Abstract
Background
Brain-derived neurotrophic factor (BDNF), tyrosine kinase receptor (trkB-TK+) and glutamic acid decarboxylase (GAD67) mRNA levels have previously been found to be reduced in the prefrontal cortex of patients with schizophrenia. To determine whether this reduction extends to other brain regions, we measured the expression levels of BDNF, trkB-TK+ and GAD67 mRNA in regions of the hippocampus, including the dentate gyrus (DG), cornu ammonis subfields (CA1–4), subiculum and entorhinal cortex (EC) of individuals with schizophrenia, bipolar disorder, major depression and unaffected controls.
Methods
In situ hybridization was performed on postmortem brain tissue obtained from the Stanley Foundation Consortium and analyzed using film-based quantification.
Results
Analyses of covariance comparing the expression of mRNA among all groups revealed a significant decrease in BDNF mRNA in CA4 in the bipolar disorder group compared with controls (33%). We found trkB-TK+ mRNA levels to be significantly reduced in CA4 in the schizophrenia group (36%) and in layer II of the EC in the bipolar disorder and major depression groups (28%, 21%, respectively) compared with controls. In addition, GAD67 mRNA levels were reduced in patients with schizophrenia in both the DG (23%) and CA4 (60%) compared with controls. Individuals with major depression also expressed significantly less GAD67 mRNA (44%) compared with controls in CA4 of the hippocampus.
Limitations
It is necessary to account for factors that influence the molecular preservation in postmortem brain tissue, including pH, postmortem interval and tissue storage time. Moreover, there are limitations to the sensitivity of the film-based method of quantification.
Conclusion
Our findings show abnormal BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders, indicating that fundamental properties of hippocampal signalling transmission, plasticity and circuitry may be affected in individuals with these major mental illnesses.
Introduction
Abnormalities of the hippocampal formation, which is involved in various forms of learning and memory as well as in more complex cognitive functions,1 have been described in association with several of the major mental illnesses. Numerous postmortem studies have shown abnormalities of histology, morphometry, neurochemistry and gene expression in the hippocampus of people with schizophrenia,1–3 bipolar disorder4–6 and depression.5,7–9 Individuals with schizophrenia have shown hippocampal hypofunctionality with simultaneous prefrontal cortex hyperactivation during episodic memory retrieval tasks.10,11 Moreover, functional neuroimaging reveals a connection between overactivation of the hippocampus and the positive psychotic symptoms of schizophrenia, such as hallucinations and delusions.10,12–14 This hyperactivity may be due to alterations involving the feedback and/or feed-forward activity of neurons within the hippocampus.15,16 This observed hippocampal overactivity may be a primary abnormality in the hippocampus of individuals with schizophrenia owing to a lack of inhibition.16 However, only limited studies have examined inhibitory neurons in the hippocampus of individuals with schizophrenia17–19 or determined whether putative reductions in inhibitory neurons are potentially linked to alterations in neurotrophin signalling, as has been found in the neocortex.20–22
Brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family, is involved in many aspects of neural development and plasticity, including regulating the maturation of inhibitory γ-aminobutyric acid (GABA) neurons through the signalling of its tyrosine kinase receptor, trkB-TK+. In vivo and in vitro studies show that BDNF induces the expression of glutamic acid decarboxylase (GAD67),20,23–26 suggesting that abnormal signalling in the BDNF/trkB-TK+ pathway may lead to abnormalities in GABAergic neurons. Furthermore, a downregulation of BDNF, trkB-TK+ and GAD67 mRNA has been identified and replicated in the prefrontal cortex of patients with schizophrenia.20–22 Thus, a goal of this study was to determine if a similar downregulation of BDNF and trkB-TK+ mRNA occurs in the hippocampus of patients with schizophrenia and if their expression levels correlate with a reduction in GAD67 mRNA expression in the hippocampus.
Brain-derived neurotrophic factor has also been implicated in the etiology of major depression. The “neurotrophic hypothesis of depression” is based on several observations, including the following: BDNF expression is decreased in patients with depression, levels of hippocampal BDNF are correlated with stress-induced behaviours and antidepressant treatments enhance the expression of BDNF.27–29 Thus, in this study, we tested whether BDNF, trkB-TK+ and GAD67 mRNA expression was altered in the hippocampal formation of individuals with schizophrenia and mood disorders and determined whether similar or distinct deficits existed across the different mental disorders. Understanding the regional and diagnostic specificity of deficits involving BDNF, trkB-TK+ and GAD67 mRNA expression may link molecular abnormalities of growth factor pathways and inhibitory neurons, thereby providing a neuroanatomic context to identify the causes of certain known pathologies. This understanding could then ultimately lead to the identification of targets for therapeutic treatments.
Methods
Tissue
Frozen coronal sections from the medial temporal lobe at the level of the midhippocampus in postmortem brain samples (2 sections per individual; 14 μm thick) were obtained from the Stanley Medical Research Institute (SMRI) Neuropathology Consortium. This cohort consists of 60 individuals in 4 groups: 15 each with schizophrenia, bipolar disorder or depression and unaffected controls (Table 1). The Office of Research at the Uniformed Services University of the Health Sciences, Maryland, reviewed and approved the collection protocol. The details of this cohort regarding brain collection, storage and postmortem diagnosis have been reported previously.30 In addition, all diagnostic groups were matched according to sex, age, race, postmortem interval (PMI) and brain pH (Table 1). The RNA integrity number (RIN) was derived from frontal pole screens of RNA quality done by SMRI; RIN values are available upon request.
Table 1.
Demographic and clinical characteristics of the Stanley Foundation neuropathology cohort
| Group; mean (SD)* |
||||
|---|---|---|---|---|
| Characteristic | Schizophrenia | Bipolar disorder | Major depression | Normal |
| Age, yr | 43 (13) | 41.9 (12) | 46.5 (9.3) | 48.1 (10.7) |
| Sex | 9M, 6F | 9M, 6F | 9M, 6F | 9M, 6F |
| Postmortem interval, h | 33.7 (15) | 32.5 (16) | 27.5 (11) | 23.7 (10) |
| Antipsychotic exposure, ×104 F-mg-Eq | 5.2 | 2.1 | NA | NA |
| Brain pH | 6.2 (0.25) | 6.2 (0.22) | 6.2 (0.05) | 6.3 (0.24) |
| Race | 13W/2A | 14W/1AA | 15W | 14W/1AA |
| RIN† | 8.17 (0.53) | 7.92 (1.12) | 7.87 (1.20) | 8.45 (0.72) |
AA = African American; A = Asian; F = female; F-mg-Eq = fluphenazine milligram equivalent dose; M = male; NA = not applicable; RIN = RNA integrity number; SD = standard deviation; W = white.
Unless otherwise indicated.
Values were derived from frontal pole screens.
Riboprobes
The human BDNF complementary DNA (cDNA) corresponding to the common protein coding exon IX 511bps that corresponds to nucleotides 704–1214 (accession #M61176) was subcloned into the Apa1 site of the 2.96 Kb Bluescript vector (Stratagene). Human TrkB-TK+ cDNA, 216 beats/s corresponding to nucleotides 1753–1969 (accession #U12140), was inserted into the TA cloning site of the 3.9 Kb pCRII vector (Invitrogen) and recognizes the full-length tyrosine kinase containing form of the receptor. The human GAD67 cDNA, 289bps corresponding to nucleotides 1678–1967 (accession #M81883), was inserted into the TA cloning site of the 3.9 Kb pCRII vector (Invitrogen).
Generation of riboprobes
Antisense and sense riboprobes for BDNF, trkB-TK+ and GAD67 were generated from complementary DNA templates using a T3, T7 or SP6 polymerase, respectively. Ribonucleic acid transcription occurred in the antisense and sense direction in the presence of 35S-UTP (Amersham) using an in vitro transcription kit as recommended by the manufacturer (Promega) and the aforementioned cDNA templates. Riboprobes with a specific activity of 2.2 × 109 were purified via ethanol precipitation.
In situ hybridization
Slides were fixed, acetylated, delipidated and dehydrated according to standard protocols31 and performed on 2 consecutive sections per individual per brain region. We added 300 μL of radiolabelled (5 ng/mL) antisense probe in hybridization cocktail to each slide.31 We then placed the sections into humidified chambers overnight at 55°C. We performed RNase digestions and stringent washes post-hybridization as previously detailed.31 BioMax MR (Kodak) film was exposed to the slide for 1 (GAD67 and trkB-TK+) to 2 weeks (BDNF). We used sense strand hybridization to determine nonspecific signalling. These sections, which were treated in the same manner as the sections receiving the anti-sense probe, showed no hybridization signal (Appendix 1, available at cma.ca/jpn).
Image analysis
The regions of the hippocampal formation that we analyzed include the dentate gyrus (DG), cornu ammonis subfields (CA1–4), subiculum and entorhinal cortex (EC) layers I–VI, as defined by Rosene and Van Hoesen32 (Appendix 2, available at cma.ca/jpn). We analyzed all autoradiograms blind to diagnosis using calibrated densitometric image analysis with a 14C standard (nCi/g; Amersham). We measured the mean optical density of 3 regions of interest (ROIs) in each hippocampal area for BDNF, trkB-TK+ or GAD67 mRNA. Data for individual cortical lamina in the caudal aspect of the EC (subdivision 28Mc)33 were obtained using the percentage of the total cortical width occupied by each layer according to the criteria of Insausti and colleagues.34 The percentages of full entorhinal cortical width corresponding to the individual lamina were as follows: I (1%–12%), II (13%–30%), III (31%–65%), V (66%–76%) and VI (77%–100%). Because GAD67 mRNA expression was homogeneously distributed within the EC, we measured 3 ROIs traversing the entire cortical area. Using radioactive standards, we then computed the densities of each probe (μg/Ci) from optical density data.
Statistical analysis
We used Statistica 6.0 (Statsoft) to determine if differences existed among the 4 diagnostic groups in the mRNA expression levels for each of the probes, BDNF, trkB-TK+ and GAD67, in each anatomically defined region of the mesial temporal lobe. Spearman rank correlations were performed to determine whether BDNF, trkB-TK+ or GAD67 mRNA expression were related to brain pH, age, PMI, RIN, storage time and/or lifetime neuroleptic drug use. If a correlation was observed, we used analysis of covariance (ANCOVA) to assess the data. However, if no correlations were detected, then we used analysis of variance (ANOVA). If ANOVA reached significance, we performed a Bonferroni post hoc test to identify the diagnostic groups responsible for the significant finding. The Grubb test was used to calculate the outliers in each diagnostic group and resulted in the removal of samples from 1–2 individuals (3%) per ROI. We also performed correlation analyses to assess correlations between GAD67 mRNA levels and BDNF, and between GAD67 and trkB-TK+ levels in each hippocampal area.
We determined if BDNF, trkB-TK+ or GAD67 mRNA varied according to descriptive categorical characteristics (i.e., cerebral hemisphere, sex, suicide, history of substance abuse or dependence, smoking) by using t tests for unequal samples. To evaluate the effect that additional medications (antidepressants or mood stabilizers) may have had on BDNF, trkB-TK+ or GAD67 mRNA expression, we divided samples from all individuals into 3 groups; those from individuals not taking the medication at the time of death (antidepressants n = 19, mood stabilizers n = 29), those taking the medication at the time of death (antidepressants n = 24, mood stabilizers n = 14) and unaffected controls (n = 15). We used analyses of variance to examine the effect of these medication categories in all hippocampal regions. If an ANOVA reached significance, we performed a post hoc least significance test (LSD) to identify the group comparisons responsible for the significant finding.
Results
Anatomic pattern of BDNF, trkB-TK+ and GAD67 mRNA expression
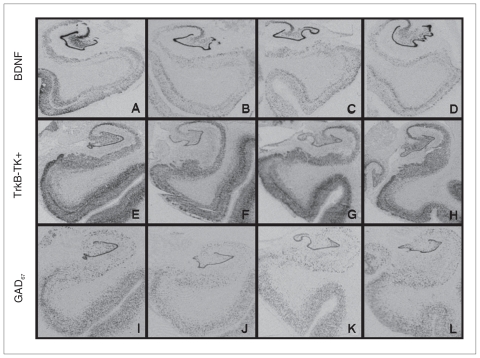
Brain-derived neurotrophic factor, trkB-TK+ and GAD67 mRNA were expressed throughout the hippocampal formation in all 4 groups analyzed (Fig. 1). Qualitatively, mRNA expression for the 3 probes was most robust in the DG compared with the other areas examined. Brain-derived neurotrophic factor and trkB-TK+ mRNA showed heavy punctate signal in CA4, whereas GAD67 mRNA displayed a somewhat lighter and even more punctate signal in this region, consistent with expression in interneurons. Brain-derived neurotrophic factor and trkB-TK+ mRNA signals in CA3 were very robust and homogeneous, whereas GAD67 mRNA expression appeared more distributed, weaker and more punctate. We observed a moderate and diffuse (trkB-TK+, BDNF) or punctate (GAD67) signal in CA1 for all 3 probes. In the subiculum, trkB-TK+ and GAD67 showed a moderate–light punctate signal, whereas BDNF mRNA signalling was more variable. We observed a strong laminar pattern for BDNF and trkB-TK+ mRNA in the EC, with a stronger signal in layers II and V; GAD67 mRNA was expressed in a more diffuse punctate pattern in the EC.
Fig. 1.
The top 4 panels show the distribution of brain-derived neurotrophic factor (BDNF) mRNA in (A) control, (B) schizophrenia, (C) bipolar disorder and (D) major depression groups. Middle panels display the distribution of tyrosine kinase receptor (trkB-TK+) mRNA in (E) control, (F) schizophrenia, (G) bipolar disorder and (H) major depression groups, and the bottom panels show the distribution of glutamic acid decarboxylase (GAD67) mRNA in (I) control, (J) schizophrenia, (K) bipolar disorder and (L) major depression groups.
BDNF, trkB-TK+ and GAD67 mRNA levels in the hippocampus vary according to diagnosis in a subregion-specific manner
Dentate gyrus
Correlations with demographic variables: BDNF mRNA expression correlated with brain pH (r = 0.29, p = 0.029; Appendix 3, available at cma.ca/jpn), whereas trkB-TK+ mRNA levels correlated with PMI, pH and RIN (r = −0.28, p = 0.040; r = 0.37, p = 0.005; r = 0.32, p = 0.019, respectively; Appendix 3). Levels of GAD67 mRNA did not correlate with any demographic variable in the dentate gyrus.
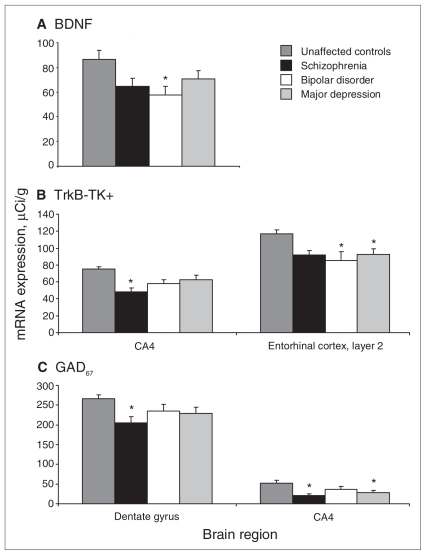
Effect of diagnosis: We did not find a significant main effect of diagnosis for BDNF or trkB-TK+ mRNA levels in the dentate gyrus (F3,50 = 1.45, p = 0.24; F3,47 = 1.33, p = 0.27; Table 2). We detected a significant main effect of diagnosis for GAD67 mRNA levels (F3,48 = 3.13, p = 0.034; Table 2) in the dentate gyrus. Bonferonni post hoc testing showed a significant reduction (22%) in GAD67 mRNA levels in the schizophrenia group (p = 0.022) compared with controls (Fig. 2A).
Table 2.
BDNF, trkB-TK+ and GAD67 mRNA expression
| Group; mean (SD)
|
|||||
|---|---|---|---|---|---|
| Marker; brain region | p value | Schizophrenia | Depression | Bipolar disorder | Control |
| BDNF | |||||
| Dentate gyrus | 0.24 | 196 (38) | 230 (67) | 216 (43) | 223 (20) |
| CA1 | 0.94 | 26 (9) | 28 (12) | 29 (8) | 29 (11) |
| CA3 | 0.35 | 131 (46) | 135 (39) | 117 (62) | 150 (40) |
| CA4* | 0.044 | 64 (25) | 71 (25) | 58 (25) | 87 (28) |
| Subiculum | 0.31 | 24 (6) | 27 (10) | 25 (7) | 29 (7) |
| Entorhinal cortex | |||||
| Layer 1 | 0.08 | 14 (5) | 10 (4) | 11 (4) | 11 (4) |
| Layer 2 | 0.90 | 45 (15) | 40 (7) | 40 (19) | 42 (11) |
| Layer 3 | 0.95 | 24 (8) | 25 (8) | 25 (9) | 24 (5) |
| Layer 5 | 0.97 | 25 (9) | 25 (9) | 24 (10) | 25 (5) |
| Layer 6 | 0.75 | 22 (7) | 24 (7) | 21 (8) | 24 (5) |
| TrkB-TK+ | |||||
| Dentate gyrus | 0.27 | 89 (21) | 100 (28) | 94 (22) | 111 (12) |
| CA1 | 0.63 | 73 (11) | 72 (11) | 75 (14) | 78 (14) |
| CA3 | 0.49 | 95 (17) | 109 (32) | 102 (29) | 115 (21) |
| CA4† | 0.025 | 48 (19) | 63 (19) | 58 (19) | 75 (10) |
| Subiculum | 0.56 | 82 (9) | 90 (23) | 79 (14) | 89 (16) |
| Entorhinal cortex | |||||
| Layer 1 | 0.05 | 58 (22) | 45 (18) | 46 (24) | 72 (20) |
| Layer 2‡ | 0.012 | 100 (29) | 93 (23) | 85 (15) | 118 (17) |
| Layer 3 | 0.48 | 84 (22) | 90 (25) | 89 (29) | 103 (15) |
| Layer 5 | 0.76 | 73 (23) | 78 (24) | 76 (25) | 88 (20) |
| Layer 6 | 0.40 | 56 (7) | 67 (17) | 62 (12) | 70 (15) |
| GAD67 | |||||
| Dentate gyrus§ | 0.034 | 205 (56) | 230 (58) | 235 (54) | 266 (36) |
| CA1 | 0.24 | 20 (12) | 24 (13) | 28 (13) | 32 (13) |
| CA3 | 0.84 | 37 (14) | 35 (16) | 39 (21) | 46 (23) |
| CA4¶ | 0.008 | 21 (16) | 29 (18) | 37 (28) | 52 (25) |
| Subiculum | 0.91 | 53 (33) | 56 (16) | 49 (24) | 63 (17) |
| Entorhinal cortex | 0.34 | 80 (53) | 72 (18) | 84 (43) | 100 (35) |
BDNF = brain-derived neurotrophic factor; CA = cornu ammonis subfield; GAD67 = glatamic acid decarboxylase; SD = standard deviation; trkB-TK+ = tyrosine kinase receptor.
Bipolar disorder < normal, p = 0.004.
Schizophrenia < normal, p < 0.001.
Major depression < normal, p = 0.006; bipolar disorder < normal, p = 0.002.
Schizophrenia < normal, p = 0.004.
Schizophrenia < normal, p < 0.001; major depression < normal, p = 0.005.
Fig. 2.
(A) Mean brain-derived neurotrophic factor (BDNF) mRNA levels in CA4 in the diagnostic groups and controls. (B) Mean tyrosine kinase receptor (trkB-TK+) mRNA levels in CA4 and layer II of the entorhinal cortex in the diagnostic groups and controls. (C) Mean glutamic acid decarboxylase (GAD67) mRNA levels in the dentate gyrus and CA4 of the diagnostic groups and controls. Standard error is represented by error bars. B = bipolar disorder; CA = cornu ammonis subfield; D = major depression; N = unaffected controls; S = schizophrenia. (*p ≤ 0.05). All reported significance values are in comparison to controls.
Cornu ammonis subfield 4
Correlations with demographic variables: BDNF mRNA expression was correlated with brain pH (r = 0.29, p = 0.030; Appendix 3), whereas trkB-TK+ mRNA levels were correlated with PMI (r = −0.38, p = 0.005) and pH (r = 0.33, p = 0.013; Appendix 3). We found that GAD67 mRNA correlated with RIN values (r = 0.30, p = 0.024; Appendix 3).
Effect of diagnosis: We found a significant effect of diagnosis on BDNF mRNA levels (F3,51 = 2.90, p = 0.044; Table 2) and on trkB-TK+ mRNA levels (F3,49 = 3.39, p = 0.025; Table 2) in the CA4 subfield of the hippocampus. Post hoc tests showed that BDNF mRNA levels were significantly reduced in the bipolar disorder group (33%, p = 0.024) compared with controls (Fig. 2B). The schizophrenia group also showed a decrease in BDNF in CA4, and the magnitude of the decrease was similar to that in the bipolar disorder group (26%); however, this only reached trend levels of significance. Post hoc testing also showed that trkB-TK+ mRNA levels were significantly reduced in the schizophrenia group (36%, p < 0.001) compared with controls (Fig. 2B). Analyses of covariance revealed a significant effect of diagnosis for GAD67 mRNA levels (F3,49 = 4.42, p = 0.008; Table 2) in CA4. Post hoc testing revealed a significant reduction in GAD67 mRNA levels in both the schizophrenia (60%, p = 0.003) and major depression (44%, p = 0.030) groups (Fig. 2B).
Cornu ammonis subfield 3
Correlations with demographic variables: Brain pH correlated with BDNF mRNA levels (r = 0.37, p = 0.006; Appendix 3), whereas PMI correlated with both trkB-TK+ (r = −0.31, p = 0.025; Appendix 3) and GAD67 (r = −0.35, p = 0.015; Appendix 3). RNA integrity also correlated with GAD67 mRNA expression (r = 0.37, p = 0.009; Appendix 3).
Effect of diagnosis: We did not detect any significant effect of diagnosis for either BDNF, trkB-TK+ or GAD67 mRNA (all p > 0.10) in CA3 (Table 2).
Cornu ammonis subfield 1
Correlations with demographic variables: BDNF and trkB-TK+ mRNA levels were not correlated with any demographic variable in CA1. However, GAD67 mRNA levels were significantly correlated with PMI and RIN (r = −0. 48, p < 0.001; r = 0.31, p = 0.019, respectively; Appendix 3).
Effect of diagnosis: There were no significant effects of diagnosis revealed by ANOVA or ANCOVA for BDNF, trkB-TK+ or GAD67 mRNA in CA1 (all p ≥ 0.10; Table 2).
Subiculum
Correlations with demographic variables: BDNF mRNA levels were significantly correlated with pH (r = 0.31, p = 0.032), whereas trkB-TK+ and GAD67 mRNA was significantly correlated with PMI (r = −0.34, p = 0.012; r = −0.40, p = 0.002; Appendix 3).
Effect of diagnosis: ANCOVA revealed no significant effect of diagnosis (all p ≥ 0.10) for BDNF, trkB-TK+ or GAD67 mRNA in the subiculum (Table 2).
Entorhinal cortex
Correlations with demographic variables: BDNF mRNA levels did not correlate with any demographic variable within the EC. Levels of trkB-TK+ mRNA were significantly correlated with PMI (layers II–VI; r −0.54, p ≤ 0.031), pH (layer III: r = 0.40, p = 0.009), RIN (layer I: r = 0.32, p = 0.039) and freezer storage time (layer I: r = −0.40, p = 0.008). Levels of GAD67 mRNA significantly correlated with PMI (r = −0.42, p = 0.003) and pH (r = 0.30, p = 0.042).
Effect of diagnosis: We did not detect a significant effect of diagnosis on BDNF mRNA levels in any layer of the EC (Table 2). However, we did detect a significant effect of diagnosis for trkB-TK+ mRNA expression in layer II (F3,35 = 4.23, p = 0.012; Table 2). Post hoc testing revealed that the bipolar disorder (28%) and major depression (21%) groups had significantly less trkB-TK+ mRNA expression in layer II compared with controls (p ≤ 0.045; Fig. 2C). There was no significant effect of diagnosis on the level of GAD67 mRNA in the EC (F3,41 = 1.14, p = 0.34; Table 2).
Analysis of noncontinuous variables on BDNF, trkB-TK+ and GAD67 mRNA levels
Levels of BDNF, trkB-TK+ and GAD67 mRNA did not typically differ according to sex, hemisphere, suicide, history of substance abuse or dependence or smoking in any of the areas examined in the overall group. However, in CA1 and layer II of the EC there was significant reduction (19% for both) in BDNF mRNA in men compared with women (p ≤ 0.034; Appendix 4, available at cma.ca/jpn), and in layer III of the EC there was a significant reduction (17%) in BDNF mRNA in the left hemisphere compared with the right (p = 0.037). Finally, individuals who committed suicide were also found to have significantly less trkB-TK+ mRNA in layer II of the EC (p = 0.017; Appendix 4) compared with those who did not commit suicide.
Effect of antidepressants and mood stabilizers on mRNA levels
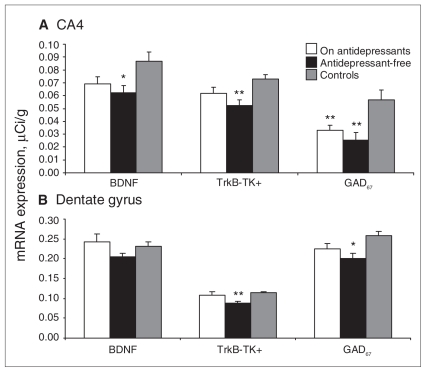
There was no correlation between the continuous variable of lifetime neuroleptic use (in fluphenazine mg equivalents) and BDNF, trkB-TK+ or GAD67 mRNA levels (Appendix 3). However, to account for the possible effect of antidepressants and mood stabilizers on BDNF, trkB-TK+ and GAD67 mRNA levels, we performed ANOVAs on all hippocampal areas comparing individuals taking antidepressants at the time of death (n = 24), patients free of antidepressants at the time of death (n = 19) and unaffected controls (n = 15). Whereas ANOVA revealed a significant effect of group in multiple areas for each probe, the most interesting results were found in 2 subregions of the hippocampus: the DG and the CA4 —the same regions where the diagnostic effects were strongest. Through LSD analysis, we found that trkB-TK+ and GAD67 levels were significantly reduced in the DG in the group without recorded use of antidepressants compared with the other 2 groups (p = 0.006 and p = 0.011; Fig. 3). Least significance testing also revealed that BDNF and trkB-TK+ mRNA levels were significantly reduced in CA4 in the group without recorded use of antidepressants compared with the other 2 groups (p = 0.012 and p = 0.003; Fig. 3). The level of GAD67 mRNA was significantly downregulated in both patient groups compared with controls (p < 0.004), suggesting that GAD67 mRNA is not changed by antidepressant treatment. Whereas individuals taking antidepressants generally had higher levels of BDNF, trkB-TK+ and GAD67 mRNA than those not taking antidepressants, they were not necessarily at the level of controls in CA4. A similar ANOVA analysis for mood stabilizers revealed that they had no effect on the expression of BDNF, trkB-TK+ or GAD67 mRNA in any of the subregions in the hippocampal formation.
Fig. 3.
Mean brain-derived neurotrophic factor (BDNF), tyrosine kinase receptor (trkB-TK+) and glutamic acid decarboxylase (GAD67) mRNA levels of individuals taking antidepressants at the time of death, patients not taking antidepressants at the time of death and unaffected controls in the dentate gyrus and cornu ammonis (CA) subfield 4. Standard error is represented by error bars (*p ≤ 0.05; **p ≤ 0.01).
Correlations between GAD67 mRNA levels and BDNF and trkB-TK+ levels
In the dentate gyrus, GAD67 mRNA levels correlated with both BDNF (r = 0.36, p = 0.011) and trkB-TK+ mRNA expression (r = 0.69, p < 0.001) across the 60 individuals. Similar correlations were found in CA4, (r = 0.51, p < 0.001; r = 0.68, p < 0.001, respectively), CA3 (r = 0.46, p = 0.003; r = 0.52, p < 0.001, respectively) and the EC (r = 0.63, p < 0.001; r = 0.40, p = 0.007; Appendix 5, available at cma.ca/jpn). In CA1 and the subiculum GAD67 mRNA was correlated with trkB-TK+ mRNA levels only (r = 0.37, p = 0.007; r = 0.41, p = 0.003, respectively; Appendix 5).
Discussion
We report a significant reduction in the expression of BDNF, trkB-TK+ and GAD67 mRNA in several temporal regions of individuals with schizophrenia and mood disorders. The most striking reductions in mRNA were found in the DG, CA4 subregions of the hippocampus and in layer II of the EC. Brain-derived neurotrophic factor mRNA was significantly reduced in CA4 (hilar region) in the bipolar disorder group, and both the major depression and bipolar disorder groups had reduced levels of trkB-TK+ mRNA in layer II of the EC. Reductions in trkB-TK+ mRNA expression were also observed in CA4 in the schizophrenia group. The schizophrenia and major depression groups also showed reductions in GAD67 mRNA expression in CA4. We found that GAD67 mRNA expression was also lower in the DG in the schizophrenia group. Thus, in the hippocampus, the abnormalities in schizophrenia occur mainly in trkB-TK+ and GAD67 mRNAs and to a lesser extent in BDNF mRNA, whereas in bipolar disorder the abnormalities are most prominent in BDNF and trkB-TK+ mRNAs. These 2 psychotic disorders appear to share some neurotrophin pathology, especially in CA4, yet they also have a somewhat different pattern of abnormalities with imprecise overlap.
The results from this study corroborate those of earlier studies that describe abnormalities in the expression of cortical BDNF and trkB-TK+ mRNA in individuals with schizophrenia while also extending these findings to a third independent cohort and additional brain regions.20,21 Studies of BDNF protein levels in the hippocampus of patients with schizophrenia have yielded inconsistent results;35 however, a recent study using this same cohort reported no abnormalities in proBDNF protein levels in the hippocampus of the schizophrenia group.36,37 Likewise, we find only a limited decrease in BDNF mRNA, restricted to CA4 in the hippocampus in the schizophrenia group. Whereas there was a reduction in BDNF mRNA in CA4 in the schizophrenia group, it did not reach a level of significance after Bonferroni testing; however, the level of BDNF in CA4 in the schizophrenia group was not significantly different from that in the bipolar disorder group, suggesting that the BDNF mRNA reduction is similar in these 2 disorders (Fig. 2A). Thus, the failure to detect a significant change in schizophrenia likely reflects the increased variability in BDNF expression in the schizophrenia group, the relatively low number of individuals with schizophrenia and the conservative statistical significance criteria rather than a true biological difference. Nevertheless, the abnormalities in BDNF mRNA and protein expression36 tend to be more pronounced in bipolar disorder. In contrast, we found significant deficits in trkB-TK+ mRNA levels in the hippocampus of the schizophrenia group, but the protein levels for trkB-TK+ did not appear to decline in schizophrenia.36
Reductions in trkB-TK+ mRNA levels were also observed within layer II of the EC in individuals with bipolar disorder and major depression but not schizophrenia. Layer II consists primarily of densely packed stellate, interneuronal and medium–large pyramidal cells,38 which project to the dentate gyrus and CA3. Qualitative microscopic analyses of morphologic abnormalities as well as quantitative measurements of neuronal density revealed no significant differences between the schizophrenia and control groups,39,40 which supports our result showing no significant differences in BDNF, trkB-TK+ or GAD67 mRNA expression between individuals with schizophrenia and unaffected controls in this region. However, a study by Pantazopoulos and colleagues41 reported a decrease in parvalbumin-immunoreactive neurons within the superficial layers of the EC in individuals with bipolar disorder but not those with schizophrenia. This calcium-binding protein delineates inhibitory interneurons within the cortex and has revealed GABAergic abnormalities in the dorso-lateral prefrontal cortex (DLPFC) and hippocampus in the major psychiatric disorders.18,42,43 Since trkB-TK+ is also associated with inhibitory neurons, the expressional deficits of trkB-TK+ mRNA in layer II could be a result of a loss of GABAergic neuronal density within this superficial layer of the EC in individuals with bipolar disorder or depression. However, additional studies will be necessary to directly correlate the parvalbumin deficit with trkB-TK+ expression.
Numerous studies have shown that BDNF and/or trkB-TK+ signalling may also be associated with depressive behaviour. Levels of BDNF and trkB-TK+ mRNA were not changed in the hippocampus of the major depression group, although there was a decrease in trkB-TK+ mRNA levels in layer II of the EC in individuals with depression. However, chronic treatment with antidepressants increases the expression of BDNF and trkB-TK+ in the hippocampus of the rat brain, and hippocampal BDNF immunoreactivity is elevated in clinically depressed patients treated with antidepressants.44 Moreover, a recent study has shown that a selective deletion of BDNF within the DG in mice results in a reduction of anti-depressant efficacy, suggesting that BDNF is essential for mediating the beneficial effects of antidepressants.45 In conjunction with these findings, we also found that compared with patients taking antidepressants at the time of death, patients free of antidepressants at the time of death expressed significantly less BDNF and trkB-TK+ mRNA in the DG and CA4 regions, where this antidepressant effect has also been found.44 This indicates that antidepressants were more likely to obscure abnormal BDNF/trkB-TK+ mRNA expression than to cause the reduction observed in this study.
Brain-derived neurotrophic factor trkB-TK+ signalling appears to regulate both excitatory and inhibitory impulses within the hippocampal formation.23,25 Multiple studies have shown that BDNF and trkB-TK+ mRNA expression levels are increased by glutamate activity,23,46–49 and that trkB-TK+ is located, although not exclusively, in GABAergic neurons,23,50 which contain the GABA-synthesizing enzyme GAD67. This suggests that BDNF/trkB-TK+ signalling participates in regulating the balance between excitation and inhibition within the hippocampus. We found that GAD67 mRNA levels were significantly reduced in the DG in the schizophrenia group and in CA4 in the schizophrenia and major depression groups. These reductions in GAD67 expression may have implications for neuroimaging studies that show an increase in hippocampal activity during the retrieval of words,10,51 auditory hallucinations13 and rest52–55 in individuals with schizophrenia. Together these results indicate that the abnormal activity seen in individuals with schizophrenia may be due to a decrease in the inhibitory tone mandatory for normal hippocampal function. Several studies have shown that there is a reduction in the density of at least a subset of the hippocampal GABAergic interneurons,17,18,56 as well as a reduction in GAD67 mRNA in the hippocampus.57,58 Thus the decrease of GAD67 mRNA observed in this study could also be due to a reduction in the density of GABAergic neurons.
Previous studies have indicated that abnormal signalling in the BDNF/trkB-TK+ pathway may lead to abnormalities in the GABAergic neurons.20,23–26 Whereas GAD67 mRNA levels were correlated with both BDNF and trkB-TK+ in almost all hippocampal areas examined, the correlations were much stronger with trkB-TK+. However, given the abnormal trkB-TK+ expression, for example, in layer II of the EC in the bipolar disorder and major depression groups, one may expect to see abnormalities of GAD67 mRNA expression in the EC or downstream in the DG or CA3 in the bipolar disorder and depression groups. In contrast to what was expected, there were no GAD67 mRNA abnormalities in the EC, and the GAD67 mRNA abnormalities in the DG actually occured in the schizophrenia group. Thus, although there may be an underlying connection between the reductions in GAD67 expression and BDNF/trkB-TK+ signalling in the hippocampus of people with mental illness, the relation is complex and cannot be definitively resolved with the current data.
Limitations
The limitations encountered in this study are those that are common to all human postmortem brain research and require accounting for various demographic and clinical variables that may influence molecular preservation in the tissue. We covaried for each variable that significantly correlated with BDNF, trkB-TK+ or GAD67 mRNA expression levels in the analysis. In addition, there are limitations to film-based densitometric measurement, which does not allow us to determine if a decrease in mRNA signal in a given area is due to less mRNA per cell, fewer expressing cells or fewer cells altogether. Nevertheless, film-based densitometric measurement is well established as an acceptable and well validated method in postmortem neuropathology research. Film-based quantification is an efficient way to assess a large number of samples. Thus, we were able to survey a larger surface area of anatomically defined tissue in a larger number of sections (n = 120 in this cohort) than would have been possible at the cellular level. In addition, to account for possible limitations due to the sensitivity of the film-based method of quantification, we used standards to calibrate the radioactivity, which is a widely accepted practice.
Conclusion
This study adds to mounting evidence that hippocampal pathology exists in schizophrenia and other psychiatric illnesses. Each major mental disorder showed some overlap but also some differences in the pattern of abnormalities in BDNF, trkB-TK+ and GAD67 mRNA expression levels depending on the probe used and area examined. It has been reported that BDNF/trkB-TK+ signalling influences the development of cortical GABA neurons and the expression of GAD67. 20,26,59,60 However, it is not clear how the GAD67 abnormalities described in this study relate to abnormalities in BDNF and trkB-TK+ given the variability in magnitude and anatomic pattern of change in the neurotrophins and the apparent lack of reduction in GAD67 mRNA despite reductions in both BDNF and trkB-TK+ in individuals with bipolar disorder. Future studies at the cellular level will give more insight into which particular cells are affected in each disorder, and may eventually lead to new targets for pharmaceutical treatments.
Acknowledgements
This work was supported by the Schizophrenia Research Institute, utilizing infrastructure funding from NSW Health, the University of New South Wales School of Psychiatry, and the Prince of Wales Medical Research Institute. This research was also funded by the Stanley Medical Research Institute.
Footnotes
Competing interests: None declared.
Contributors: Drs. Weickert and Webster designed the study. Mr. Wyatt acquired the data, which all authors analyzed. Drs. Thompson Ray and Webster wrote the article. All authors reviewed it and approved publication.
References
- 1.Weinberger DR. Cell biology of the hippocampal formation in schizophrenia. Biol Psychiatry. 1999;45:395–402. doi: 10.1016/s0006-3223(98)00331-x. [DOI] [PubMed] [Google Scholar]
- 2.Harrison PJ. The hippocampus in schizophrenia: a review of the neuropathological evidence and its pathophysiological implications. Psychopharmacology (Berl) 2004;174:151–62. doi: 10.1007/s00213-003-1761-y. [DOI] [PubMed] [Google Scholar]
- 3.Altar CA, Jurata LW, Charles V, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 4.Rajkowska G. Cell pathology in bipolar disorder. Bipolar Disord. 2002;4:105–16. doi: 10.1034/j.1399-5618.2002.01149.x. [DOI] [PubMed] [Google Scholar]
- 5.Harrison PJ. The neuropathology of primary mood disorder. Brain. 2002;125:1428–49. doi: 10.1093/brain/awf149. [DOI] [PubMed] [Google Scholar]
- 6.Strasser HC, Lilyestrom J, Ashby ER, et al. Hippocampal and ventricular volumes in psychotic and nonpsychotic bipolar patients compared with schizophrenia patients and community control subjects: a pilot study. Biol Psychiatry. 2005;57:633–9. doi: 10.1016/j.biopsych.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Rusch BD, Abercrombie HC, Oakes TR, et al. Hippocampal morphometry in depressed patients and control subjects: relations to anxiety symptoms. Biol Psychiatry. 2001;50:960–4. doi: 10.1016/s0006-3223(01)01248-3. [DOI] [PubMed] [Google Scholar]
- 8.Sequeira A, Klempan T, Canetti L, et al. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–55. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- 9.Videbech P, Ravnkilde B. Hippocampal volume and depression: a meta-analysis of MRI studies. Am J Psychiatry. 2004;161:1957–66. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
- 10.Heckers S, Rauch SL, Goff D, et al. Impaired recruitment of the hippocampus during conscious recollection in schizophrenia. Nat Neurosci. 1998;1:318–23. doi: 10.1038/1137. [DOI] [PubMed] [Google Scholar]
- 11.Weiss AP, Schacter DL, Goff DC, et al. Impaired hippocampal recruitment during normal modulation of memory performance in schizophrenia. Biol Psychiatry. 2003;53:48–55. doi: 10.1016/s0006-3223(02)01541-x. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki Y, Maeda Y, Sakai N, et al. Regional cerebral blood flow in patients with schizophrenia: relevance to symptom structures. Psychiatry Res. 1996;67:49–58. doi: 10.1016/0925-4927(96)02685-6. [DOI] [PubMed] [Google Scholar]
- 13.Silbersweig DA, Stern E, Frith C, et al. A functional neuroanatomy of hallucinations in schizophrenia. Nature. 1995;378:176–9. doi: 10.1038/378176a0. [DOI] [PubMed] [Google Scholar]
- 14.Gur RE, Grath C, Chan RM, et al. An fMRI study of facial emotion processing in patients with schizophrenia. Am J Psychiatry. 2002;159:1992–9. doi: 10.1176/appi.ajp.159.12.1992. [DOI] [PubMed] [Google Scholar]
- 15.Benes FM, Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsycho-pharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 16.Lisman JE, Coyle JT, Green RW, et al. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008;31:234–42. doi: 10.1016/j.tins.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benes FM, Kwok EW, Vincent SL, et al. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biol Psychiatry. 1998;44:88–97. doi: 10.1016/s0006-3223(98)00138-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZJ, Reynolds GP. A selective decrease in the relative density of parvalbumin-immunoreactive neurons in the hippocampus in schizophrenia. Schizophr Res. 2002;55:1–10. doi: 10.1016/s0920-9964(01)00188-8. [DOI] [PubMed] [Google Scholar]
- 19.Heckers S, Stone D, Walsh J, et al. Differential hippocampal expression of glutamic acid decarboxylase 65 and 67 messenger RNA in bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2002;59:521–9. doi: 10.1001/archpsyc.59.6.521. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto T, Bergen SE, Nguyen QL, et al. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25:372–83. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weickert CS, Hyde TM, Lipska BK, et al. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 22.Weickert CS, Ligons DL, Romanczyk T, et al. Reductions in neurotrophin receptor mRNAs in the prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2005;10:637–50. doi: 10.1038/sj.mp.4001678. [DOI] [PubMed] [Google Scholar]
- 23.Bolton MM, Pittman AJ, Lo DC. Brain-derived neurotrophic factor differentially regulates excitatory and inhibitory synaptic transmission in hippocampal cultures. J Neurosci. 2000;20:3221–32. doi: 10.1523/JNEUROSCI.20-09-03221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto T, Lewis DA. BDNF Val66Met polymorphism and GAD67 mRNA expression in the prefrontal cortex of subjects with schizophrenia. Am J Psychiatry. 2006;163:534–7. doi: 10.1176/appi.ajp.163.3.534. [DOI] [PubMed] [Google Scholar]
- 25.Marty S, Wehrle R, Sotelo C. Neuronal activity and brain-derived neurotrophic factor regulate the density of inhibitory synapses in organotypic slice cultures of postnatal hippocampus. J Neurosci. 2000;20:8087–95. doi: 10.1523/JNEUROSCI.20-21-08087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada MK, Nakanishi K, Ohba S, et al. Brain-derived neurotrophic factor promotes the maturation of GABAergic mechanisms in cultured hippocampal neurons. J Neurosci. 2002;22:7580–5. doi: 10.1523/JNEUROSCI.22-17-07580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–27. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–93. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- 29.Dwivedi Y. Brain-derived neurotrophic factor: role in depression and suicide. Neuropsychiatr Dis Treat. 2009;5:433–49. doi: 10.2147/ndt.s5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrey EF, Webster M, Knable M, et al. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–5. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 31.Whitfield HJ, Jr, Brady LS, Smith MA, et al. Optimization of cRNA probe in situ hybridization methodology for localization of glucocorticoid receptor mRNA in rat brain: a detailed protocol. Cell Mol Neurobiol. 1990;10:145–57. doi: 10.1007/BF00733641. [DOI] [PubMed] [Google Scholar]
- 32.Rosene DL, Van Hoesen GW. The hippocampal formation of the primate brain: a review of some comparative aspects of cytoarchitecture and connections. In: Jones EG, Peters A, editors. Cerebral cortex Vol 6 Further aspects of cortical function, including hippocampus. New York (NY): Plenum; 1987. [Google Scholar]
- 33.Krimer LS, Hyde TM, Herman MM, et al. The entorhinal cortex: an examination of cyto- and myeloarchitectonic organization in humans. Cereb Cortex. 1997;7:722–31. doi: 10.1093/cercor/7.8.722. [DOI] [PubMed] [Google Scholar]
- 34.Insausti R, Tunon T, Sobreviela T, et al. The human entorhinal cortex: a cytoarchitectonic analysis. J Comp Neurol. 1995;355:171–98. doi: 10.1002/cne.903550203. [DOI] [PubMed] [Google Scholar]
- 35.Durany N, Michel T, Zöchling R, et al. Brain-derived neurotrophic factor and neurotrophin 3 in schizophrenic psychoses. Schizophr Res. 2001;52:79–86. doi: 10.1016/s0920-9964(00)00084-0. [DOI] [PubMed] [Google Scholar]
- 36.Dunham JS, Deakin JF, Miyajima F, et al. Expression of hippocampal brain-derived neurotrophic factor and its receptors in Stanley consortium brains. J Psychiatr Res. 2009;43:1175–84. doi: 10.1016/j.jpsychires.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Iritani S, Niizato K, Nawa H, et al. Immunohistochemical study of brain-derived neurotrophic factor and its receptor, TrkB, in the hippocampal formation of schizophrenic brains. Prog Neuropsycho-pharmacol Biol Psychiatry. 2003;27:801–7. doi: 10.1016/S0278-5846(03)00112-X. [DOI] [PubMed] [Google Scholar]
- 38.Canto CB, Wouterlood FG, Witter MP. What does the anatomical organization of the entorhinal cortex tell us? Neural Plast. 2008;2008:381243. doi: 10.1155/2008/381243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akil M, Lewis DA. Cytoarchitecture of the entorhinal cortex in schizophrenia. Am J Psychiatry. 1997;154:1010–2. doi: 10.1176/ajp.154.7.1010. [DOI] [PubMed] [Google Scholar]
- 40.Krimer LS, Herman MM, Saunders RC, et al. A qualitative and quantitative analysis of the entorhinal cortex in schizophrenia. Cereb Cortex. 1997;7:732–9. doi: 10.1093/cercor/7.8.732. [DOI] [PubMed] [Google Scholar]
- 41.Pantazopoulos H, Lange N, Baldessarini RJ, et al. Parvalbumin neurons in the entorhinal cortex of subjects diagnosed with bipolar disorder or schizophrenia. Biol Psychiatry. 2007;61:640–52. doi: 10.1016/j.biopsych.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torrey EF, Barci BM, Webster MJ, et al. Neurochemical markers for schizophrenia, bipolar disorder, and major depression in postmortem brains. Biol Psychiatry. 2005;57:252–60. doi: 10.1016/j.biopsych.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 43.Knable MB, Barci BM, Webster MJ, et al. Molecular abnormalities of the hippocampus in severe psychiatric illness: postmortem findings from the Stanley Neuropathology Consortium. Mol Psychiatry. 2004;9:609–20. 544. doi: 10.1038/sj.mp.4001471. [DOI] [PubMed] [Google Scholar]
- 44.Chen B, Dowlatshahi D, MacQueen GM, et al. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry. 2001;50:260–5. doi: 10.1016/s0006-3223(01)01083-6. [DOI] [PubMed] [Google Scholar]
- 45.Adachi M, Barrot M, Autry AE, et al. Selective loss of brain-derived neurotrophic factor in the dentate gyrus attenuates antidepressant efficacy. Biol Psychiatry. 2008;63:642–9. doi: 10.1016/j.biopsych.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falkenberg T, Ernfors P, Persson H, et al. Cortical transynaptic activation of tyrosine kinase receptor trkB messenger RNA expression in rat hippocampus. Neuroscience. 1992;51:883–9. doi: 10.1016/0306-4522(92)90527-9. [DOI] [PubMed] [Google Scholar]
- 47.Falkenberg T, Metsis M, Timmusk T, et al. Entorhinal cortex regulation of multiple brain-derived neurotrophic factor promoters in the rat hippocampus. Neuroscience. 1993;57:891–6. doi: 10.1016/0306-4522(93)90034-d. [DOI] [PubMed] [Google Scholar]
- 48.Lindefors N, Ernfors P, Falkenberg T, et al. Septal cholinergic afferents regulate expression of brain-derived neurotrophic factor and beta-nerve growth factor mRNA in rat hippocampus. Exp Brain Res. 1992;88:78–90. doi: 10.1007/BF02259130. [DOI] [PubMed] [Google Scholar]
- 49.Zafra F, Castren E, Thoenen H, et al. Interplay between glutamate and gamma-aminobutyric acid transmitter systems in the physiological regulation of brain-derived neurotrophic factor and nerve growth factor synthesis in hippocampal neurons. Proc Natl Acad Sci U S A. 1991;88:10037–41. doi: 10.1073/pnas.88.22.10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zachrisson O, Falkenberg T, Lindefors N. Neuronal coexistence of trkB and glutamic acid decarboxylase67 mRNAs in rat hippocampus. Brain Res Mol Brain Res. 1996;36:169–73. doi: 10.1016/0169-328x(95)00281-v. [DOI] [PubMed] [Google Scholar]
- 51.Heckers S, Goff D, Schacter DL, et al. Functional imaging of memory retrieval in deficit vs nondeficit schizophrenia. Arch Gen Psychiatry. 1999;56:1117–23. doi: 10.1001/archpsyc.56.12.1117. [DOI] [PubMed] [Google Scholar]
- 52.Friston KJ, Liddle PF, Frith CD, et al. The left medial temporal region and schizophrenia. A PET study. Brain. 1992;115:367–82. doi: 10.1093/brain/115.2.367. [DOI] [PubMed] [Google Scholar]
- 53.Gur RE, Mozley PD, Resnick SM, et al. Resting cerebral glucose metabolism in first-episode and previously treated patients with schizophrenia relates to clinical features. Arch Gen Psychiatry. 1995;52:657–67. doi: 10.1001/archpsyc.1995.03950200047013. [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki Y, Suzuki M, Maeda Y, et al. Regional cerebral blood flow in patients with schizophrenia. A preliminary report. Eur Arch Psychiatry Clin Neurosci. 1992;241:195–200. doi: 10.1007/BF02190252. [DOI] [PubMed] [Google Scholar]
- 55.Liddle PF, Friston KJ, Frith CD, et al. Patterns of cerebral blood flow in schizophrenia. Br J Psychiatry. 1992;160:179–86. doi: 10.1192/bjp.160.2.179. [DOI] [PubMed] [Google Scholar]
- 56.Heckers S, Konradi C. Hippocampal neurons in schizophrenia. J Neural Transm. 2002;109:891–905. doi: 10.1007/s007020200073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benes FM, Lim B, Matzilevich D, et al. Circuitry-based gene expression profiles in GABA cells of the trisynaptic pathway in schizophrenics versus bipolars. Proc Natl Acad Sci U S A. 2008;105:20935–40. doi: 10.1073/pnas.0810153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benes FM, Lim B, Matzilevich D, et al. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104:10164–9. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcántara S, Frisén J, del Río JA, et al. TrkB signaling is required for postnatal survival of CNS neurons and protects hippocampal and motor neurons from axotomy-induced cell death. J Neurosci. 1997;17:3623–33. doi: 10.1523/JNEUROSCI.17-10-03623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones KR, Farinas I, Backus C, et al. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;76:989–99. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]