Abstract
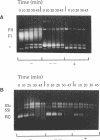
We have studied the fate of covalently-closed circular DNA in the temperature range from 95 to 107 degrees C. Supercoiled plasmid was not denatured up to the highest temperature tested. However, it was progressively transformed into open DNA by cleavage and then denatured. Thermodegradation was not dependent on the DNA supercoiling density. In particular, DNA made positively supercoiled by an archaeal reverse gyrase was not more resistant to depurination and thermodegradation than negatively supercoiled DNA. Thermodegradation was similar in aerobic or anaerobic conditions but strongly reduced in the presence of physiological concentrations of K+ or Mg2+. These results indicate that the major problem faced by covalently closed DNA in hyperthermophilic conditions is not thermodenaturation, but thermodegradation, and that intracellular salt concentration is important for stability of DNA primary structure. Our data suggest that reverse gyrase is not directly required to protect DNA against thermodegradation or thermodenaturation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almagor M., Cole R. D. A high melting (105 degrees C) form of chromatin characterizes the potential of cells for mitosis. J Biol Chem. 1987 Nov 5;262(31):15071–15075. [PubMed] [Google Scholar]
- Barzilai R. SV40 DNA: quantitative conversion of closed circular to open circular form by an ethidium bromide-restricted endonuclease. J Mol Biol. 1973 Mar 15;74(4):739–742. doi: 10.1016/0022-2836(73)90062-4. [DOI] [PubMed] [Google Scholar]
- Bouthier de la Tour C., Portemer C., Huber R., Forterre P., Duguet M. Reverse gyrase in thermophilic eubacteria. J Bacteriol. 1991 Jun;173(12):3921–3923. doi: 10.1128/jb.173.12.3921-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier F., Erauso G., Barbeyron T., Prieur D., Forterre P. Evidence that a plasmid from a hyperthermophilic archaebacterium is relaxed at physiological temperatures. J Bacteriol. 1992 Oct;174(19):6103–6108. doi: 10.1128/jb.174.19.6103-6108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier F., Forterre P. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J Bacteriol. 1994 Mar;176(5):1251–1259. doi: 10.1128/jb.176.5.1251-1259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguet M. The helical repeat of DNA at high temperature. Nucleic Acids Res. 1993 Feb 11;21(3):463–468. doi: 10.1093/nar/21.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EIGNER J., BOEDTKER H., MICHAELS G. The thermal degradation of nucleic acids. Biochim Biophys Acta. 1961 Jul 22;51:165–168. doi: 10.1016/0006-3002(61)91028-9. [DOI] [PubMed] [Google Scholar]
- Freifelder D., Dewitt R. Thermal hydrolysis as a means of opening supercoiled circles of DNA. Gene. 1977 Jul;1(5-6):385–387. doi: 10.1016/0378-1119(77)90042-7. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993 Apr 22;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Rosa S., Fortini P., Karran P., Bignami M., Dogliotti E. Processing in vitro of an abasic site reacted with methoxyamine: a new assay for the detection of abasic sites formed in vivo. Nucleic Acids Res. 1991 Oct 25;19(20):5569–5574. doi: 10.1093/nar/19.20.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandman K., Krzycki J. A., Dobrinski B., Lurz R., Reeve J. N. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5788–5791. doi: 10.1073/pnas.87.15.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci U S A. 1981 Jan;78(1):224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G., Malcom A. D. Production of positively supercoiled DNA by netropsin. J Mol Biol. 1983 Jun 15;167(1):211–216. doi: 10.1016/s0022-2836(83)80043-6. [DOI] [PubMed] [Google Scholar]
- Stein D. B., Searcy D. G. Physiologically important stabilization of DNA by a prokaryotic histone-like protein. Science. 1978 Oct 13;202(4364):219–221. doi: 10.1126/science.694528. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J Mol Biol. 1974 Aug 25;87(4):797–816. doi: 10.1016/0022-2836(74)90085-0. [DOI] [PubMed] [Google Scholar]
- Zhou W., Doetsch P. W. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6601–6605. doi: 10.1073/pnas.90.14.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]