Abstract
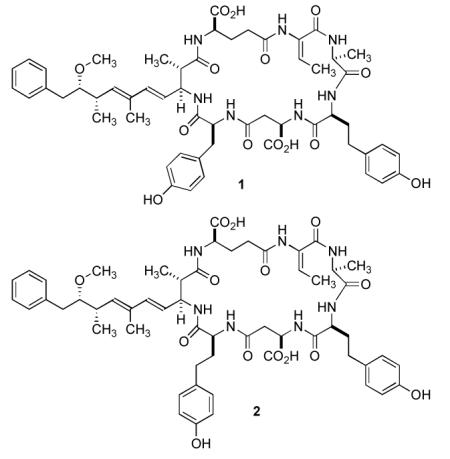
Microcystins (MCs) are toxic heptapeptides found in cyanobacteria and share the common structure cyclo(-d-Ala1-l-X2-d-isoMeAsp3-l-Z4-Adda5-d-isoGlu6-Mdha7). The letters X and Z in the general formula above represent a wide range of L-amino acids that occupy positions 2 and 4, respectively. In general the variation in structural variants is due to the exchange of amino acids in position 7, 2, and 4. In the present work we report two homotyrosine (Hty)-containing microcystin variants, [d-Asp3,(E)-Dhb7]-MC-HtyY (1) and [d-Asp3,(E)-Dhb7]-MC-HtyHty (2), which were isolated from strain No80 of Planktothrix rubescens. Their structures were elucidated using amino acid analysis as well as 1D- and 2D NMR techniques. The adenylation domains of McyABC involved in amino acid activation in positions 7, 2, and 4 of the microcystin molecule, respectively, were compared with corresponding genes of Planktothrix strain CYA126/8 producing [d-Asp3,Mdha7]-MC-RR and [d-Asp3,Mdha7]-MC-LR. While the adenylation domain comparison of McyAB between the two Planktothrix strains revealed considerable DNA recombination, the adenylation domain of McyC showed only a single amino acid substitution that was correlated with the replacement of Arg by Hty in position 4 of the microcystin molecule.
Microcystins (MC) are cyclic heptapetides produced by cyanobacteria, most prominently by the genera Anabaena, Microcystis, and Planktothrix. MCs are known to be toxic to aquatic biota, livestock and humans.1 MCs have the common structure cyclo(-d-Ala1-l-X2-d-isoMeAsp3-l-Z4-Adda5-d-isoGlu6-Mdha7), where the upper case numbers indicate the position of an amino acid according to the accepted numbering system.2 Structural diversification is the result of amino acid substitutions in positions 2, 3, 4, and 7. The changes in positions 3 and 7 are semiconservative. Thus, either d-aspartic acid (d-Asp) or d-erythro-β-methylaspartic acid (d-MeAsp) may occupy position 3, whereas either N-methyl-dehydroalanine (Mdha), dehydroalanine (Dha), 2-amino-2-butenoic acid (Dhb) or another dehydroamino acid (Dhaa), is incorporated into position 7. The letters X and Z in the general formula above represent a wide range of l-amino acids that occupy positions 2 and 4, respectively. These latter substitutions are important in MC nomenclature as the identity of these amino acids is indicated as an appendix using the one letter code. Thus, MC-LR refers to a microcystin bearing leucine and arginine in the variable positions 2 and 4, respectively. The d-alanine in position 1 and Adda ((2S,3S,8S,9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid) in position 5 are mostly conserved in this class of compounds, while the d-glutamic acid residue in position 6 is strictly conserved.2 MCs are synthesized by the thiotemplate mechanism like other nonribosomal peptides produced by bacteria and fungi.3 The enzymes catalyzing these reactions have a modular structure, each module containing specific functional domains for activation, aminoacyl adenylation (adenylation (A) domains), and thioesterification (thiolation domains) of the amino acid substrate and for the elongation (condensation domains) of the growing peptide. Adenylation domains show a high degree of conservation in their core motifs, thus enabling the definition of rules for the structural basis of substrate recognition in A-domains of nonribosomal peptide synthetases.4-6 Our understanding of substrate specificity is based on the analysis of the X-ray crystal structure of the A-domain of gramicidin synthetase (GrsA).7 Mutational investigations of the A-domain of the peptide synthetase (GrsA) activating phenylalanine revealed significant changes in enzyme activity after mutation of the critical amino acids within the A3 to A6 region.4 The structural organization of the mcy gene cluster encoding MC biosynthesis has been elucidated, and it has been proposed that McyA, McyB and McyC (the McyABC cluster) are responsible for the co-linear activation and incorporation of Mdha7, d-Ala1, l-X2, d-MeAsp3, l-Z4 during biosynthesis.8 For Planktothrix spp., it has been shown recently that specific genotypes of McyAA1 and McyBA1 correlate with the presence of specific amino acids in positions 2 and 7 of the MC molecules produced by these strains.9 In the present study, the structures of two new microcystins containing tyrosine or homotyrosine instead of arginine in position 4 of the sequence are reported: [d-Asp3,(E)-Dhb7]-MC-HtyY (1) and [d-Asp3,(E)-Dhb7]-MC-HtyHty (2) isolated from strain No80 of P. rubescens. In addition, DNA sequence comparisons are provided for adenylation domains within the mcyABC cluster between strains No80 and CYA126/8, a P. agardhii strain producing [d-Asp3,Mdha7]-MC-RR and [d-Asp3,Mdha7]-MC-LR.
Results and Discussion
In an earlier study it was reported that seven isolates of P. rubescens obtained from a specific lake (Schwarzensee) in the Austrian Alps (Upper Austria), contained exclusively unknown microcystin variants, all of which eluted more than three minutes after [d-MeAsp3,Mdha7]-MC-LR during HPLC analysis under standardized conditions.10 In P. rubescens strain No80, the two most abundant MC variants, 1 (tR = 24.0 min) and 2 (tR = 23.5 min), contributed 76±5% (SE) and 7±4%, respectively, of the total microcystin content, whereas P. agardhii strain CYA126/8 produced mainly [d-Asp3,Mdha7]-MC-RR (tR = 13.9 min, 92±1%) and [d-Asp3,Mdha7]-MC-LR (tR = 19.5 min, 7±1%).
P. rubescens strain No80 was grown in laboratory culture under standard conditions and the crude peptide fraction was isolated from the dried cell mass after aqueous methanolic extraction. Microcystins 1 and 2 were obtained pure in milligram amounts after pre-purification on C18 solid-phase extraction cartridges followed by preparative HPLC.
The planar structure of 1 was elucidated by analysis of spectroscopic data. FABMS analysis gave a series of pseudomolecular ions at 1074 and 1090 amu for [M+Na]+ and [M+K]+, respectively. A [M+Na]+ ion at m/z 1074.4815 during HR-ESI-TOF-MS analysis suggested a molecular formula of C55H69N7O14Na (calc 1074.4800, Δ −1.5 mmu).
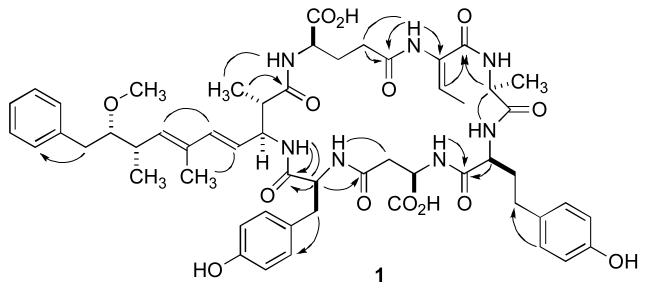
The 1H NMR spectrum of 1 in CD3OH (600 MHz and 800 MHz) displayed the typical profile of a peptide. The satisfactory separation of the NH resonances in the 1H NMR spectrum allowed the identification of individual spin systems on the basis of 1D-TOCSY data (Table 1). These were expanded and confirmed by analysis of gCOSY, gHSQC, gHMBC, and ROESY data as well as by amino acid analysis (Figure 1). In this fashion, the alanine, aspartate, glutamate, tyrosine and homotyrosine residues were readily identified. The Adda unit was assembled in straightforward fashion on the basis of the gCOSY, gHSQC, and gHMBC data. ROESY data supported the assignment and suggested the conventional all-E configuration of the trisubstituted double bonds. Subsequently, starting from a methyl doublet and a quartet due to an olefinic proton resonating at δ 1.94 and at δ 5.77, respectively, a 2-amino-2-butenoic acid (Dhb) residue was assembled on the basis of the 2D-NMR data. This suggests that 1 bears this residue instead of a dehydroalanine (Dha) or a N-methyl-dehydroalanine (Mdha) commonly found in other MCs.
Table 1.
1H and 13C NMR Chemical Shifts of Microcystin 1 Measured in CD3OH (600 MHz, 25 °C) (A part of the HMBC spectrum was additionally recorded at 800 MHz).
| MC1 | |||||
|---|---|---|---|---|---|
| Unit | C/H no | δ C | δH (J in Hz) | HMBC a | ROESY |
| Adda | 1 | 176.2 | |||
| 2 | 45.6 | 2.65, m | 1, 3, 11 | ||
| 3 | 55.4 | 4.64, m | 2, 4, 5 | 2 | |
| 4 | 125.5 | 5.41, dd (15.6, 8.8) | 3, 6 | 2, 3, 11, 12 | |
| 5 | 138.9 | 6.31, d (15.6) | 3, 6, 7, 12 | 2, 3, 4, 7, 11, 12 | |
| 6 | 133.2 | ||||
| 7 | 137.1 | 5.48, d (9.7) | 5, 8, 9, 12, 13 | 8, 9, 13 | |
| 8 | 37.2 | 2.62, m | 6, 7, 9, 13 | 12 | |
| 9 | 87.9 | 3.27, m | 7, 8, 13, 14, 15 | ||
| 10a | 38.5 | 2.82, dd (14.0, 4.8) | 8, 9, 15, 16/20 | 13 | |
| 10b | 2.67, m | 8, 9, 15, 16/20 | 11 | ||
| 11 | 15.9 | 1.09, d (6.8) | 1, 2, 3 | ||
| 12 | 12.3 | 1.62, s | 6, 7 | ||
| 13 | 16.0 | 1.03, d (6.8) | 7, 8, 9 | ||
| 14 | 58.2 | 3.24, s | 9 | ||
| 15 | 140.0 | ||||
| 16/20 | 130.2 | 7.19, m | 10, 16/20, 18 | ||
| 17/19 | 128.9 | 7.22, m | 15, 17/19 | 9, 10b | |
| 18 | 126.6 | 7.17, m | 16/20 | ||
| NH | 7.28, m | Tyr-1 | 2, 4, Tyr-2 | ||
| Glu | 1 | 174.2 | |||
| 2 | 52.6 | 4.46, m | 1, 3 | 3a/b, 4a/b | |
| 3a | 28.9 | 1.77, m | |||
| 3b | 2.08, m | ||||
| 4a 4b |
33.6 | 2.44, m 2.32, m |
5 | 3a/b 3a/b |
|
| 5 | 174.0 | ||||
| NH | 7.22, m | 2, 3a, Adda-11 | |||
| Dhb | 1 | 166.0 | |||
| 2 | 131.5 | ||||
| 3 | 123.5 | 5.77, q (7.3) | 1, 2, 4 | 4 | |
| 4 | 13.0 | 1.94, q (7.3) | 1, 2, 3 | ||
| NH | 9.93, s | 1, Glu-5 | 3, 4, Tyr-2, Glu-4a/b | ||
| Ala | 1 | 175.3 | |||
| 2 | 49.1 | 4.54, m | 1, 3, Dhb-1 | 3 | |
| 3 | 16.5 | 1.01, d (7.5) | 1, 2 | ||
| NH | 7.54, br | 2, 3 | |||
| Hty1 | 1 | 174.3 | |||
| 2 | 55.7 | 4.08, m | 1 | 3a/b, 4a/b | |
| 3a | 33.8 | 2.13, m | |||
| 3b | 2.01, m | ||||
| 4a | 31.9 | 2.71, m | 2, 3 | 3a/b | |
| 4b | 2.49, m | ||||
| 5 | 132.6 | ||||
| 6/10 | 130.3 | 7.00, d (8.4) | 4a, 7/9, 6/10, 8 | 2, 3a/b, 4a/b | |
| 7/9 | 115.8 | 6.65, d (8.4) | 7/9, 5, 8 | ||
| 8 | 156.2 | ||||
| NH | 8.63, br | 2, 3a/b, 4a/b, Asp-NH, Ala-NH, Ala-2, Ala-3, Asp-NH |
|||
| Asp | 1 | 175.1 | |||
| 2 | 49.7 | 4.77, m | 1, 4 | ||
| 3a | 35.9 | 2.74, m | |||
| 3b | 2.12, m | ||||
| 4 | 173.9 | ||||
| NH | 8.39, d (8.8) | 3a, Hty-1 | 1, 3a/b, Ala-NH, Hty-2, Hty-3b, Glu-3a, |
||
| Tyr | 1 | 171.3 | |||
| 2 | 55.4 | 4.63, m | 1, 3, Asp-4, | 3b | |
| 3a | 36.6 | 3.38, m | 5/9 | ||
| 3b | 2.48, m | 5/9 | |||
| 4 | 129.2 | ||||
| 5/9 | 130.6 | 6.96, d (8.4) | 7, 5/9, 6/8 | 2, 3a/b, Ala-3 | |
| 6/8 | 116.0 | 6.58, d (8.4) | 4, 6/8, 7 | 3a, Ala3, Dhb-4 | |
| 7 | 156.9 | ||||
| NH | 8.81, br | 2, 3b, Adda-NH, Asp-3a/b | |||
HMBC correlations are given from proton(s) stated to the indicated carbon atom.
Figure 1.
Key HMBC (arrows) and ROESY (solid lines) correlations for 1.
After all of the subunits have been assembled, it was important to place the tyrosine and the homotyrosine residues into the correct positions within the cyclic heptapeptide. As a starting hypothesis, it was assumed that these two amino acids would occupy the variable sites 2 and 4 in the accepted nomenclature system. For this task the ROESY data proved to be extremely valuable. The α- and β-protons of the D-Ala unit resonating at 4.54 ppm and 1.01 ppm showed cross-correlations to the exchangeable NH proton of the homotyrosine unit. The assignment of the latter to the homotyrosine rests on a COSY correlation to the well-separated aminomethine resonance at 4.08 ppm, which is part of the C-2, C-3, and C-4 spin system as demonstrated by 1D-TOCSY.
A similar argument was constructed for the placement of the tyrosine residue in position 4, between the aspartate and the Adda unit. Thus, a ROESY correlation was observed between the NH proton of the Adda unit resonating at 7.28 ppm and the α-proton of the tyrosine unit at 4.63 ppm. The latter resonance also showed a ROESY correlation to the resonances for the H-5/9 protons of the tyrosine aromatic ring at 6.96 ppm, thereby confirming the assignments for the tyrosine based on analysis of gCOSY and gHMBC data. In addition, a HMBC correlation between the NH of Adda to the carbonyl C atom of the Tyr residues secured the sequence assignment.
Lastly, the configuration of the Dhb residue remained to be assigned. No relevant cross-peaks could be observed in the ROESY spectra, so we had to resort to a chemical shift correlation. The higher field chemical shift (δ 5.77) of H-3 in 1 when compared to the chemical shift of the corresponding proton of the (Z)-Dhb unit in nodularin (δ 6.90-6.94) suggested that the geometry of the Dhb unit in 1 is E. This analysis is in agreement with the assignment of the stereochemistry of the Dhb residue in [d-Asp3,(E)-Dhb7]-MC-LR and MC-HtyR isolated from Oscillatoria agardhii CCAP1459/14.11 The olefinic proton of the Dhb unit in these compounds resonates at δ 5.69 and 5.73, respectively.
The assignment of the structure of 2 followed a similar approach. FABMS analysis of 2 suggested the presence of an additional methylene group as the pseudomolecular ions for [M+Na]+ and [M+K]+ were observed at m/z 1088 and m/z 1104, respectively. This was confirmed by HR-ESI-TOF-MS, which yielded a pseudomolecular ion [M+Na]+ at m/z 1088.4960, suggesting a molecular formula of C56H71N7O14Na (calc 1088.4957, Δ −0.3 mmu). The appearance of the 1H NMR spectrum of 2 suggested that it was closely related to 1 since many signals indicative of the characteristic structural elements present in 1 were observed. The location of the additional methylene group became apparent during the amino acid analysis of the hydrolysate of 2 when two equivalents of Hty were observed and tyrosine was not found. Further analysis of 1D-TOCSY, gCOSY, gHMBC and ROESY data confirmed that the structure of 1 and 2 were identical but for the incorporation of a second homotyrosine in lieu of the tyrosine residue present in 1 (Table 2).
Table 2.
1H and 13C NMR Chemical Shifts of Microcystin 2 Measured in CD3OH (600 MHz, 25 °C)(A Part of the HMBC spectrum was additionally recorded at 800 MHz).
| MC2 | |||||
|---|---|---|---|---|---|
| Unit | C/H no | δ C | δH (J in Hz) | HMBC a | ROESY |
| Adda | 1 | 176.9 | |||
| 2 | 45.2 | 2.91, m | 11 | ||
| 3 | 55.8 | 4.60, m | 2, 4, 5 | ||
| 4 | 126.2 | 5.46, dd (15.4, 8.5) | 6 | 3, 11, 12 | |
| 5 | 138.3 | 6.24, d (15.4) | 3, 6, 7, 12 | 3, 4, 7, 11, 12 | |
| 6 | 133.6 | ||||
| 7 | 136.9 | 5.43, d (9.8) | 5, 12 | 8, 13 | |
| 8 | 37.4 | 2.58, m | 12, 13 | ||
| 9 | 88.1 | 3.23, m | |||
| 10a | 38.7 | 2.79, dd (14.0, 4.9) | 9, 15, 16/20 | 13 | |
| 10b | 2.65, dd (14.0, 7.4) | 8, 9, 15, 16/20 | |||
| 11 | 15.9 | 1.05, d (6.8) | 1, 2, 3 | ||
| 12 | 12.5 | 1.61, s | 5, 6, 7 | ||
| 13 | 16.1 | 0.99, d (6.7) | 7, 8, 9 | ||
| 14 | 58.4 | 3.22, s | 9 | ||
| 15 | 140.3 | ||||
| 16/20 | 130.5 | 7.17, m | 18, 10 | 10a/b | |
| 17/19 | 128.9 | 7.23, m | 15, 17/19 | ||
| 18 | 126.7 | 7.16, m | 16/20 | ||
| NH | 7.68, d (8.6) | 2, 3, 4, Hty2-2 | |||
| Glu | 1 | 175.9 | |||
| 2 | 54.1 | 4.32, m | 1, 3 | 3b, 4b | |
| 3a | 28.3 | 2.07, m | |||
| 3b | 1.97, m | ||||
| 4a | 33.8 | 2.43, m | |||
| 4b | 2.29, m | ||||
| 5 NH |
174.6 |
n.d. |
|||
| Dhb | 1 | 166.5 | |||
| 2 | 131.5 | ||||
| 3 | 124.3 | 5.74, q (7.4) | 1, 2 | 4 | |
| 4 | 13.2 | 1.93, d (7.4) | 2, 3 | ||
| NH | 9.84, s | 1, Glu-5 | 3, Glu-4a/b | ||
| Ala | 1 | 175.4 | |||
| 2 | 49.4 | 4.60, m | 1, 3, Dhb-1 | 3 | |
| 3 | 17.0 | 1.34, d (7.2) | 1, 2 | ||
| NH | 8.04, d (7.9) | 2, 3, Hty2-6/10 | |||
| Hty1 | 1 | 174.2 | |||
| 2 | 55.9 | 4.16, m | 1 | 3a/b, 4b | |
| 3a | 34.0 | 2.25, m | |||
| 3b | 2.10, m | ||||
| 4a | 32.1 | 2.74, m | 6/10 | 4b | |
| 4b | 2.54, m | 6/10 | |||
| 5 | 132.9 | ||||
| 6/10 | 130.6 | 7.02, d (8.5) | 4, 6/10, 8 | 2, 3a/b, 4a/b | |
| 7/9 | 116.0 | 6.65, d (8.5) | 5, 7/9, 8 | ||
| 8 | 156.3 | ||||
| NH | 8.74, d (7.1) | 1, Ala-1 | 2, 3a/b, 4a/b, Ala-2, Ala-NH, Asp-NH | ||
| Asp | 1 | n.d. | |||
| 2 | 51.1 | 4.77, m | |||
| 3a | 37.1 | 2.94, m | 3b | ||
| 3b | 2.31, m | ||||
| 4 | 175.0 | ||||
| NH | 8.17, d (9.1) | 3b | |||
| Hty2 | 1 | 172.7 | |||
| 2 | 53.2 | 4.36, m | 1, Asp-4 | 3a/b, 4b | |
| 3a | 34.0 | 2.26, m | 3b | ||
| 3b | 1.74, m | ||||
| 4a | 32.0 | 2.54, m | 5 | 3a/b | |
| 4b | 2.45, m | 5, 6/10 | 3a/b | ||
| 5 6/10 |
132.8 130.1 |
6.94, d (8.5) |
6/10, 8 |
2, 3a/b, 4a, Ala-3 |
|
| 7/9 | 116.0 | 6.64, d (8.5) | 5, 7/9, 8 | ||
| 8 | 156.5 | ||||
| NH | 8.64, d (8.8) | Asp-4 | |||
HMBC correlations are given from proton(s) stated to the indicated carbon atom.
Surprisingly differences in chemical shifts were observed between 1 and 2: In 2 the amide and the methyl proton resonances of alanine are significantly shifted when compared to those in 1 (1: δ (NH) = 7.54, δ (Me) = 1.01; 2: δ (NH) = 8.04, δ (Me) = 1.34). This can be rationalized on the basis of the model of Trogen et al., who proposed a saddle-shaped form for MC-RR on the basis of NMR data and molecular dynamics calculation.12,13 The longer alkyl chain of Hty could allow the aromatic ring to be spatially close to the alanine residue in 2 whereas the arm is not sufficiently long for the tyrosine phenyl ring present in 1 to be placed similarly close to the alanine residue. However, the close spatial proximity of the tyrosine and alanine residues in 1 is indicated by a ROESY correlation between the alanine methyl resonance and the H-6/8 protons of the aromatic ring of the tyrosine, suggesting additional conformational consequences upon replacement of a Tyr by a Hty residue.
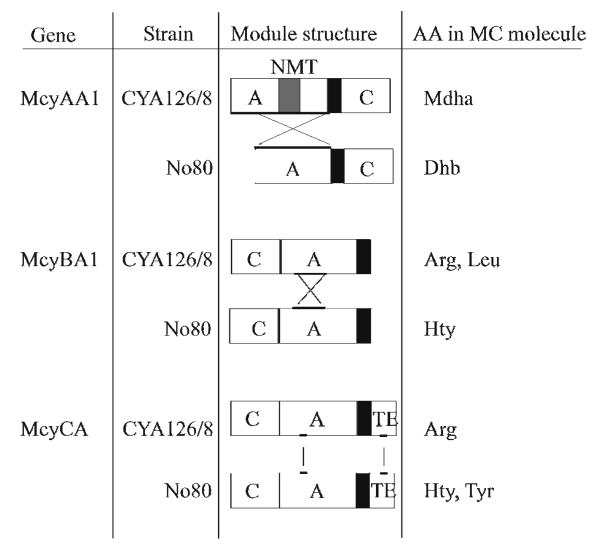
According to the model proposed by Tillett et al., McyAA1 is responsible for the selection of l-serine for incorporation into position 7 of the molecule.8 In a reaction that is not well understood, this residue is subsequently transformed by dehydration into N-methyl-dehydroalanine. l-Threonine is thought to be dehydrated into 2-amino-2-butenoic acid in an analogous process.14 In Planktothrix spp., genomic sequence analysis of mcyAA1 allows the prediction of the presence of Mdha7 or Dhb7 in the microcystin molecule, i.e., genotypes with a short mcyAA1 (1,692 bp), such as P. rubescens No80, lack the N-methyl transferase (NMT) and produce MCs with Dhb7, whereas genotypes with a long mcyAA1 (3,022 bp), such as CYA126/8, possess a NMT and produce MCs bearing a Mdha7 residue.9 Outside the NMT region the McyAA1 genotype of strain No80 and of CYA126/8 differ in amino acid composition by about 45%. This is not surprising since the two domains must select different amino acids, threonine and serine, respectively, in order to produce these different MCs. The current predictive structure-based model requires the presence of ten critical amino acids (so called “signature sequences”) in the putative binding pockets of adenylation domains that activate these two building blocks. 4-6 The sequences for these signature sequences of strain No80, DFWNIGMVHK and of CYA126/8, DVWHISLIDK, respectively, are in perfect agreement with this model.
According to the analysis of Tillett et al.,8 McyBA1 is encoding the protein for the incorporation of the variable amino acid in position 2. Our sequence analysis suggests that the difference in mcyBA1 (1,539 bp) is 4% between the two strains, No80 and CYA126/8. Some of these differences affect the putative binding pocket between conserved motives A3 to A6.3 Notably, the McyBA1 genotype of strain No80 shows the signature sequence DALLFGFVAK and matches exactly the unique McyBA1 genotype reported to contain MCs bearing homotyrosine and leucine in position 2 but no trace of metabolites with arginine in that position.9 In contrast, strain CYA126/8, showing the signature sequence DALFFGLVDK, falls into a group of genotypes elaborating MCs bearing arginine, arginine and leucine or arginine, leucine and homotyrosine in position 2.9 For both adenylation domains, McyAA1 and McyBA1, it was shown that recombination of DNA fragments caused the alteration leading to the observed new substrate specificities (Figure 2).9,15
Figure 2.
Scheme Illustrating the Two Types of Adenylation (A) Domain Alterations Leading to New Substrate Specificities in Planktothrix spp.: Recombination of Whole Domains (McyAA1)9 or Shorter DNA Fragments (McyBA1)15 or Point Mutations (McyCA). C, condensation domain, NMT, N-methyl transferase, TE, thioesterase. Thick bars indicate the gene regions affected by mutation.
In contrast, the entire mcyC gene (3,897 bp) of strain No80 showed only five nucleotide substitutions when compared to that of mcyC from CYA126/8. Two substitutions are located within mcyCA (1,344 bp), equivalent to a sequence difference of only 0.15%, of which one results in the replacement of an aspartate by asparagine (position 2,326 in AJ749287), part of the signature sequence DPWGFGLVNK vs. DPWGFGLVDK of McyCA in CYA126/8. Both signature sequences had no precedent in the database.6 Within the type I thioesterase part of mcyC, one non-synonymous substitution results in the replacement of a threonine by alanine (position 3,520). Considering that the major MCs of strain CYA126/8 have arginine in position 4 whereas the MCs of strain No80 bear homotyrosine or tyrosine at that site, these sequence differences are surprisingly minor given the differences in charge and polarity between the amino acids. First attempts to reconstitute the mutation in vivo within McyCA in the genetic manipulable model strain CYA126/8 were unsuccessful.
Experimental Section
General Experimental procedures
One and two-dimensional NMR spectra were recorded on a Bruker DRX-600 equipped with a cryoprobe and a Bruker AVII-800 equiped with a cryoprobe. For NMR analysis, 1 and 2 were dissolved in CD3OH (methyl-d3 alcohol) and 1H and 13C NMR spectra were recorded at 25 °C. HR-ESI-TOF-MS were recorded on an Agilent LC-MSDTOF instrument. For amino acid analysis a Hitachi L-8500A amino acid analyzer was used. The determination of the amino acids enantiomers was carried out on a GC-MS instrument (Fison Instruments, GC 8000 Top, MD 800). HPLC separations were performed on an Agilent HP1100 system equipped with diode array detection.
Culture of Planktothrix
Strain No80 has been isolated during the study of Kurmayer et al. from Lake Schwarzensee (Upper Austria).10 It has been deposited under this accession number in the culture collection of the Institute for Limnology in Mondsee, Austria. The strain CYA126/8 (Lake Langsjθn, Finland) was kindly provided by Kaarina Sivonen (Helsinki University, Finland). According to PCR analysis and sequencing of various marker genes, strain No80 was classified as P. rubescens.16 Both strains were cultivated in modified BG11 medium17 containing 2 mM NaNO3 + 10 mM NaHCO3 at 15 °C under continuous light (5-10 μmol m−2 s−1, Osram Type L30W/77 Fluora).
Extraction and Isolation
Cells of No80 were harvested from mass cultures in BG11 and lyophilized. Cells were ultrasonicated and extracted with 50% MeOH (v/v) on a shaker (250 rpm) for 30 min. The extract was centrifuged at 16,000 g for 10 min twice and cleaned up by solid-phase extraction using Sep-Pak ® tC18 cartridges (Waters, Vienna, Austria). The two major MC variants (1, 2) were preparatively separated on a LiChrosper® 100, ODS, 5 μm, LiChroCART® 250-4 cartridge system (Merck, Darmstadt, Germany) using a linear gradient of aqueous CH3CN (with 0.05% v/v TFA) starting with 30% CH3CN (v/v) and increasing up to 44% CH3CN within 10 min and ending after 15 min with 49.5% CH3CN with a flow of 1 mL min−1. The elution times were 17.0 min (2) and 17.5 min (1). Both fractions were colorless oils and HPLC analysis of the purified compound did not show any UV absorbing contaminants. From compound 1 2,214 μg and from compound 2 735 μg were obtained.
Amino Acid Analysis
For amino acid analysis, 10 – 100 μg each of pure MCs were heated at 110 °C for 16 h in 0.5 mL of 6 M HCl containing 1% phenol. The reaction mixture was dried, dissolved in 100 μL of 0.02 M HCl, and subjected to amino acid analysis. Retention times (min) of the amino acid derivatives were as follows: 1: Asp (10.18), Glu (19.81), Ala (34.48), Tyr (49.33), Hty (60.13); 2: Asp (10.32), Glu (19.86), Ala (34.48), Hty (60.05). To determine the enantiomers of the amino acids, 100 μg of each highly purified microcystin (1 and 2) and equivalent amounts of d- and l- amino acid standards were esterified with 100 μL 3 M HCl gas in MeOH (Supelco, Bellefonte, PA) at 110 °C for 24 h. The reaction mixtures were dried and subsequently trifluoroacetylated with trifluoroacetic acid anhydride (TFAA; Fluka, Switzerland) at 80 °C for 1 h. Analyses were conducted on a Chirasil Val column (Permabond® - l - Chirasil-Val; 25 m × 0.25 mm; Macherey-Nagel, Düren, Germany) under the following separation conditions: 2 min at 80 °C, 80 °C to 180 °C at the rate of 8 °C min−1, and 20 min at 180 °C. The retention times (min) of enantiomeric amino acids were: d-Ala (5.22), l-Ala (5.39), d-Asp (11.05), l-Asp (11.09), d-Glu (13.59), l-Glu (13.66), d-Tyr (17.83), l-Tyr (17.95), and l-Hty (23.73). l-Hty was obtained after isolation and purification of anabaenopeptin B (m/z 837 [M+H+]), subsequently followed by acid hydrolosis and esterification as described above.18
Quantitative Microcystin Analysis
Strains were grown in parallel and analyzed independently three times. Three weeks after inoculation, cells were filtered onto glass fiber filters (GF/C, Comesa, Vienna, Austria) and MCs were extracted using 75% (w/v) aqueous MeOH and the extracts were analyzed for MC by HPLC-DAD using the cartridge system as described above using a linear gradient of aqueous CH3CN (with 0.05% v/v TFA) starting with 30% CH3CN (v/v) and ending after 42 min with 70 % CH3CN with a flow of 1 mL min−1.19 MC variants were identified at 240 nm by their characteristic absorption spectra (original spectrum and first order derivative) and retention times.9 [MeAsp3, Mdha7]-MC-RR, MC-YR and MC-LR were used as external standards (Calbiochem, Schwalbach, Germany).
Genetic Analysis
For DNA extraction 2 mL of culture were incubated for 1 h on ice and centrifuged at 16,000 g for 10 min, and the pellet was lyophilized in a vacuum centrifuge at 30 °C. DNA was extracted using a previously described protocol.20 For PCR, DNA extracts were diluted one hundred-fold and 1.0 μL of the sample was pipetted into reaction tubes and incubated as described below. PCR amplifications were performed in a volume of 20 μL, containing 2 μL of Qiagen PCR buffer (Qiagen, VWR, Austria), 1.2 μL MgCl2 (25 mM, Qiagen), 0.6 μL deoxynucleotide triphosphates (10 μM each, MBI Fermentas, St. Leon-Rot, Germany), 1 μL of each primer (10 pmol μL−1), 0.1 μL Taq DNA polymerase (Qiagen), 13.1 μL sterile Millipore water, and 1.0 μL of the sample. Primers used for PCR and sequencing and all thermal cycling protocols have been published previously.9 The sequences have been submitted to the DDBJ/ EMBL/GenBank databases under accession numbers (AJ749254, AJ749278, AJ749287) and compared with strain CYA126/8, which was sequenced for the mcy gene cluster AJ441056.21
Supplementary Material
Acknowledgment
We thank J. Schmidt and J. Knoblechner for culturing of cyanobacterial strains and microcystin purification in the laboratory. We thank M. Meixner for sequencing. K. Ishida performed the amino acid analysis. This study was supported by the Austrian Science Funds (P18185). Work in Hawaii was supported by NSF grant OCE04-32479 and PHS grant P50ES012740. K.G. is a European Young Investigator (EURYI) and thanks the SNF for financial support (PE002-117136/1). J. B. thanks J. Pernthaler for his support. The work in Kilchberg was supported by SNF grant 3100A0-112106 /1.
Footnotes
Supporting Information Available. Copies of 1H NMR spectra are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chorus I, Bartram J. Toxic Cyanobacteria in Water:A Guide to their Public Health Consequences, Monitoring and Management. WHO, E & FN Spon; London: 1999. p. 416. [Google Scholar]
- 2.Diehnelt CW, Dugan NR, Peterman SM, Budde WL. Anal. Chem. 2006;78:501–512. doi: 10.1021/ac051556d. [DOI] [PubMed] [Google Scholar]
- 3.Marahiel MA, Stachelhaus T, Mootz HD. Chem. Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 4.Stachelhaus T, Mootz HD, Marahiel MA. Chem. Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 5.Challis GL, Ravel J, Townsend CA. Chem. Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 6.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson D. Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti E, Stachelhaus T, Marahiel MA, Brick P. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tillett D, Dittmann E, Erhard M, von Döhren H, Börner T, Neilan BA. Chem. Biol. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- 9.Kurmayer R, Christiansen G, Gumpenberger M, Fastner J. Microbiol. 2005;151:1525–1533. doi: 10.1099/mic.0.27779-0. [DOI] [PubMed] [Google Scholar]
- 10.Kurmayer R, Christiansen G, Fastner J, Börner T. Environ. Microbiol. 2004;6:831–841. doi: 10.1111/j.1462-2920.2004.00626.x. [DOI] [PubMed] [Google Scholar]
- 11.Sano T, Kaya K. Tetrahedron. 1998;54:463–470. [Google Scholar]
- 12.Trogen G-B, Annila A, Eriksson J, Kontteli M, Meriluoto J, Sethson I, Zdunek J, Edlund U. Biochemistry. 1996;35:3197–3205. doi: 10.1021/bi952368s. [DOI] [PubMed] [Google Scholar]
- 13.Trogen G-B, Edlund U, Larsson G, Sethson I. Eur. J. Biochem. 1998;258:301–312. doi: 10.1046/j.1432-1327.1998.2580301.x. [DOI] [PubMed] [Google Scholar]
- 14.Rinehart KL, Namikoshi M, Choi BW. J. Appl. Phycol. 1994;6:159–176. [Google Scholar]
- 15.Kurmayer R, Gumpenberger M. Mol. Ecol. 2006;15:3849–3861. doi: 10.1111/j.1365-294X.2006.03044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christiansen G, Molitor C, Philmus B, Kurmayer R. Mol. Biol. Evol. 2008;25:1695–1704. doi: 10.1093/molbev/msn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rippka R. Meth. Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 18.Grach-Pogrebinsky O, Sedmak B, Carmeli S. Tetrahedron. 2003;59:8329–8336. [Google Scholar]
- 19.Lawton LA, Edwards C, Codd GA. Analyst. 1994;119:1525–1530. doi: 10.1039/an9941901525. [DOI] [PubMed] [Google Scholar]
- 20.Kurmayer R, Christiansen G, Chorus I. Appl. Environ. Microbiol. 2003;69:787–795. doi: 10.1128/AEM.69.2.787-795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christiansen G, Fastner J, Erhard M, Börner T, Dittmann E. J. Bacteriol. 2003;185:564–572. doi: 10.1128/JB.185.2.564-572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.