Abstract
Background
α2A-Adrenoceptors (α2A-ARs) have important roles in sympathetic cardiovascular regulation. Variants of ADRA2A affect gene transcription and expression and are associated with insulin release and risk for type 2 diabetes. We examined whether ADRA2A variants are also associated with cardiovascular responses to the selective α2-AR-agonist, dexmedetomidine.
Methods and Results
73 healthy subjects participated in a placebo-controlled single-blind study. After 3 infusions of placebo, subjects received 3 incremental infusions of dexmedetomidine (cumulative dose, 0.4 mcg/kg). Primary outcomes were changes in systolic blood pressure (SBP) and plasma norepinephrine concentrations, measured as difference of the area-under-the-curve during placebo and dexmedetomidine infusions (ΔAUC). We used multiple linear regression analysis to examine the associations between 9 ADRA2A tagging variants and 5 inferred haplotypes and ΔAUC after adjustment for covariates. Homozygous carriers of rs553668, and the corresponding haplotype 4, previously associated with increased α2A-AR expression, had a 2.2-fold greater decrease in AUCSBP after dexmedetomidine (adjusted P=0.006); similarly, the maximum decrease in SBP was 24.7±8.1 mmHg compared to 13.6±5.9 mmHg in carriers of the wildtype allele (P=0.007). Carriers of haplotype 3, previously associated with reduced α2A-AR expression, had a 44% smaller decrease in AUCSBP (P=0.013). Haplotype information significantly improved the model predicting the decrease in SBP (P<0.001). There were similar but non-significant trends for diastolic blood pressure and heart rate. Genotypes were not significantly associated with norepinephrine responses.
Conclusion
Common ADRA2A variants are associated with the hypotensive response to dexmedetomidine. Effects of specific variants/haplotypes in vivo are compatible with their known effects on gene expression in vitro.
Keywords: receptors, adrenergic, alpha, genetic polymorphism, pharmacogenetics, receptor, variability in drug response
Introduction
α2A-Adrenoceptors (α2A-ARs) are important regulators of sympathetic tone through central and peripheral (presynaptic) sympathetic inhibition. They are also directly involved in several homeostatic functions, including contraction and relaxation of vascular smooth muscle, control of vigilance, anxiety and stress-related behaviors, pain perception, platelet aggregation, lipolysis, and insulin release. Genetically engineered mice that do not express the α2A-AR gene (ADRA2A) have a hyper-adrenergic phenotype with increased blood pressure, heart rate, and plasma norepinephrine concentrations, and develop cardiac hypertrophy and heart failure.1–4 Moreover, ADRA2A knock-out mice do not decrease blood pressure in response to an α2-AR-agonist such as clonidine.1
We and others have systematically defined genetic variation and the haplotype structure of ADRA2A.5, 6 Some of the variants identified have distinct functional effects on receptor expression or function in vitro.6–8 However, studies to define the functional significance of ADRA2A genetic variation in cardiovascular regulation in vivo have yielded inconsistent results. Some studies have found associations between ADRA2A variants and hypertension,9–11 while others, including two genome-wide association studies, have not.12–15
Blood pressure is regulated by many factors and mechanisms, and the contributions of single genetic variants to blood pressure regulation may be difficult to detect in population-based studies where many confounding variables remain uncontrolled. A more sensitive approach to defining the functional effects of genetic variants in membrane-bound receptors is the administration of a pharmacological agonist or antagonist in a controlled experimental setting.16–18 Indeed, Sober et al. recently hypothesized that functional ADRA2A variants could affect the pharmacological responses to α2-AR agonists such as clonidine.11
Thus, we examined the hypothesis that ADRA2A variants affect cardiovascular responses to α2-AR-agonists by administering dexmedetomidine, a selective α2-AR-agonist, to healthy volunteers in a highly controlled setting.
Methods
Subjects
The Institutional Review Board of Vanderbilt University Medical Center approved the study protocol, and subjects gave written informed consent. Details of the study methods and subjects have been published previously.19, 20 In brief, we studied unrelated healthy American black and white subjects, aged 18–45 years, without clinically significant abnormal findings on medical history, physical examination, or routine laboratory testing. For 5 days prior to the study, subjects were on an alcohol- and caffeine-free diet (providing 150 mmol of sodium, 70 mmol of potassium, and 600 mmol of calcium daily). All subjects were free of medications and dietary supplements for at least 2 weeks.
Study procedure
The study was placebo-controlled, single-masked, and was performed at the Vanderbilt University Clinical Research Center. After an overnight fast, intravenous cannulae were placed in antecubital veins bilaterally, one for blood collection and the other for drug infusion. After a 30-minute supine resting period, blood pressure (BP) and heart rate were obtained from the left brachial artery with a semi-automated device (Dinamap MPS; GE Medical Systems, Waukesha, WI, USA), and a blood sample was taken for DNA extraction and measurement of baseline plasma catecholamine concentrations. All heart rate and blood pressure measurements were performed twice and averaged for analysis. After baseline measures, six infusions (each infusion lasted 10 minutes and was followed by a 20 minute observation period) were administered (Figure S1, online data supplement). Placebo [normal saline] was infused during cycles 1–3, and dexmedetomidine [Precedex®, Abbott Laboratories, Abbott Park, IL, USA] at doses of 0.1, 0.15, and 0.15 mcg/kg body weight, respectively, during cycles 4–6 (cumulative dose, 0.4 mcg/kg). Ten minutes after each infusion, heart rate and blood pressure were measured and a blood sample was drawn for measurement of plasma catecholamine and dexmedetomidine concentrations.
Genotyping
We genotyped nine ADRA2A tag single nucleotide polymorphisms (tagSNPs) representing common genetic variations in black and white Americans5 (Table S1 in the online data supplement). The selection of tagSNPs was based on our previous study of 135 demographically similar black and white residents of Nashville, in whom we sequenced ADRA2A (including approximately 2.2 kb of 5′-flanking and 0.1 kb of 3′-flanking regions) and found 41 SNPs, among which we selected 9 tagSNPs by LD-based binning (binning criterion, r2≥0.4) and prevalence criteria (MAF≥5%) that optimally captured genetic variability of ADRA2A.5 We genotyped the 9 tagSNPs by allelic discrimination with TaqMan 5′-nuclease assays21 on an ABI 7900 HT real-time PCR system (Applied Biosystems, Foster City, CA) using validated TaqMan probes. For genotyping quality control, we included 6 samples with known genotypes previously determined by direct sequencing,5 and all control samples had concordant genotypes.
Haplotype assignment
Haplotypes were inferred from the 9 tagSNPs using an expectation-maximization algorithm implemented in the software Powermarker.5 Since subtypes of haplotypes 4 and 5 were expected to be infrequent, we grouped these subtypes into haplotype families for analysis as previously defined.5 The variant rs553668 was a tagSNPs for haplotype 4, and rs553668 genotype groups therefore comprised the same patients as corresponding HT4 haplotype groups. Only haplotypes that could be inferred with ≥85% probability were assigned; haplotypes with <85% probability were handled as missing data.
Plasma catecholamine determination
Blood was collected into cooled heparinized tubes which were immediately placed on ice until centrifuged at 4 °C for 10 minutes at 3,000 rpm. Plasma was separated and stored at −20 °C in tubes containing 40 μL of reduced glutathione (6%) until assayed. Norepinephrine and epinephrine concentrations were measured by high-performance liquid chromatography using electrochemical detection with dihydroxybenzylamine as internal standard.22
Plasma dexmedetomidine determination
Plasma dexmedetomidine concentrations were determined by reversed-phase high-performance liquid chromatography with tandem mass spectrometric detection23 (PE Sciex API4000, PE Sciex, Foster City, CA) as previously described.23, 24
Data and statistical analysis
Data are expressed as means and standard deviations (SD) or 95% confidence intervals (CI). Genotype distribution was tested for deviation from Hardy-Weinberg equilibrium with the use of a chi-square test with one degree of freedom. To generate a summary variable representing the overall response, we plotted outcomes (systolic and diastolic blood pressure [BP], heart rate, and plasma catecholamine concentrations) against time for each subject, and determined the area under the curve (AUC) for the 1.5 hour periods of placebo (AUCPLAC) and dexmedetomidine infusions (AUCDEX), respectively, by the trapezoidal rule, assuming linear changes between the measurements. AUC data was normally distributed, and dexmedetomidine responses were assessed as the difference between AUCDEX and AUCPLAC for each outcome variable by paired t-test. In a sensitivity analysis, we also assessed the decrease in the outcome measure between the last placebo infusion and the last dexmedetomidine infusion. Since low doses of dexmedetomidine preferentially reduce systolic blood pressure and plasma norepinephrine concentrations,5, 24–26 changes in these variables were the primary outcomes, and changes in heart rate and plasma epinephrine concentrations were secondary outcomes. For single-marker analysis of the nine tagSNPs, we used one-way analysis of variance (ANOVA) to compare outcomes by number of variant alleles. When there were two or fewer subjects homozygous for the variant allele, we grouped them with heterozygous carriers for statistical analysis. To adjust for potential covariates (age, sex, race, plasma dexmedetomidine concentration, body mass index [BMI], and AUCPLAC for the corresponding outcome), we performed linear regression analyses, assuming an additive mode of inheritance for the SNP. For each of the five haplotypes, we created biallelic genotypes by treating the haplotype as a variant allele and grouping the other haplotypes together as the other allele, and repeated the above analyses. In addition, we also performed multiple linear regression analyses including all covariates and all five haplotypes with additive haplotypic effects. All tests were two-tailed. In these exploratory analyses, we did not adjust for multiple comparisons, and P-values of <0.05 were considered statistically significant. Analyses were performed with the statistical software packages R (www.r-project.org) and SPSS (SPSS v.15.0, SPSS Inc., Chicago, IL, USA).
Results
Subjects
We studied 73 healthy subjects; their demographic characteristics and baseline measures are shown in Table 1.
Table 1.
Demographic and baseline characteristics (n=73).
| Parameter | n (%) or mean ± SD |
|---|---|
| Female sex | 32 (43.8%) |
| Age (years) | 25.4 ± 4.6 |
| Race | |
| White | 37 (50.7%) |
| Black | 36 (49.3%) |
| Body mass index (kg/m2) | 25.3 ± 3.7 |
| Systolic blood pressure (mmHg) | 113.6 ± 9.2 |
| Diastolic blood pressure (mmHg) | 69.0 ± 6.8 |
| Heart rate (bpm) | 64.5 ± 7.7 |
| Norepinephrine plasma concentration (pg/mL) | 230.0 ± 95.0 |
| Epinephrine plasma concentration (pg/mL) | 21.8 ± 17.0 |
ADRA2A genotypes and haplotypes
Genotypes of the 9 ADRA2A variants could be determined in 94.5%–100% of the subjects (mean call rate, 98.2%) and are shown in Table S1 (Online Data Supplement). Minor allele frequencies were in the expected range in both ethnic groups.6, 27 All genotypes conformed to Hardy-Weinberg equilibrium in each ethnic group (all P>0.10). Haplotype families could be assigned to 99.3% of haplotypes with >85% probability, and the haplotype distribution was in the expected range (Table S2, online data supplement).5
Determinants of outcomes at baseline and during placebo
Male sex (+8.9 mmHg, 95% CI, 5.3 to 12.4 mmHg; P<0.001), black ethnicity (+4.6 mmHg; 95% CI, 1.1 to 8.1 mmHg; P=0.010), and a higher BMI (per kg/m2 unit, +0.6 mmHg; 95% CI, 0.05 to 1.1 mmHg; P=0.033) were individually associated with higher systolic BP at baseline, and similarly, during placebo infusions. After adjustment for these covariates, none of the ADRA2A tagSNPs or haplotypes was associated with systolic BP or any of the other outcomes (diastolic BP, plasma norepinephrine and epinephrine concentrations, and heart rate) at baseline or during the placebo infusions (all P>0.07).
ADRA2A variants and systolic BP responses to dexmedetomidine
During the dexmedetomidine infusions, the AUC for all outcome measures decreased significantly compared to that during placebo infusions (Table 2; all P≤0.028; Figure S2, online data supplement). For the primary outcome, systolic blood pressure, a higher AUCSBP during placebo was associated with a greater decrease in AUCSBP during dexmedetomidine (P=0.007), but other covariates (age, sex, race, BMI, dexmedetomidine concentration) did not affect the reduction. Table 3 shows ΔAUCSBP in genotype groups by single marker analysis assuming an additive model of inheritance. Three subjects homozygous for rs553668 had a 115% (2.2-fold; 95% CI, 1.5 to 2.8-fold; P=0.001) greater decrease in AUCSBP than carriers of one of more copies of the major allele, and the difference remained significant after adjustment for covariates (9.6 mmHg; 95% CI, 2.8 to 16.4 mmHg*hr; P=0.006; Figure 1). A similar trend for rs2484516 did not reach statistical significance (adjusted P=0.07; Table 3).
Table 2.
Cardiovascular outcome measures during placebo and dexmedetomidine infusions.
| Placebo | Dexmedetomidine | ΔAUC | P-value | |
|---|---|---|---|---|
| AUCSBP, mmHg*hr | 171.3±14.1 | 160.2±12.4 | −11.1±6.3 | <0.001 |
| AUCDBP, mmHg*hr | 103.7±9.1 | 95.7±8.0 | −8.0±5.8 | <0.001 |
| AUCHR, bpm*hr | 97.5±12.1 | 95.2±12.4 | −2.3±5.6 | 0.001 |
| AUCNE, pg*hr/mL | 346.8±125.1 | 247.9±108.2 | −98.9±76.0 | <0.001 |
| AUCEpi, pg*hr/mL | 35.8±21.9 | 32.9±20.2 | −3.0±10.7 | 0.028 |
AUC=area under the curve; SBP=systolic blood pressure, DBP=diastolic BP, HR=heart rate, NE=norepinephrine, Epi=epinephrine, ΔAUC=difference in AUC between dexmedetomidine and placebo infusions.
Table 3. Single-marker analysis of decrease in systolic blood presssure (SBP) and norepinephrine plasma concentrations (NE) after dexmedetomidine.
Decrease of systolic BP and NE was assessed as difference between the area under the curve for the respective variables between placebo infusions and dexmedetomidine infusions (ΔAUCSBP and ΔAUCNE, respectively). Uncorrected p-values are from multiple linear regression analyses assuming an additive model of inheritance. Adjusted p-values are after adjustment for the covariates AUCSBP or AUCNE during placebo, respectively, dexmedetomidine plasma concentrations, age, sex, race, and BMI.
| ΔAUCSBP, mmHg* hr (mean±SD) | ΔAUCNE, pg/mL* hr (mean±SD) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Number of minor alleles | P-value | Adjusted P-value | Number of minor alleles | P-value | Adjusted P-value | |||||
| 0 | 1 | 2 | 0 | 1 | 2 | ||||||
| 1 | rs11195418 | −11.0±6.2 n=65 |
−10.1±4.1 n=5 |
−12.5 n=1 |
0.92 | 0.90 | −104.5±76.5 n=65 |
−29.1±54.5 n=5 |
−66.3 n=1 |
0.15 | 0.72 |
| 2 | rs1800544 | −9.5±5.0 n=23 |
−12.3±6.1 n=32 |
−11.1±6.3 n=18 |
0.26 | 0.49 | −119.2±80.2 n=23 |
−86.1±63.6 n=31 |
−95.1±88.3 n=18 |
0.28 | 0.12 |
| 3 | rs2484516 | −10.5±5.8 n=64 |
−13.6±7.5 n=4 |
−25.3 n=1 |
0.06* | 0.07* | −101.3±76.4 n=64 |
−99.9±75.1 n=4 |
−153.5 n=1 |
0.79 | 0.46 |
| 4 | rs1800545 | −11.4±6.5 n=42 |
−10.8±6.4 n=28 |
−10.8±4.9 n=3 |
0.93 | 0.13 | −106.3±81.2 n=42 |
−89.6±71.5 n=27 |
−79.3±19.7 n=3 |
0.61 | 0.41 |
| 5 | rs1800035 | −10.8±6.3 n=67 |
−15.3±5.5 n=6 |
n.a. | 0.090 | 0.18 | −97.2±77.8 n=67 |
−118.2±52.8 n=6 |
n.a. | 0.52 | 0.53 |
| 6 | rs1800038_ | −10.9±6.1 n=67 |
−12.8±4.1 n=3 |
n.a. | 0.58 | 0.73 | −98.4±78.4 n=67 |
−127.1±47.7 n=3 |
n.a. | 0.53 | 0.99 |
| 7 | rs34303217 | −10.8±6.1 n=69 |
−17.1±8.7 n=4 |
n.a. | 0.051 | 0.53 | −99.4±77.7 n=69 |
−91.1±42.2 n=4 |
n.a. | 0.83 | 0.81 |
| 8 | rs553668 | −10.1±5.8 n=51 |
−12.1±5.8 n=19 |
−22.8±7.4 n=3 |
0.002 | 0.028 | −104.6±77.1 n=51 |
−88.6±69.7 n=19 |
−69.3±111.4 n=3 |
0.59 | 0.17 |
| 9 | rs3750625 | −11.5±6.4 n=53 |
−9.1±4.6 n=15 |
−9.9±6.5 n=2 |
0.38 | 0.052 | −100.9±85.6 n=53 |
−97.1±47.1 n=15 |
−85.9±22.8 n=2 |
0.96 | 0.82 |
The single subject homozygous for the variant allele was grouped with the heterozygotes for statistical analysis.
SNP=single nucleotide polymorphism. n.a.=genotype not represented in cohort.
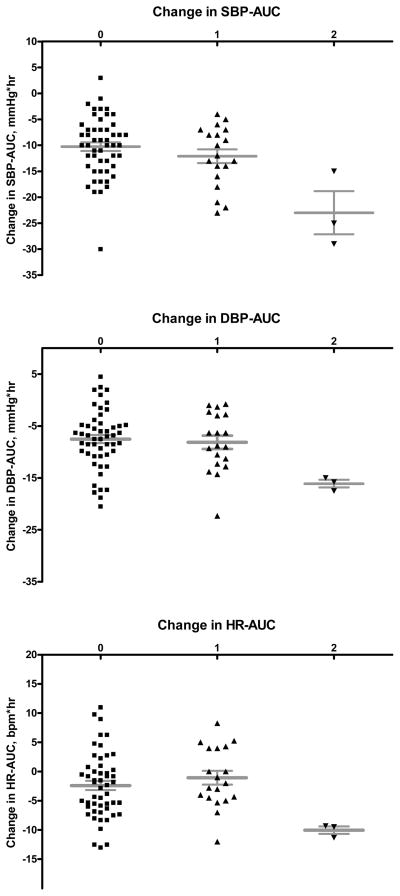
Figure 1. Differences in AUC (ΔAUC) between placebo and dexmedetomidine infusions for different outcomes by haplotype 4.
Subjects homozygous for haplotype 4 (defined by rs553668) had a greater reduction in systolic BP (upper panel; adjusted P=0.006), diastolic BP (middle panel; adjusted P=0.007), and heart rate (lower panel; adjusted P=0.035) compared to carriers of 0 or 1 copy. Horizontal lines represent means, whiskers the standard error of the mean.
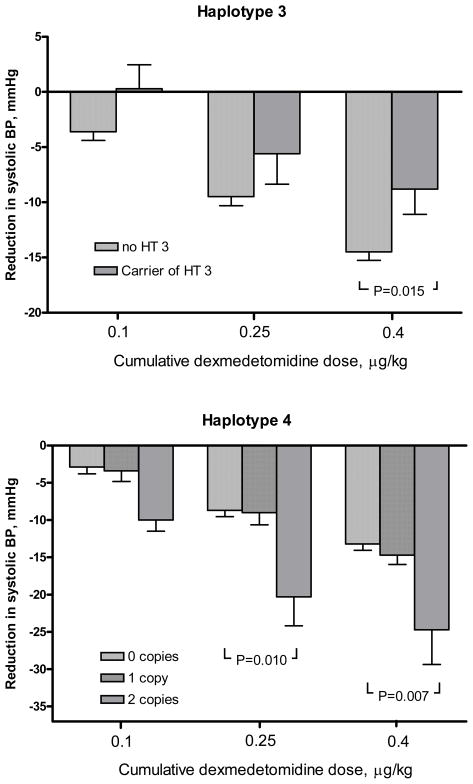
Table 4 shows ΔAUCSBP by haplotypes in single haplotype analyses. Homo- or heterozygous carriers of haplotype 3 (HT3) had a 44% smaller reduction in AUCSBP (i.e., a smaller hypotensive response; adjusted P=0.008), while the three subjects with two copies of HT4 (already defined above by the rs553668 variant) had 2.2-fold greater reductions in AUCSBP (i.e. a larger hypotensive response) than carriers of one or no copy (adjusted P=0.006; Figure 1). When analyzing races separately in a sensitivity analysis, white and black homozygotes for rs553668/HT4 had a similarly larger ΔAUCSBP compared to the other genotypes (2.3-fold and 2.1-fold for whites and blacks, respectively); however, genotype differences were statistically significant only in whites (unadjusted P=0.003), likely reflecting the small sample size for black homozygotes (n=1).
Table 4. Single-haplotype analysis of decrease in systolic BP and norepinephrine plasma concentrations (NE) after dexmedetomidine.
Decrease of systolic BP and NE was assessed as difference between the area under the curve for the respective variables between placebo infusions and dexmedetomidine infusions (ΔAUCSBP and ΔAUCNE, respectively). Uncorrected p-values are from multiple linear regression analysis assuming an additive model of inheritance. Adjusted p-values are after adjustment for the covariates AUCSBP or AUCNE during placebo, respectively, dexmedetomidine plasma concentrations, age, sex, race, and BMI. Except placebo AUCSBP and placebo AUCNE, respectively (P<0.021 in all analyses), no other covariate was significantly associated with the outcomes.
| ΔAUCSBP, mmHg* hr (mean±SD) | ΔAUCNE, pg/mL* hr (mean±SD) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Number of haplotype copies | P-value | Adjusted P-value | Number of haplotype copies | P-value | Adjusted P-value | ||||
| 0 | 1 | 2 | 0 | 1 | 2 | |||||
| HT1 | −12.1±7.7 n=23 |
−11.5±6.2 n=25 |
−9.5±5.0 n=23 |
0.17 | 0.77 | −107.8±74.7 n=23 |
−77.6±65.7 n=24 |
−119.2±80.2 n=23 |
0.60 | 0.46 |
| HT2 | −10.6±6.4 n=65 |
−15.3±5.5 n=6 |
n.a. n=0 |
0.09 | 0.18 | −99.6±76.6 n=65 |
−118.2±52.8 n=6 |
n.a. n=0 |
0.56 | 0.52 |
| HT3 | −11.6±6.2 n=63 |
−6.1±7.2 n=7 |
−9.5 n=1 |
0.034* | 0.008* | −101.2±71.8 n=63 |
−73.9±70.6 n=7 |
−291.3 n=1 |
0.99* | 0.81* |
| HT4 | −10.1±5.8 n=51 |
−12.1±5.8 n=19 |
−22.8±7.4 n=3 |
0.002 | 0.028 | −104.6±77.1 n=50 |
−88.6±69.7 n=19 |
−69.3±111.4 n=3 |
0.44 | 0.18 |
| HT5 | −11.2±6.6 n=40 |
−10.8±6.4 n=28 |
−10.8±4.9 n=3 |
0.79 | 0.16 | −110.6±79.0 n=40 |
−89.6±71.5 n=27 |
−79.3±19.7 n=3 |
0.49 | 0.35 |
For statistical analysis, the single subject homozygous for HT3 was grouped with the 7 heterozygous carriers.
HT=haplotype, n.a.= not represented in cohort.
Adding all ADRA2A haplotypes into a model predicting ΔAUCSBP that included all other covariates improved the model significantly, increasing the coefficient of determination R2 (the percent of the variability of ΔAUCSBP explained by the model) from 19.2% to 32.2% (P<0.001). In this model, HT3 carrier status was associated with a smaller (P=0.013) and HT4 with a larger hypotensive SBP response (P=0.047) to dexmedetomidine (Table S3, online data Supplement). When HT4 was modeled as a recessive trait, taking into account the post hoc observation that only homozygous HT4 carriers had greater hypotensive responses, the association between HT4 and ΔAUCSBP was stronger (P=0.007).
In a sensitivity analysis, as an additional assessment of responses to dexmedetomidine, we analyzed the reduction in systolic blood pressure between the last placebo infusion and the last dexmedetomidine infusions (ΔSBP). In this analysis, haplotype 4 was also associated with a greater hypotensive response, and haplotype 3 with a smaller response (Figure 2), with a magnitude similar to that observed with the summary measure ΔAUCSBP. Carriers of HT3, compared to non-carriers, had a 39% (0.61-fold) smaller hypotensive response (ΔSBP=8.8±6.5 mmHg and 14.5±6.1 mmHg, respectively; P=0.015), and subjects homozygous for HT4 had an 82% (1.8-fold) greater hypotensive response (ΔSBP=24.7±8.1 mmHg compared to 13.6±5.9 mmHg; P=0.007; Figure 2) compared to carriers of other haplotypes.
Figure 2. Reduction in systolic blood pressure during dexmedetomidine.
The bar graphs depict the reduction in systolic BP from the last placebo infusion after each of three dexmedetomidine infusions, stratified by number of copies of haplotype 3 (upper panel) and haplotype 4 (or rs553668 minor alleles; lower panel). For statistical analysis, the single subject homozygous for HT3 was grouped with the heterozygous carriers. Error bars represent standard errors of the mean.
For the second main outcome, change in plasma norepinephrine concentrations between placebo and dexmedetomidine infusions, ΔAUCNE was not affected by ADRA2A variants in single marker (all P-values > 0.17; Table 3) or haplotype analyses (all P-values > 0.46; Table 4). Accordingly, adding all haplotypes to a model with ΔAUCNE as outcome and all other covariates did not improve model fit (P=0.66).
ADRA2A variants and secondary outcomes
For secondary outcomes, in single marker analyses, the same markers associated with a greater hypotensive response for SBP (rs553668 and HT4) were also associated with a greater decrease in diastolic blood pressure (ΔAUCDBP; P=0.040) and heart rate (ΔAUCHR; P=0.033; Figure 1). However, after adjustment for covariates, these associations were weakened and remained significant only under a recessive mode of inheritance (P= 0.007 and P=0.035 for ΔAUCDBP and ΔAUCHR, respectively). HT3, the haplotype associated with a smaller decrease in SBP after dexmedetomidine, was also weakly associated with a smaller decrease in DBP (ΔAUCDBP; P=0.15; adjusted P=0.042) but not heart rate response (ΔAUCHR; P=0.12; adjusted P=0.11) in single haplotype analyses. None of the variants or haplotypes was associated with the decrease in plasma epinephrine concentrations (ΔAUCEpi).
Discussion
In this study, we systematically defined the effects of common ADRA2A variants on cardiovascular responses to the selective α2-AR agonist, dexmedetomidine. Our main findings are that ADRA2A variants previously associated with changes in α2A-AR expression contribute to the inter-individual variability in blood pressure and heart rate responses to dexmedetomidine, with particular genotypes or haplotypes being associated with approximately 40% smaller and 100% greater hypotensive responses, respectively.
The 3′-UTR SNP rs553668 (G>A), formerly identified as the DraI restriction fragment length polymorphism, defines the haplotype 4 family and has a minor allele frequency of approximately 15% in white and 20–30% in black populations.5, 6 Some earlier studies reported a higher prevalence of the rs553668 variant allele in various populations with high blood pressure but several recent studies, including two large genome-wide analyses, did not confirm these findings. The molecular mechanisms for potential phenotypic effects of this variant have been partially elucidated.
In cell lines transfected with different ADRA2A haplotypes, three of four haplotypes containing the variant rs553668 allele were associated with increased α2A-AR mRNA transcription.6 The rs553668 variant also resulted in increased α2A-AR expression on pancreatic islet cell membranes, and thus decreased insulin secretion; moreover, it is associated with an increased prevalence of type 2 diabetes mellitus.8 Thus, the observation that rs553668/HT4 are associated with increased cell surface α2A-AR expression is biologically concordant with our finding of increased responses to the α2-AR agonist dexmedetomidine. Interestingly, beyond systolic BP, reductions in the secondary outcomes diastolic BP and heart rate were also more pronounced in subjects carrying this variant.
ADRA2A haplotype 3 is characterized by a minor allele at a single SNP (rs1800544) in the absence of other SNPs. This haplotype occurs in 8–11% of black Americans, but has not been described in white populations.5, 6 In the present study, carriers of haplotype 3 had significantly smaller hypotensive responses to dexmedetomidine. Interestingly, in a study with haplotype-transfected cell lines, Small et al. found this same haplotype (designated as haplotype 1 in their nomenclature) to have the lowest transcription (about 60% of the α2A-mRNA of the wild-type haplotype) and receptor expression (about 20% of the wild-type haplotype) compared to other haplotypes.6 Thus, the smaller hypotensive response to dexmedetomidine that we observed in carriers of haplotype 3 is concordant with its in vitro effects of lower α2A-AR expression.
We did not find an association between ADRA2A variants and the reduction in plasma norepinephrine concentrations after dexmedetomidine. Possible explanations include the lack of statistical power in view of our sample size, and the fact that in non-steady state conditions, such as occur after dexmedetomidine administration, plasma norepinephrine concentrations are not an ideal measure of sympathetic activity. Norepinephrine spill-over studies would be a more precise way of assessing sympathetic activity, but because they involve administration of radiolabeled norepinephrine in order to measure clearance, they are technically challenging and not feasible for larger study cohorts.
Our findings have several implications. First, variants and haplotypes predicted to affect α2A-AR expression have functional effects on cardiovascular regulation in vivo, concordant with previous findings in vitro. Since the effect size was considerable (with mean differences in hypotensive responses ranging from 0.6-fold to 2.2-fold for particular ADRA2A haplotypes), ADRA2A genetic variation may contribute to interindividual differences in blood pressure regulation, especially after physiological (stress-induced) or pharmacological activation (by α2-AR agonists such as clonidine and dexmedetomidine). Clinically, α2-agonists are used to date primarily as antihypertensive agents (clonidine) or for sedation and anesthesia during surgery or in the intensive care setting (dexmedetomidine), and predicting interindividual variability in hypotensive blood pressure response (either as therapeutic goal or adverse drug effect) could be of significant clinical importance. Additionally, since ADRA2A variants have in vivo functional effects on cardiovascular outcomes, other α2A-AR-mediated outcomes such as platelet function, metabolic regulation, or various central nervous system functions may be similarly affected. In fact, with respect to the α2A-AR-mediated inhibition of insulin secretion from islet cells, a recent study confirmed the association of ADRA2A rs553668 with impaired insulin secretion and type 2 diabetes mellitus, and a meta-analysis of genome-wide association studies (GWAS) identified a strong association of rs10885122, a variant approximately 200 kb downstream of ADRA2A, with higher fasting glucose.8, 28 In addition, a recent GWAS identified one SNP each in whites and blacks downstream ADRA2A (about 63–70 kb) to be associated with decreased epinephrine-induced platelet aggregation.29 In contrast, despite the role of α2A-ARs in spinal and central nervous system pain processing, in a previous study we did not find any association between ADRA2A variants and pain perception in an experimental pain model.20
Our study had several strengths and limitations. We used the highly selective α2-AR agonist, dexmedetomidine, to avoid cardiovascular effects mediated by α1-ARs, as occurs with mixed agonists, for example clonidine. To account for inter-individual differences in dexmedetomidine pharmacokinetics, we measured plasma dexmedetomidine concentrations and adjusted for these in our analyses. We studied healthy subjects in a highly controlled study setting to avoid the confounding effects of environmental factors, disease, and concomitant drugs. This design increased our ability to isolate the functional effects of genetic ADRA2A variants; however, our findings cannot necessarily be extrapolated to patients with hypertension or other diseases. Other limitations include the small sample size, resulting in some small genotype groups and thus wide confidence intervals around the estimates of effect sizes. Thus, our findings are preliminary and require validation in larger, clinical cohorts. ADRA2A variants and haplotype structure vary between whites and blacks,5, 6, 29 but our cohort was underpowered for meaningful separate analyses by race. Nevertheless, the trend toward increased blood pressure reduction in subjects homozygous for rs553668 was similar in magnitude in both races. Indeed, consistency of an association between a variant and a given outcome across multiple ethnic groups with differences in genetic diversity and haplotype structure can be helpful in differentiating truly causal variants from those merely associated with such causative variants by linkage disequilibrium.30 Additionally, the tagSNPs we used represent well the genetic variability of ADRA2A and its 3′-flanking region (approximately 2.2 kb) in our study population, but we did not cover more distant intergenic variants. This may be significant in view of the recently reported association of reduced epinephrine-induced platelet aggregation with two more distant variants (rs4311994 in whites and rs869244 in blacks) approximately 63–70 kb downstream ADRA2A,29 and another report of a distant downstream variant (202 kb; rs10885122) associated with increased fasting glucose.28 It is unclear whether these functional associations with distant downstream variants represent linkage disequilibrium with causal variants in closer proximity to or within ADRA2A, or long-range regulatory elements.29 Using data available on Hapmap, we found that none of these variants is in significant linkage disequilibrium with rs553668, the variant associated with increased dexmedetomidine responsiveness in our study (all r2≤0.125 for both Caucasians with European ancestry and blacks from Yoruba, Nigeria). Thus, multiple variants within and around ADRA2A may be associated with diverse α2A-AR- mediated responses, possibly reflecting tissue-specific effects of specific genetic variants or study-related methodological differences.
In conclusion, this translational study provides evidence that genetic variants in ADRA2A are associated with different blood pressure responses to the selective α2-selective agonist, dexmedetomidine. Variants or haplotypes previously linked with greater or lesser gene transcription/expression were associated with approximately 100% greater and 40% lesser responses to the drug, respectively. Future studies of these variants in other cardiovascular settings, e.g. stress responses, and other physiological functions mediated by the α2A-AR will be of interest.
Supplementary Material
Acknowledgments
Funding Sources: This work was supported by P01 HL56693 from the National Institutes of Health; the Vanderbilt CTSA grant from the National Center for Research Resources [1 UL1 RR024975]; and a Pharmacogenetics Research Network Grant (U01 HL65962).
Footnotes
Conflicts of interest: The laboratory of Dr. Scheinin has contract research relationships with Orion Corporation (Espoo, Finland) and Hospira (Lake Forest, IL, USA). Hospira has a license agreement with Orion Corporation concerning dexmedetomidine (PrecedexR). Dr. Scheinin has received speaker fees and consulting fees from Orion Corporation. None of the other authors has a conflict of interest relevant to the work presented.
References
- 1.Altman JD, Trendelenburg AU, MacMillan L, Bernstein D, Limbird L, Starke K, Kobilka BK, Hein L. Abnormal regulation of the sympathetic nervous system in alpha2A-adrenergic receptor knockout mice. Mol Pharmacol. 1999;56:154–61. doi: 10.1124/mol.56.1.154. [DOI] [PubMed] [Google Scholar]
- 2.Hein L, Altman JD, Kobilka BK. Two functionally distinct alpha2-adrenergic receptors regulate sympathetic neurotransmission. Nature. 1999;402:181–4. doi: 10.1038/46040. [DOI] [PubMed] [Google Scholar]
- 3.Knaus AE, Muthig V, Schickinger S, Moura E, Beetz N, Gilsbach R, Hein L. Alpha2-adrenoceptor subtypes--unexpected functions for receptors and ligands derived from gene-targeted mouse models. Neurochem Int. 2007;51:277–81. doi: 10.1016/j.neuint.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 4.Makaritsis KP, Johns C, Gavras I, Altman JD, Handy DE, Bresnahan MR, Gavras H. Sympathoinhibitory function of the alpha(2A)-adrenergic receptor subtype. Hypertension. 1999;34:403–7. doi: 10.1161/01.hyp.34.3.403. [DOI] [PubMed] [Google Scholar]
- 5.Kurnik D, Muszkat M, Li C, Sofowora GG, Solus J, Xie HG, Harris PA, Jiang L, McMunn C, Ihrie P, Dawson EP, Williams SM, Wood AJ, Stein CM. Variations in the alpha2A-adrenergic receptor gene and their functional effects. Clin Pharmacol Ther. 2006;79:173–85. doi: 10.1016/j.clpt.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Small KM, Brown KM, Seman CA, Theiss CT, Liggett SB. Complex haplotypes derived from noncoding polymorphisms of the intronless alpha2A-adrenergic gene diversify receptor expression. Proc Natl Acad Sci U S A. 2006;103:5472–7. doi: 10.1073/pnas.0601345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small KM, Forbes SL, Brown KM, Liggett SB. An asn to lys polymorphism in the third intracellular loop of the human alpha 2A-adrenergic receptor imparts enhanced agonist-promoted Gi coupling. J Biol Chem. 2000;275:38518–23. doi: 10.1074/jbc.M004550200. [DOI] [PubMed] [Google Scholar]
- 8.Rosengren AH, Jokubka R, Tojjar D, Granhall C, Hansson O, Li DQ, Nagaraj V, Reinbothe TM, Tuncel J, Eliasson L, Groop L, Rorsman P, Salehi A, Lyssenko V, Luthman H, Renstrom E. Overexpression of alpha2A-adrenergic receptors contributes to type 2 diabetes. Science. 2010;327:217–20. doi: 10.1126/science.1176827. [DOI] [PubMed] [Google Scholar]
- 9.Lockette W, Ghosh S, Farrow S, MacKenzie S, Baker S, Miles P, Schork A, Cadaret L. Alpha 2-adrenergic receptor gene polymorphism and hypertension in blacks. Am J Hypertens. 1995;8:390–4. doi: 10.1016/0895-7061(95)00024-j. [DOI] [PubMed] [Google Scholar]
- 10.Svetkey LP, Timmons PZ, Emovon O, Anderson NB, Preis L, Chen YT. Association of hypertension with beta2- and alpha2c10-adrenergic receptor genotype. Hypertension. 1996;27:1210–5. doi: 10.1161/01.hyp.27.6.1210. [DOI] [PubMed] [Google Scholar]
- 11.Sober S, Org E, Kepp K, Juhanson P, Eyheramendy S, Gieger C, Lichtner P, Klopp N, Veldre G, Viigimaa M, Doring A, Putku M, Kelgo P, Shaw-Hawkins S, Howard P, Onipinla A, Dobson RJ, Newhouse SJ, Brown M, Dominiczak A, Connell J, Samani N, Farrall M, Caulfield MJ, Munroe PB, Illig T, Wichmann HE, Meitinger T, Laan M. Targeting 160 candidate genes for blood pressure regulation with a genome-wide genotyping array. PLoS One. 2009;4:e6034. doi: 10.1371/journal.pone.0006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel MC, Plogmann C, Philipp T, Brodde OE. Functional correlates of alpha(2A)-adrenoceptor gene polymorphism in the HANE study. Nephrol Dial Transplant. 1999;14:2657–63. doi: 10.1093/ndt/14.11.2657. [DOI] [PubMed] [Google Scholar]
- 13.Li JL, Canham RM, Vongpatanasin W, Leonard D, Auchus RJ, Victor RG. Do allelic variants in alpha2A and alpha2C adrenergic receptors predispose to hypertension in blacks? Hypertension. 2006;47:1140–6. doi: 10.1161/01.HYP.0000217972.80731.ef. [DOI] [PubMed] [Google Scholar]
- 14.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der HP, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di GA, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dishy V, Sofowora GG, Xie HG, Kim RB, Byrne DW, Stein CM, Wood AJ. The effect of common polymorphisms of the beta2-adrenergic receptor on agonist-mediated vascular desensitization. N Engl J Med. 2001;345:1030–5. doi: 10.1056/NEJMoa010819. [DOI] [PubMed] [Google Scholar]
- 17.Kurnik D, Li C, Sofowora GG, Friedman EA, Muszkat M, Xie HG, Harris PA, Williams SM, Nair UB, Wood AJ, Stein CM. Beta-1-adrenoceptor genetic variants and ethnicity independently affect response to beta-blockade. Pharmacogenet Genomics. 2008;18:895–902. doi: 10.1097/FPC.0b013e328309733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez MPEd. The adrenergic receptors in the 21st century. 1. Totowa, NJ: Humana Press; 2005. [Google Scholar]
- 19.Kurnik D, Muszkat M, Sofowora GG, Friedman EA, Dupont WD, Scheinin M, Wood AJ, Stein CM. Ethnic and genetic determinants of cardiovascular response to the selective alpha 2-adrenoceptor agonist dexmedetomidine. Hypertension. 2008;51:406–11. doi: 10.1161/HYPERTENSIONAHA.107.098939. [DOI] [PubMed] [Google Scholar]
- 20.Kohli U, Muszkat M, Sofowora GG, Harris PA, Friedman EA, Dupont WD, Scheinin M, Wood AJ, Stein CM, Kurnik D. Effects of variation in the human alpha2A- and alpha2C-adrenoceptor genes on cognitive tasks and pain perception. Eur J Pain. 2010;14:154–9. doi: 10.1016/j.ejpain.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ. SNP genotyping by the 5′-nuclease reaction. Methods Mol Biol. 2003;212:129–47. doi: 10.1385/1-59259-327-5:129. [DOI] [PubMed] [Google Scholar]
- 22.He HB, Deegan RJ, Wood M, Wood AJ. Optimization of high-performance liquid chromatographic assay for catecholamines. Determination of optimal mobile phase composition and elimination of species-dependent differences in extraction recovery of 3,4-dihydroxybenzylamine. J Chromatogr. 1992;574:213–8. [PubMed] [Google Scholar]
- 23.Ji QC, Zhou JY, Gonzales RJ, Gage EM, El Shourbagy TA. Simultaneous quantitation of dexmedetomidine and glucuronide metabolites (G-Dex-1 and G-Dex-2) in human plasma utilizing liquid chromatography with tandem mass spectrometric detection. Rapid Commun Mass Spectrom. 2004;18:1753–60. doi: 10.1002/rcm.1548. [DOI] [PubMed] [Google Scholar]
- 24.Snapir A, Posti J, Kentala E, Koskenvuo J, Sundell J, Tuunanen H, Hakala K, Scheinin H, Knuuti J, Scheinin M. Effects of low and high plasma concentrations of dexmedetomidine on myocardial perfusion and cardiac function in healthy male subjects. Anesthesiology. 2006;105:902–10. doi: 10.1097/00000542-200611000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Ebert TJ, Hall JE, Barney JA, Uhrich TD, Colinco MD. The effects of increasing plasma concentrations of dexmedetomidine in humans. Anesthesiology. 2000;93:382–94. doi: 10.1097/00000542-200008000-00016. [DOI] [PubMed] [Google Scholar]
- 26.Angst MS, Ramaswamy B, Davies MF, Maze M. Comparative analgesic and mental effects of increasing plasma concentrations of dexmedetomidine and alfentanil in humans. Anesthesiology. 2004;101:744–52. doi: 10.1097/00000542-200409000-00024. [DOI] [PubMed] [Google Scholar]
- 27.Belfer I, Buzas B, Hipp H, Phillips G, Taubman J, Lorincz I, Evans C, Lipsky RH, Enoch MA, Max MB, Goldman D. Haplotype-based analysis of alpha 2A, 2B, and 2C adrenergic receptor genes captures information on common functional loci at each gene. J Hum Genet. 2005;50:12–20. doi: 10.1007/s10038-004-0211-y. [DOI] [PubMed] [Google Scholar]
- 28.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le BO, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van HM, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson AD, Yanek LR, Chen MH, Faraday N, Larson MG, Tofler G, Lin SJ, Kraja AT, Province MA, Yang Q, Becker DM, O’Donnell CJ, Becker LC. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42:608–13. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaitlen N, Pasaniuc B, Gur T, Ziv E, Halperin E. Leveraging genetic variability across populations for the identification of causal variants. Am J Hum Genet. 2010;86:23–33. doi: 10.1016/j.ajhg.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.