzymatic activity [24, 25]. Activated PARPs generate PAR, a unique, highly electronegative biopolymer, by cleavage of the nicotinamide-ribose bond of NAD+ and subsequent polymerization of the ADP-ribose units. PAR formation is an important step in DNA base excision and strand break repair pathways. Automodification of PARPs with PAR inactivates the enzymes and allows them to dissociate from the break. Dissociation is facilitated by electrostatic repulsion of the negatively charged PAR, which is covalently attached to the PARP protein, from the DNA [26]. PAR has a short half-life of only a few minutes because of its quick and specific degradation by PAR glycohydrolase (PARG), an essential and ubiquitous enzyme. The coordinated actions of PARPs and PARG result in a local, rapid, and transient cyclic metabolism of PAR in response to DNA strand breaks. PAR can be covalently attached to histones and other target proteins, but many of the effects exerted by the polymer are based on noncovalent interactions with other proteins through binding to conserved PAR-binding motifs [27, 28], specialized macrodomains [29], or a PAR-binding zinc finger motif [30]. Local chromatin decondensation resulting from eviction of H1 and core histones from damaged sites and recruitment of DNA repair factors appear to be major roles of PARP1 and PARP2 in DNA repair [31, 32].
Our laboratory recently showed that PAR metabolism facilitates histone H1 removal in spermatids and that genetic or pharmacological PARP inhibition during spermiogenesis results in abnormal chromatin condensation and histone retention in mature sperm, which is associated with poor sperm chromatin integrity, reduced spermatid elongation, and subfertility in mice [33, 34]. We hypothesized that PARP activation is dependent on DNA strand breaks generated by TOP2B and that abnormal chromatin condensation caused by PARP inhibition may therefore be, at least partially, due to perturbed functional interaction with TOP2B. To investigate such functional correlations between PAR metabolism and TOP2B activity, in vitro assays involving purified TOP2B, PAR, PARP1, PARP2, and PARG were performed. In addition, covalent DNA cross-linking of TOP2B as a measure of TOP2B activity was quantified in mouse spermatids in vivo. The results demonstrate that PARP1 and PARP2 inhibit TOP2B activity in vitro and in vivo. In the context of work published by us and others, the data suggest that the activities of the DNA-relaxing enzyme TOP2B and the DNA strand break-dependent enzymes PARP1 and PARP2 may be able to directly and dynamically regulate each other via the formation of DNA strand breaks and PAR to mediate simultaneous DNA relaxation and histone H1 removal as essential steps of spermatid chromatin remodeling necessary for sperm function.
MATERIALS AND METHODS
Mouse Models
Parp1 gene-disrupted mice, that is, Parp1tm1Zqw [35] (referred to here as Parp1−/−), as well as Parg gene-disrupted mice, that is, Parg(110)−/− [36], were maintained and used according to the guidelines of the University of Pennsylvania Institutional Animal Care and Use Committee. Mouse strains were maintained both as heterozygous Parg(110)−/+ and Parg(110)−/− homozygous lines in a 129SvEv (129S6/SvEvTac) background (Taconics Inc., Hudson, NY). Wild-type controls used in the described studies, designated as 129SVE for simplicity in this report, were siblings of knockout mice from heterozygous parents from a larger, heterozygous but highly inbred Parp1−/−/Parg(110)−/− breeding colony [33]. Parg(110)−/− mice have a targeted deletion of exons 2 and 3 in the Parg gene, which leads to ablation of the three large PARG protein isoforms of 110, 102, and 98 kDa, but the two smaller ones of 63 kDa (ubiquitous) and 58 kDa (mitochondrial) [36–38] are still expressed. These animals were shown to have high steady-state levels of PARP1 and PARP2 automodification—and hence self-inactivation—in spermatids. This phenotype therefore resembles the functional inhibition of PARP1 and PARP2 by an inhibitor such as PJ34 [34]. In contrast, mice homozygous for a complete knockout of Parg show an early embryonic lethal phenotype [39]. Parp1−/− gene-disrupted mice do not express PARP1, but they have an intact gene for PARP2, which has overlapping functions with PARP1. Both enzymes are highly expressed in spermatids [33, 40]. Deletion of both Parp1 and Parp2 genes is embryonic lethal [41].
Analyses of In Vivo TOP2 Binding (TARDIS Assays)
Male mice of the different genotypes, that is, wild-type 129SVE, Parg(110)−/−, or Parp1−/− knockout, were closely age-matched for the studies (79–81 days old), and their genotypes confirmed by PCR. Groups of 3–6 male mice were each injected i.p. either with a specific TOP2 inhibitor, that is, 80 mg/kg etoposide (ETO; Sigma, St. Louis, MO), or with the highly specific and potent PARP inhibitor N-(6-oxo-5,6-dihydro-phenanthridin-2-yl)-N,N-dimethylacetamide HCl [42] (PJ34; Axxora, San Diego, CA) at a concentration of 10 mg/kg in a volume of 100 μl saline diluent per 20 g of body weight, or both. Animals were euthanized 2 h after the injection, their testes were removed, and tubule segments corresponding to stages IX–XII were prepared in PBS (catalog no. 14190-136; Invitrogen, Carlsbad, CA) supplemented with either 10 μM ETO, 3 μM PJ34, or a combination of 10 μM ETO with 3 μM PJ34, corresponding to the treatment the animals had received, according to a published method [43]. Spermatid steps contained in the dissected tubule sections are illustrated in Figure 1. Approximately 20–50 tubule sections of 1–5 mm in length/testis were collected, gently squashed between a slide and coverslip in a small volume of supplemented PBS, and the resulting intact cell suspension was collected. Parallel slides were used to confirm developmental stages of spermatids in each preparation. TARDIS (trapped in agarose DNA immunostaining) assays were performed essentially as described [44]. Briefly, cells were embedded in low melting point agarose on microscopic slides and lysed in 80 mM potassium phosphate buffer (pH 6.5) containing 1% SDS, 10 mM ethylenediaminetetraacetic acid, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, 2 μg/ml pepstatin, and 1 mM dithiothreitol (DTT) (all the chemicals, Sigma). Unbound proteins were then extracted by washing the slides in 1 M NaCl with protease inhibitors prior to standard immunofluorescence detection using two alternative specific TOP2B rabbit polyclonal antibodies (Topogen, Port Orange, FL, diluted 1:40; or 18513β antibody [44] diluted 1:150, which gave the same results) in combination with fluorescein isothiocyanate (FITC)-coupled goat anti-rabbit antibodies (Jackson Immunochemicals, West Grove, PA). Slides were finally stained with 10 μM Hoechst 33258 (Sigma) in PBS and mounted.
Immunofluorescence Staining of Tissue Sections, Fluorescence Microscopy, and Data Quantification
Immunostaining using rabbit anti-TOP2B antibodies (see above) or mouse monoclonal antibodies to phosporylated H2AFX was performed as described before [14]. Fluorescence microscopy was performed using a Nikon TE 2000-U equipped with monochrome CCD camera (Photometrics Coolsnap; Photometrics, Tucson, AZ) connected to a computer workstation using ImagePro Plus version 5.1.2 software (Media Cybernetics, Bethesda, MD) for documentation and data analyses. Automated analysis of large numbers of pictures was performed to measure areas of stained nuclei and to quantify fluorescence signal intensity (i.e., the density) in TARDIS assays as described before [45]. Background correction was performed using slides that were stained with secondary antibody only to correct for nonspecific FITC fluorescence caused by the secondary antibody and endogenous tissue autofluorescence. Hoechst 33258 images and TOP2B-FITC images were taken together to first create a nuclear outline (Hoechst) that was then used to create a mask that was used to measure nuclear FITC signal intensities.
Protein Extracts and Immunoblotting
SDS extracts of whole testis were made from frozen testes by weighing unthawed decapsulated tissue in precooled, preweighed 1.5-ml reaction tubes. Three volumes (v/w) of precooled 1× RIPA buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 1% nonyl phenoxypolyethoxylethanol, also known as NP-40, 1% deoxycholic acid, and 0.1% SDS) (all the chemicals, Sigma) prepared with complete protease inhibitor cocktail (Roche, Indianapolis, IN) were added to allow for homogenization using 1.5-ml reaction tubes with tight-fitting micropestles (VWR, Radnor, PA). Then, six volumes (v/w) of 2× Laemmli sample buffer (161-0737; BioRad, Hercules, CA) were added to give a final concentration of 100 ng/μl total testis fresh weight.
Testis lysates in Laemmli buffer were separated by 8% or 15% SDS-PAGE, transferred to PVDF membranes (Immobilon, Millipore, Billerica, MA), and subjected to antibody detection using rabbit anti-PAR (LP96–10, 1:2000; BD Bioscience, San Diego, CA), mouse anti-ACTB (AC-15, 1:5000; Sigma) rabbit anti-TOP2B (1:2000, Topogen), and horseradish peroxidase-coupled donkey anti-rabbit secondary antibodies (Jackson Immunolabs), according to standard procedures [33]. PAR signals were quantified using the ImageJ software package (Wayne Rasband, National Institutes of Health; http://rsb.info.nih.gov/ij/).
Topoisomerase Assays
Purified, active human topoisomerase II alpha (TOP2A) was purchased (∼15 U/μl initially, current activity was determined at the respective day of the experiments; Topogen) as well as purified PARP1 (specific activity = 22 222 U/mg, 10 U/μl stock solution, 1 U = 10 pmol PAR/30 min at 30°C; Trevigen, Gaithersburg, MD), PARP2 (specific activity = 380 U/mg, 1 μg/ml stock solution, 1 U = synthesis of 1 nmol PAR/min at 25°C; ENZO Life Sciences International, Plymouth Meeting, PA), and PARG (specific activity = ∼10 000 U/mg, 10 mU/μl; ENZO Life Sciences International). Catenated kinetoplast DNA (kDNA) and size markers for decatenated or cleaved kDNA (M2 and M3, respectively) were purchased from Topogen. Recombinant purified TOP2B was prepared as reported before [46–48]. Standard decatenation reactions (10 μl) contained 100 ng kDNA and 1.5 U TOP2A or TOP2B, where 1 U was defined as the activity that decatenates 200 ng kDNA/30 min at 37°C in freshly prepared reaction buffer (0.5 M Tris HCl, pH 8, 150 mM NaCl, 100 mM MgCl2, 5 mM DTT, 300 μg/ml bovine serum albumin, 2 mM ATP). PARP1, PARP2, or PARG was prediluted in 1× TOP2 reaction buffer and added to the reaction, keeping the total reaction volume constant at 10 μl. Similarly, NAD+ (Sigma) was diluted in water and added to give a final concentration of 1 mM. Reactions were stopped after 30 min by the addition of 5 μl of stop buffer (5% sarkosyl, 25% sucrose, 4 mg/L bromophenol blue) and resolved on 1% agarose gel with 0.5 μg/ml ethidium bromide. In experiments testing the effects of free PAR (Trevigen), 5 ng of PAR (corresponding to ∼1 μM or 1 pmol/sample) were added. For each sample, intensities of DNA bands representing unprocessed kDNA and the respective decatenation products were both determined by densitometric quantification using ImageJ software and used for calculation of the decatention rates.
RESULTS
PARP1 and PARP2 Activity Inhibits TOP2B Activity In Vitro
PARP1 and PARP2 each efficiently inhibited TOP2B (Fig. 2) in a dose-dependent manner in vitro. Because a potential function of TOP2A in spermatids could not be completely ruled out at this point, we also investigated PARP-dependent TOP2A inhibition, which gave similar results (Fig. 3). While the results indicate that PARP-dependent inhibition occurs on both TOP2A and TOP2B, we did not see any TOP2A signals in spermatids in the TARDIS assay (data not shown), which is consistent with published data [15, 19].
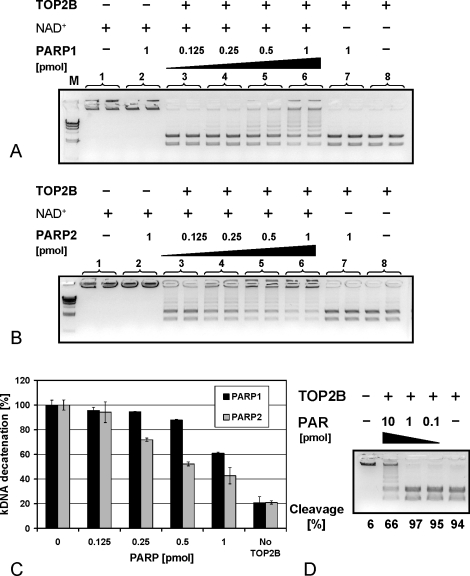
FIG. 2.
TOP2B is inhibited by PARP1 and PARP2 activity in an NAD+-dependent manner in vitro. TOP2B activity was measured by quantifying the conversion rate of high molecular kDNA, which in its native catenated form does not enter the agarose gel, into two low molecular weight bands. A) TOP2B activity is inhibited by PARP1 in the presence, but not in the absence, of NAD+ in an enzyme concentration-dependent manner. B) Like PARP1, PARP2 inhibits TOP2B activity in a comparable manner. C) Quantification of conversion rates from two independent experiments performed in duplicate. The small amount of decatenation in the absence of topoisomerase (right column) represents trace background amounts of kDNA degradation already present in the kDNA substrate. Error bars indicate the standard deviations. D) TOP2B activity is inhibited by purified PAR, albeit only at relatively high levels (1 pmol/μl), indicating that TOP2B may be able to interact with PAR and that covalent posttranslational modification with PAR is not necessary for TOP2B inhibition in vitro.
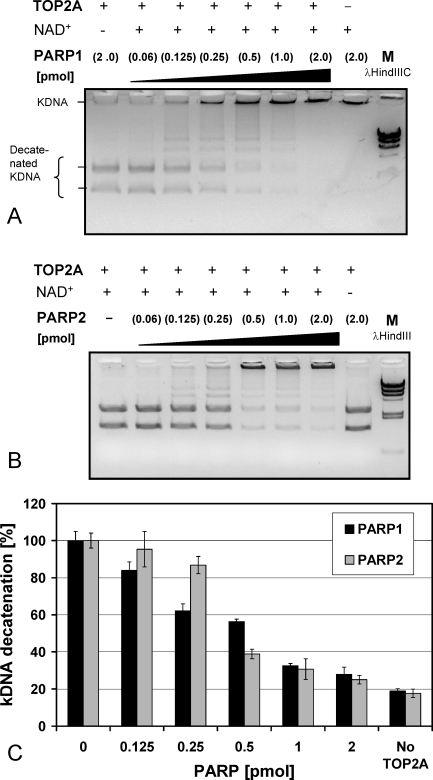
FIG. 3.
TOP2A is inhibited by PARP1 and PARP2 activity. A) PARP1 inhibited TOP2A activity in the presence, but not in the absence, of NAD+ in an enzyme concentration-dependent manner. PARP1 itself exhibited no decatenating activity (lane 8). B) Similarly, PARP2 inhibited TOP2A activity in the presence, but not absence, of NAD+ in an enzyme concentration-dependent manner. C) TOP2A-dependent kDNA conversion rates from two independent experiments performed in duplicate are shown. Error bars indicate the standard deviations.
PARP enzymes are essentially enzymatically inactive in the absence of DNA strand breaks. Only when PARPs bind to DNA strand breaks do they become highly enzymatically activated and rapidly consume NAD+ to synthesize large amounts of PAR. Generally, TOP2 inhibition by PARP was strictly dependent on the presence of NAD+, demonstrating that the inhibitory effect requires activation of PARP enzymes. Importantly, this finding also implies that TOP2 activity in vitro generates DNA strand breaks that are able to activate PARP enzymes.
Incubation of TOP2B with highly purified free PAR, albeit at a relatively high concentration, inhibited the reaction by ∼35% (Fig. 2D). This suggests that TOP2 inhibition can also be achieved by a noncovalent interaction with PAR without covalent modification of TOP2 with PAR (i.e., PARsylation).
PARG Blocks PARP-Dependent TOP2B Inhibition
Recombinant, highly purified PARG, which represents the catabolic arm of PAR metabolism, partly reversed TOP2B inhibition by the activities of PARP1 or PARP2 in a dose-dependent manner (Fig. 4A), while the presence of PARG alone did not affect TOP2B activity and by itself did not cause DNA unknotting or cleavage (Fig. 4A, far right panel). Compared to PARP1, PARG has been shown to have an almost 100-fold higher specific activity in vivo and in vitro [49], which is also reflected by the finding that a relatively small amount (44 fmol) of purified recombinant PARG was able to counteract the activity of 1 pmol PARP1 or PARP2. Similar results were obtained for assays involving TOP2A (Supplemental Fig. S1; all supplemental data are available online at www.biolreprod.org).
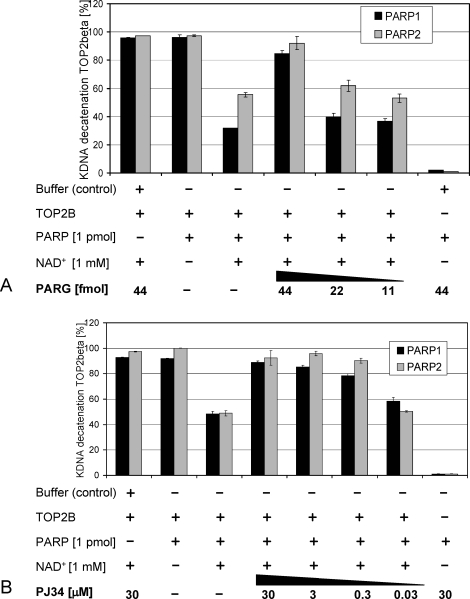
FIG. 4.
PARG activity and inhibition of PARPs by PJ34 restore TOP2B-mediated kDNA decatenating activity in a dose-dependent manner in vitro. A) Addition of PARG to reactions where TOP2B is inhibited by the addition of 1 pmol PARP1 or PARP2 in the presence of 1 mM NAD+ reduced the inhibition in a PARG enzyme concentration-dependent fashion. PARG itself had no decatenating activity and no inhibitory effect on TOP2B activity. B) Addition of PJ34 to inhibit PARP1 and PARP2 activity prevented the inhibition of TOP2B in a dose-dependent manner but by itself did not inhibit or activate TOP2B. Similarly, PARP enzymes did not cause kDNA decatenation or DNA strand breakage. Error bars indicate the standard deviations.
PJ34, a Pharmacological PARP Inhibitor, Prevents PARP-Dependent TOP2B Inhibition
PARP1- and PARP2-dependent inhibition of TOP2B was dose-dependently suppressed by addition of the PARP inhibitor PJ34 to the reaction. The drug, which has an approximate IC50 value of 20 nM for PARP1, blocked PARP1- and PARP2-dependent TOP2B inhibition in the presence of NAD+ (Fig. 4B). PJ34 alone did not inhibit TOP2A or TOP2B activity even when tested over a large dose range (0.3–30 μM PJ34, data not shown). These results indicate that the enzymatic activities of both PARP1 and PARP2 negatively regulate TOP2B activity, which may be an overlapping function of these PARPs. Furthermore, the results demonstrate that regulation of PARP1 or PARP2 activity by either PARG or PJ34 in vitro indirectly regulates TOP2B activity because these counteracted the inhibitory effects mediated by PARP1 and PARP2 in the presence of NAD+.
These data led us to hypothesize that the chromatin condensation defect observed in step 12 spermatids of PJ34-treated mice and of Parg(110)−/− mice that we reported previously [33, 34] may be partly attributed to inappropriately increased TOP2B activity due to blocking of negative regulation normally provided by PARP1 and PARP2. To further test this hypothesis, TOP2B expression was analyzed by immunofluorescence staining of testis sections using TOP2B-specific antibodies (Supplemental Fig. S2). The results confirmed that in testis TOP2B expression is strongly and selectively upregulated in spermatid cells [15–19].
In Vivo PARP Inhibition Promotes Increased TOP2B DNA Binding in Step 9–12 Spermatids
To test the hypothesis that TOP2B activity is modulated by PARP activity in vivo, we assessed TOP2B activity in testicular cells of 129SVE, Parp1−/−, and Parg(110)−/− mice using a method that measures TOP2B activity through quantification of covalently DNA-bound TOP2B (i.e., TARDIS assay) [44]. Because of the transient nature of PAR metabolism and TOP2B reactions, time-consuming cell separation techniques to isolate the relevant stages of spermatid development could not be used. Instead, elongating spermatids of steps 9–12 were isolated by microdissection of so-called pale zones from testis tubules in prewarmed PBS (Fig. 1), which can be quickly achieved within a few minutes [43]. In this assay, we quantified specific anti-TOP2B antibody signals solely within the perimeters of cell nuclei (Fig. 5A) that varied in intensity depending on their cell-type developmental stage and treatment regimen (Fig. 5A).
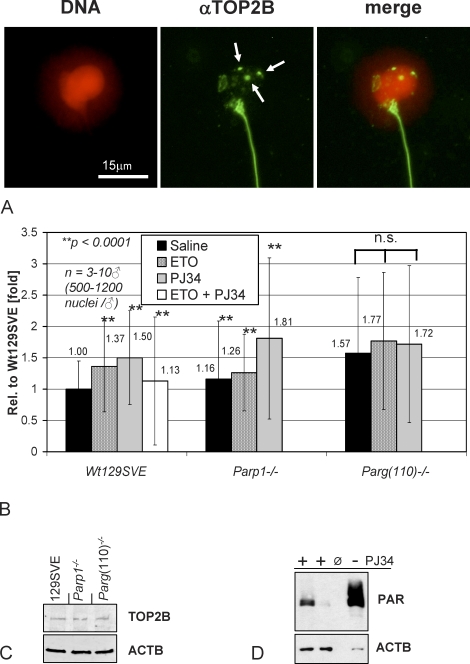
FIG. 5.
PARP inhibition increases testicular TOP2B activity in spermatids in vivo. Tubule sections enriched for stages that contain condensing spermatids (see Fig. 1) were analyzed for TOP2B activity using the TARDIS assay method. Live cells from stages IX–XII testis tubules isolated from wild-type mice (i.e., Wt129SV) or mice with genetically altered PAR pathways were embedded in low melting point agarose and lysed in the presence of protease inhibitors; proteins that were not covalently bound to the DNA were extracted using the high salt buffer. A) TOP2B bound to the DNA (red pseudocolored Hoechst 33258 dye) of decondensed spermatid nuclei was stained by indirect immunofluorescence detection (FITC, green) and photographed for digital quantification of signal intensities. Salt extraction-resistant TOP2B is detected as diffuse punctate staining and additional distinct foci restricted to the nucleus (arrows); developing spermatid flagella and midpieces are also stained, indicating the possible presence of TOP2B in these extranuclear cell compartments. Secondary antibody controls were completely negative (data not shown). Immunostaining of epididymal sperm using TOP2B antibodies confirmed the presence of TOP2B in the sperm midpiece (data not shown). Bar = 15 μm. B) TARDIS assay quantification of TOP2B immunofluorescence signals of spermatids isolated from Wt129SVE, Parp1−/−, or Parg(110)−/− mice that had been previously treated with ETO (80 mg/kg) or PJ34 (10 mg/kg) or both for 2 h. For each data point, 3–10 males were used and 500–1200 nuclei were measured per mouse. Each of the separate experiments was designed to include saline- and PJ34-treated wild-type controls. Statistical analyses were done using Student t-test. Data sets highly significant from untreated wild-type controls (P < 0.0001) are indicated by two asterisks (**). Treatment groups within the Parg(110)−/− genotype were not statistically significant from each other (n.s.) but highly significantly different from the wild-type controls. Error bars indicate the standard error of the mean. C) Immunoblot analysis of testicular SDS extracts confirming comparable TOP2B expression levels in 129SVE, Parp1−/−, and Parg(110)−/− mice. D) Testicular PAR steady-state levels are reduced after PJ34 injection of 129SVE mice (left two lanes) compared to saline-injected control mice, as used in the TARDIS assays. Whole testis SDS lysates were used for immunoblotting with PAR-specific antibodies. The loading control was β-actin (ACTB).
While generally a portion of extraction-resistant TOP2B signals were seen as intensely stained foci per nucleus, overall TOP2B signals were diverse in size and also included small dot-like staining. To quantify the overall amount of nuclear cross-linked TOP2B fluorescence signals, it was therefore decided to determine fluorescence intensities expressed as arbitrary units. That way, weaker and more diffuse TOP2B fluorescence signals could be included in the measurements, adding to the accuracy of the measurements. Depending on individual sets of experiments, average fluorescence units (AU) in untreated 129SVE varied slightly with an average of 1370 ± 234 AU (three independent experiments). Only nuclei from spermatids (DNA content correlating to a haploid genome, 1C) were measured (Fig. 5B); nuclei from spermatocytes (diploid genome, 4C) or Sertoli cells (diploid genome, 2C), which were excluded based on their DNA content, were not measured. Because the isolated cell population contained spermatids ranging from step 9 to step 12 (corresponding to tubule stages IX–XII), TOP2B staining intensity of individual spermatid nuclei varied with the exact individual developmental step, with signals being most prevalent in step 10 spermatids.
ETO is a type II topoisomerase poison that stabilizes the DNA cleavage complex and thus causes prolonged covalent linkage of TOP2B to the DNA [50, 51]. As expected, ETO injection resulted in measurably increased TOP2B bound to the DNA, confirming that experimental conditions allowed for sensitive measurement of alterations in TOP2 DNA binding and hence TOP2 activity (Fig. 5B). Consistent with the results of the in vitro assays, TOP2B DNA binding was highly significantly increased by ∼50% in 129SVE mice in the presence of the PARP inhibitor PJ34 (P < 0.0001, Student t-test). Notably, treatment of mice with both ETO and PJ34 did not have additive or synergistic effects (Fig. 5B). However, because ETO is a direct poison of TOP2 enzymes whereas PJ34 indirectly acts as an activating drug by blocking the inhibitory effects of PARPs, results of such dual treatments may be difficult to interpret.
In the Parp1−/− mouse, baseline TOP2B binding to DNA was elevated by ∼16% compared to wild-type siblings, which may be interpreted as a mild but significant (P < 0.0001) effect of the Parp1 gene deletion, where Parp2 is still expressed. It can therefore be assumed that PARP1 and PARP2 have overlapping functions in TOP2B regulation, which is also supported by the in vitro data (Figs. 2–4). The presence of one of the two enzymes may be sufficient to appropriately attenuate TOP2B. In Parp1−/− mice, PJ34 treatment resulted in a significant increase of TOP2B activity due to inhibition of PARP2 (Fig. 5B, middle panels). In Parg(110)−/− mice, average baseline TOP2B activity in spermatids was significantly elevated by ∼57% compared to wild-type controls, likely because of reduced PARP1 and PARP2 activities related to the prolonged automodification and therefore more sustained inactivation of PARPs in this mouse mutant (Fig. 5B, right panels) [34]. While TOP2B signals appeared to be slightly elevated by ETO or PJ34 in Parg(110)−/− animals, the increase did not reach statistical significance within three independent experiments using three animals each.
Immunoblot analyses confirmed that TOP2B expression was not different in the three mouse genotypes (Fig. 5C). PAR detection by immunoblotting using cell lysates from the second testis of each wild-type male used in the TARDIS assays (Fig. 5D) showed the efficiency of the PJ34 treatment but with some individual variation in PJ34-mediated PARP inhibition.
Taken together, the data demonstrate that TOP2B generates DNA strand breaks that activate PARP1 and PARP2 to form PAR, which in turn strongly inhibits TOP2B activity. Furthermore, this PARP activity-dependent inhibition of TOP2B is reversible by PARG activity and prevented by PARP inhibition using PJ34. Finally, the finding that TOP2B activity in intact mice can be altered by modulation of the PAR pathway demonstrates the potential relevance of these findings for spermatid chromatin remodeling.
DISCUSSION
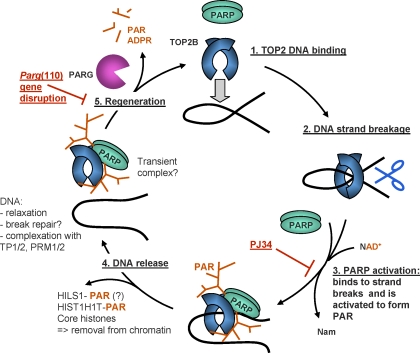
DNA strand breaks as they appear during midspermiogenesis in all spermatids are puzzling because they are potentially dangerous if they remain unrepaired, and it seems clear that they should be carefully regulated. The two main results of the present study are 1) that it provides a potential regulatory mechanism that limits the number of naturally occurring endogenous DNA strand breaks in spermatids and 2) that it proposes a mechanism in which DNA strand breaks and local chromatin decondensation facilitating nucleoprotein exchange are coordinated by PAR formation and degradation. We model this interaction of TOP2B and PARP1 as well as PARP2 and PARG in Figure 6. TOP2B binds to DNA (Fig. 6, step 1) and introduces a DNA strand break (step 2) to which PARP1 and/or PARP2 are recruited, similar to what was described elsewhere [52, 53]. This assumption is supported by the current result that PARP1 and PARP2 are indirectly activated by TOP2B in vitro because PARP activation to form PAR requires both the presence of NAD+ and the formation of DNA strand breaks by TOP2. PAR formation then inhibits TOP2 (Fig. 2). Activation of PARPs results in local formation of large chains of PAR, which then leads to automodification (i.e., inactivation) of the activated PARP bound to the DNA break (Fig. 6, step 3). This entails dissociation of adjacent proteins (i.e., PARP, TOP2B, H1 proteins, and core histones) from the DNA (Fig. 6, step 4) because these proteins lose their ability to bind DNA as a result of their interaction with the highly negatively charged polymer [26, 53, 54]. Local removal of H1 linker proteins and core histones from the DNA causes local DNA decondensation, which should facilitate further exchange of histones for transition proteins TP1 and TP2 as well as protamines. In support of this view, mice with genetically or pharmacologically impaired PAR metabolism show abnormally high retention of H1 and core histones in sperm [34]. Until PARG removes the inactivating PAR and thus allows PARPs and TOP2B to enter a new cycle of enzymatic activity (Fig. 6, step 5), the proteins may remain complexed with PAR for some time.
FIG. 6.
Proposed model of TOP2B and PARP1/2 activation coordinating spermatid DNA relaxation with nucleoprotein exchange. TOP2B, and potentially TOP2A, binds to target DNA (1) and generates a DNA strand break (2). Such DNA strand breaks trigger PARP activation and consequently PAR synthesis (3). Modification with PAR leads to catalytic inactivation and release of the topoisomerase-PARP complex from the DNA, along with PAR acceptor proteins, for example, HILS1, HIST1H1T, and core histones (4). Hypothetically, decatenated DNA is now accessible for binding to other available DNA-binding proteins, for example, transition proteins (TP1/2) and protamines (PRM1/2). The PARsylated topoisomerase-PARP complex is released from the DNA, and degradation of PAR by PARG is essential to restore enzyme activities (5). After removing histones and relieving DNA supercoiling, regenerated PARP and TOP2B enzymes can then move to another site along the chromatin and perform the same actions (1). Pharmacological PARP inhibition with PJ34 inhibits PARP activation (red block at step 3), while PAR accumulation observed in the Parg gene-disrupted mouse model indicates partially compromised enzyme regeneration (red block at step 5). ADPR, adenosine diphosphate ribose; Nam, nicotinamide.
Relevant to this model, reduced PARG activity (e.g., as observed in the Parg(110)−/− animals [36]) will inhibit PARP function through prolonged auto-PARsylation of PARPs (as illustrated in Fig. 6 by the red block at step 5), preventing their ability to undergo repeated cycles of DNA binding, automodification with PAR, and dissociation from the DNA. The overall reduction in so-called activatable PARP enzymes available to respond to TOP2-mediated DNA strand breaks therefore leads to reduced downregulation of TOP2B activity by PARsylation. Direct inhibition of PARP with PJ34 was shown to upregulate TOP2B activity in a similar manner, but in our model is expected to block progression at step 3.
The negative feedback mechanism provided by PARP may prevent TOP2B hyperactivity and thus limit or attenuate the rate at which potentially deleterious DNA strand breaks are generated. This notion is also in line with a report of spermatogenic and spermiogenic defects in Parg(110)−/− [33] and Parp2−/− mice [40].
DNA strand break-dependent shuttling of H1 from the DNA by auto-PARsylation of PARP1 has been explained by the ability of PAR, which carries two negative charges per adenine residue instead of only one as in RNA or DNA, to compete with DNA for binding to the positively charged histones and histone linker proteins [26, 54]. PAR metabolism thus acts as a biochemical mechanism that facilitates the removal of histones and histone H1-like proteins to promote opening of condensed, inaccessible chromatin domains [55–57]. Core histones and the testicular histone H1 variant H1t, official symbol HIST1H1T, are known acceptor proteins for PAR [58, 59].
Recently, a PARP1/TOP2B complex that is involved in transcriptional activation of promoters by nuclear receptors in MCF-7 cells has been reported, supporting the idea that TOP2B acts as a DNA nicking or breaking entity that is able to trigger PARP1 activation [53]. Specifically, in these cases, PAR formation through PARP1 removes H1 from promoters that are silenced by chromatin condensation in the form of a 30-nm fiber and allows for replacement of H1 by high mobility group B proteins. This alteration of chromatin structure to a more open state facilitates nuclear receptor-dependent transcription, recruitment of DNA-dependent protein kinase for DNA repair, as well as modulation of chromatin structure during transcription initiation [60–62].
The results of the present study support that view. However, the data presented show that local chromatin opening mediated by TOP2B/PARP is not limited to a role in transcription regulation but may represent a much more general means of chromatin reorganization such as that found in spermatids, which undergo global nuclear restructuring but do not perform gene transcription. Moreover, PAR-mediated modulation of TOP2B as a consequence of PARP activation by the topoisomerase as shown in the present report represents an added, novel layer of regulation.
Pioneering early efforts showed that PARsylation is in principle able to inhibit TOP2 in vitro by drastically reducing its affinity to bind DNA [63]. However, no well-defined or even recombinant PARP or topoisomerase enzymes were available at the time of those studies, which were done using biochemical cell fractions, and neither TOP2B nor PARP2 had been identified at that time [47, 64, 65]. Direct evidence of an interaction of PAR with TOP2B was also provided by a recent proteomic screening study aimed at the identification of all in vivo PARsylated proteins [28].
In summary, our data support, and at the same time significantly expand, current views that PAR metabolism is involved in local chromatin decondensation in concert with TOP2 enzymes by showing for the first time in vitro and in vivo that TOP2B-mediated DNA strand breaks induce PAR formation through activating both PARP1 and also PARP2. In steps 1 and 2 of our model (Fig. 6), we assume that TOP2B creates the DNA strand break, but the initial trigger in spermatids is not known yet. In somatic cells, it has been recently proposed for estrogen receptor-dependent transcription initiation that histone H3K9me2 demethylation by lysine-specific demethylase LSD1 (now called K-demethylase 1) causes local DNA oxidation and results in recruitment of 8-oxoguanine-DNA glycosylase 1 and TOP2B to the damaged site [66]. Interestingly, hyperacetylation of H4 has been identified as a prerequisite for TOP2B-mediated DNA strand break formation during spermiogenesis [19]. This notion emphasizes a potentially very important role of histone modifications in the generation and regulation of DNA strand breaks in spermatids that will require further investigations.
Supplementary Material
Acknowledgment
We thank Ian Cowell for critical reading of the manuscript.
Footnotes
Supported by grants from the National Institutes of Health (NIH R01 HD48837; R.G.M.) and the Mari Lowe Center for Comparative Oncology at the University of Pennsylvania (R.G.M.). This work was partially supported by an NIEHS funded Environmental Health Sciences Core Center grant P30-ES013508.
REFERENCES
- Zhao M, Shirley CR, Hayashi S, Marcon L, Mohapatra B, Suganuma R, Behringer RR, Boissonneault G, Yanagimachi R, Meistrich ML. Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis 2004; 38: 200 213 [DOI] [PubMed] [Google Scholar]
- Li Y, Lalancette C, Miller D, Krawetz SA. Characterization of nucleohistone and nucleoprotamine components in the mature human sperm nucleus. Asian J Androl 2008; 10: 535 541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 2009; 19: 1338 1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460: 473 478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 2010; 17: 679 687 [DOI] [PubMed] [Google Scholar]
- Evenson DP, Wixon R. Data analysis of two in vivo fertility studies using sperm chromatin structure assay-derived DNA fragmentation index vs. pregnancy outcome. Fertil Steril 2008; 90: 1229 1231 [DOI] [PubMed] [Google Scholar]
- Sharma RK, Said T, Agarwal A. Sperm DNA damage and its clinical relevance in assessing reproductive outcome. Asian J Androl 2004; 6: 139 148 [PubMed] [Google Scholar]
- Agarwal A, Said TM. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum Reprod Update 2003; 9: 331 345 [DOI] [PubMed] [Google Scholar]
- Aitken RJ, De Iuliis GN, McLachlan RI. Biological and clinical significance of DNA damage in the male germ line. Int J Androl 2009; 32: 46 56 [DOI] [PubMed] [Google Scholar]
- Aitken RJ, De Iuliis GN. On the possible origins of DNA damage in human spermatozoa. Mol Hum Reprod 2010; 16: 3 13 [DOI] [PubMed] [Google Scholar]
- Sakkas D, Manicardi G, Bianchi PG, Bizzaro D, Bianchi U. Relationship between the presence of endogenous nicks and sperm chromatin packaging in maturing and fertilizing mouse spermatozoa. Biol Reprod 1995; 52: 1149 1155 [DOI] [PubMed] [Google Scholar]
- Smith A, Haaf T. DNA nicks and increased sensitivity of DNA to fluorescence in situ end labeling during functional spermiogenesis. Biotechniques 1998; 25: 496 502 [DOI] [PubMed] [Google Scholar]
- Marcon L, Boissonneault G. Transient DNA strand breaks during mouse and human spermiogenesis: new insights in stage specificity and link to chromatin remodeling. Biol Reprod 2004; 70: 910 918 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Scherthan H, Burkle A, Meyer RG. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma 2005; 114: 67 74 [DOI] [PubMed] [Google Scholar]
- Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod 2008; 78: 324 332 [DOI] [PubMed] [Google Scholar]
- McPherson SM, Longo FJ. Nicking of rat spermatid and spermatozoa DNA: possible involvement of DNA topoisomerase II. Dev Biol 1993; 158: 122 130 [DOI] [PubMed] [Google Scholar]
- Roca J, Mezquita C. DNA topoisomerase II activity in nonreplicating, transcriptionally inactive, chicken late spermatids. EMBO J 1989; 8: 1855 1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Longo FJ. Expression and localization of DNA topoisomerase II during rat spermatogenesis. Mol Reprod Dev 1996; 45: 61 71 [DOI] [PubMed] [Google Scholar]
- Laberge RM, Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol Reprod 2005; 73: 289 296 [DOI] [PubMed] [Google Scholar]
- Shaman JA, Prisztoka R, Ward WS. Topoisomerase IIB and an extracellular nuclease interact to digest sperm DNA in an apoptotic-like manner. Biol Reprod 2006; 75: 741 748 [DOI] [PubMed] [Google Scholar]
- Wang JC. A journey in the world of DNA rings and beyond. Annu Rev Biochem 2009; 78: 31 54 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Ward WS. Topoisomerase II-mediated breaks in spermatozoa cause the specific degradation of paternal DNA in fertilized oocytes. Biol Reprod 2007; 76: 666 672 [DOI] [PubMed] [Google Scholar]
- Boaz SM, Dominguez K, Shaman JA, Ward WS. Mouse spermatozoa contain a nuclease that is activated by pretreatment with EGTA and subsequent calcium incubation. J Cell Biochem 2008; 103: 1636 1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J Mol Biol 2005; 348: 609 615 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 2006; 70: 789 829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, Realini CA. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem 1994; 138: 53 59 [DOI] [PubMed] [Google Scholar]
- Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol 2005; 83: 354 364 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 2008; 36: 6959 6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J 2005; 24: 1911 1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 2008; 451: 81 85 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Jacobson EL, Jacobson MK. Poly(ADP-ribose) polymerases: managing genome stability. Int J Biochem Cell Biol 2005; 37: 920 926 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 2008; 13: 3046 3082 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar J, Credidio C, Ihara M, Li Y, Wang ZQ, Meyer RG. Disruption of poly(ADP-Ribose) homeostasis affects spermiogenesis and sperm chromatin integrity in mice. Biol Reprod 2009; 81: 46 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Ihara M, Lonchar JD, Meistrich ML, Austin CA, Min W, Wang ZQ, Meyer RG. Poly(ADP-ribose) metabolism is essential for proper nucleoprotein exchange during mouse spermiogenesis. Biol Reprod 2010; 84: 218 228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 1995; 9: 509 520 [DOI] [PubMed] [Google Scholar]
- Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol 2004; 24: 7163 7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res 2007; 313: 2920 2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatcott CJ, Meyer-Ficca ML, Meyer RG, Jacobson MK. A specific isoform of poly(ADP-ribose) glycohydrolase is targeted to the mitochondrial matrix by a N-terminal mitochondrial targeting sequence. Exp Cell Res 2009; 315: 3477 3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A 2004; 101: 17699 17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci U S A 2006; 103: 14854 14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 2003; 22: 2255 2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelkarim GE, Gertz K, Harms C, Katchanov J, Dirnagl U, Szabo C, Endres M. Protective effects of PJ34, a novel, potent inhibitor of poly(ADP-ribose) polymerase (PARP) in in vitro and in vivo models of stroke. Int J Mol Med 2001; 7: 255 260 [PubMed] [Google Scholar]
- Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch JL, Davidson I, Parvinen M, Sassone-Corsi P. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods 2004; 1: 249 254 [DOI] [PubMed] [Google Scholar]
- Willmore E, Frank AJ, Padget K, Tilby MJ, Austin CA. Etoposide targets topoisomerase IIalpha and IIbeta in leukemic cells: isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol Pharmacol 1998; 54: 78 85 [DOI] [PubMed] [Google Scholar]
- Padget K, Carr R, Pearson AD, Tilby MJ, Austin CA. Camptothecin-stabilised topoisomerase I-DNA complexes in leukaemia cells visualised and quantified in situ by the TARDIS assay (trapped in agarose DNA immunostaining). Biochem Pharmacol 2000; 59: 629 638 [DOI] [PubMed] [Google Scholar]
- Austin CA, Marsh KL, Wasserman RA, Willmore E, Sayer PJ, Wang JC, Fisher LM. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J Biol Chem 1995; 270: 15739 15746 [DOI] [PubMed] [Google Scholar]
- Austin CA, Sng JH, Patel S, Fisher LM. Novel HeLa topoisomerase II is the II beta isoform: complete coding sequence and homology with other type II topoisomerases. Biochim Biophys Acta 1993; 1172: 283 291 [DOI] [PubMed] [Google Scholar]
- Marsh KL, Meczes EL, Thorn R, Marshall R, Austin CA. Site-directed mutagenesis of human DNA topoisomerase II beta. Biochem Soc Trans 1997; 25: S638 [DOI] [PubMed] [Google Scholar]
- Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res 2001; 268: 7 13 [DOI] [PubMed] [Google Scholar]
- Pommier Y, Schwartz RE, Kohn KW, Zwelling LA. Formation and rejoining of deoxyribonucleic acid double-strand breaks induced in isolated cell nuclei by antineoplastic intercalating agents. Biochemistry 1984; 23: 3194 3201 [DOI] [PubMed] [Google Scholar]
- Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 2010; 17: 421 433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Rosenfeld MG. A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle 2006; 5: 2557 2560 [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006; 312: 1798 1802 [DOI] [PubMed] [Google Scholar]
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter P, Realini C. Histone shuttle driven by the automodification cycle of poly(ADP-ribose)polymerase. Environ Mol Mutagen 1993; 22: 278 282 [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 2003; 299: 560 562 [DOI] [PubMed] [Google Scholar]
- Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol 2003; 56: 55 83 [DOI] [PubMed] [Google Scholar]
- Wacker DA, Frizzell KM, Zhang T, Kraus WL. Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: possible targets for drug-based therapies. Subcell Biochem 2007; 41: 45 69 [DOI] [PubMed] [Google Scholar]
- Faraone Mennella MR, Quesada P, Farina B, Leone E, Jones R. Purification of non-histone acceptor proteins for ADP-ribose from mouse testis nuclei. Biochem J 1984; 221: 223 233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanga M, Atorino L, Tramontano F, Farina B, Quesada P. Poly(ADP-ribose) binding properties of histone H1 variants. Biochim Biophys Acta 1998; 1399: 154 160 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 2010; 39: 736 749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 2010; 39: 8 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol 2008; 20: 294 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby MK, Schmitt B, Jongstra-Bilen J, Vosberg HP. Inhibition of calf thymus type II DNA topoisomerase by poly(ADP-ribosylation). EMBO J 1985; 4: 2129 2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics 1999; 57: 442 445 [DOI] [PubMed] [Google Scholar]
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 1999; 274: 17860 17868 [DOI] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 2008; 319: 202 206 [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma 2003; 111: 483 488 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.