Abstract
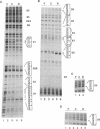
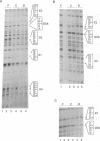
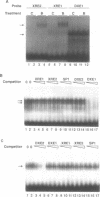
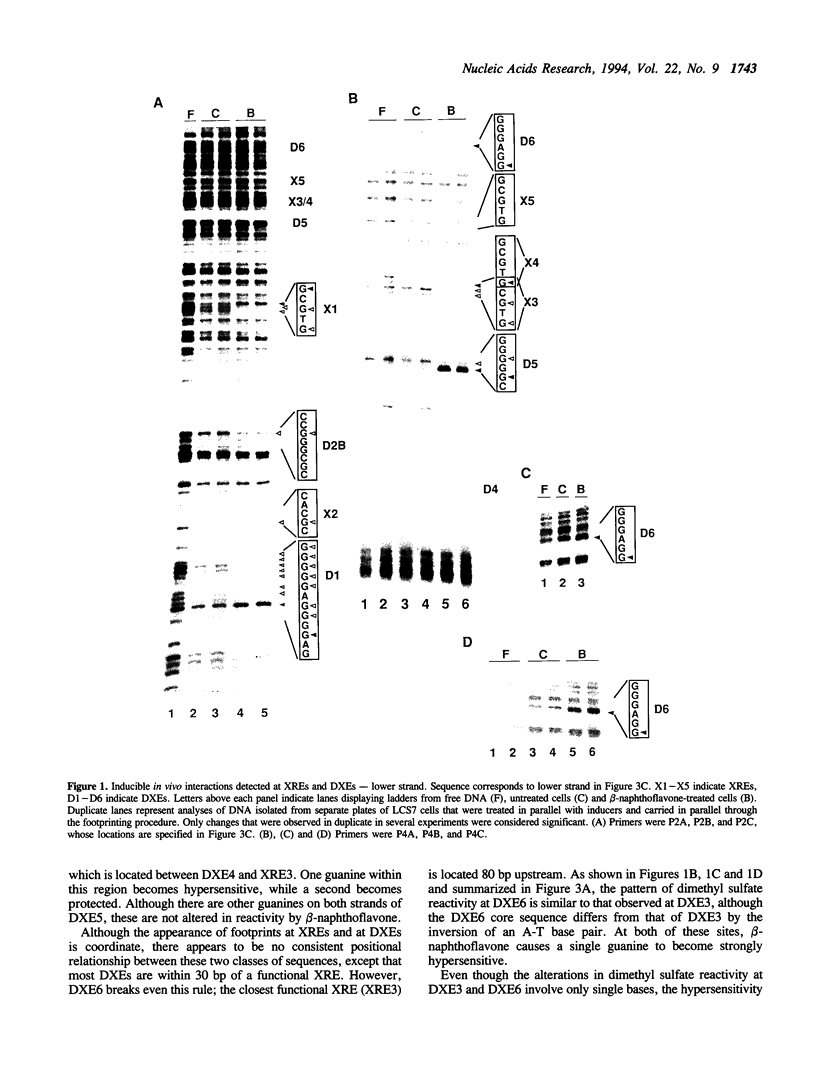
In vivo footprinting experiments, augmented with gel shift and transfection analyses suggest that activation of the CYP1A1 gene by aryl hydrocarbons may be a multicomponent process. During the first 30 minutes of exposure to aryl hydrocarbon carcinogens and environmental contaminants, in vivo footprints appear at nine distinct sites within a 281 bp region centered 950 bp upstream of the CYP1A1 transcription start site. Six of these sites are unrelated in sequence to the three xenobiotic response elements (XREs) within this region, at which the aryl hydrocarbon (AH) receptor is known to bind. These six display a variety of footprint patterns, are diverse in sequence and range in G-C content from 60 to 75%. This diversity suggests that multiple nuclear factors may be responsible for these six in vivo footprints. These observations are consistent with competition gel shift experiments showing that the nuclear factors binding at two of these sites are different from each other, as well as from the AH receptor. Gel shifts also indicate that the sequence-specific factors binding at these sites are expressed constitutively. This is consistent with a model in which in vivo footprints are induced at these six sites, not through direct activation or de novo synthesis of DNA-binding factors, but through a two phase mechanism in which binding of the nuclear AH receptor complex to XREs facilitates the binding of constitutive factors at these sites. This facilitation could be mediated either through specific protein-protein interactions or through alterations in chromatin structure that make these sites accessible to constitutive nuclear factors. A function for the sequences at which aryl hydrocarbons induce in vivo footprints is suggested by transfection experiments showing that one of these sequences cooperates with a weak XRE to confer on a reporter gene responsiveness to aryl hydrocarbons.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer T. K., Lefebvre P., Wolford R. G., Hager G. L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992 Mar 20;255(5051):1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- Burbach K. M., Poland A., Bradfield C. A. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I. L., Kelly S. E. Probing the nature of chromosomal DNA-protein contacts by in vivo footprinting. Biotechniques. 1991 Aug;11(2):188-90, 192-4, 196 passim. [PubMed] [Google Scholar]
- Chou J. Y., Schlegel-Haueter S. E. Study of liver differentiation in vitro. J Cell Biol. 1981 May;89(2):216–222. doi: 10.1083/jcb.89.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J. Y. Temperature-sensitive adult liver cell line dependent on glucocorticoid for differentiation. Mol Cell Biol. 1983 Jun;3(6):1013–1020. doi: 10.1128/mcb.3.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987 Jan 30;48(2):261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Darrow A. L., Rickles R. J., Pecorino L. T., Strickland S. Transcription factor Sp1 is important for retinoic acid-induced expression of the tissue plasminogen activator gene during F9 teratocarcinoma cell differentiation. Mol Cell Biol. 1990 Nov;10(11):5883–5893. doi: 10.1128/mcb.10.11.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Fisher J. M., Whitlock J. P., Jr The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J Biol Chem. 1988 Nov 25;263(33):17221–17224. [PubMed] [Google Scholar]
- Durrin L. K., Whitlock J. P., Jr 2,3,7,8-Tetrachlorodibenzo-p-dioxin-inducible aryl hydrocarbon receptor-mediated change in CYP1A1 chromatin structure occurs independently of transcription. Mol Cell Biol. 1989 Dec;9(12):5733–5737. doi: 10.1128/mcb.9.12.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin L. K., Whitlock J. P., Jr In situ protein-DNA interactions at a dioxin-responsive enhancer associated with the cytochrome P1-450 gene. Mol Cell Biol. 1987 Aug;7(8):3008–3011. doi: 10.1128/mcb.7.8.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S. Modularity in promoters and enhancers. Cell. 1989 Jul 14;58(1):1–4. doi: 10.1016/0092-8674(89)90393-0. [DOI] [PubMed] [Google Scholar]
- Fisher J. M., Wu L., Denison M. S., Whitlock J. P., Jr Organization and function of a dioxin-responsive enhancer. J Biol Chem. 1990 Jun 15;265(17):9676–9681. [PubMed] [Google Scholar]
- Fujisawa-Sehara A., Sogawa K., Yamane M., Fujii-Kuriyama Y. Characterization of xenobiotic responsive elements upstream from the drug-metabolizing cytochrome P-450c gene: a similarity to glucocorticoid regulatory elements. Nucleic Acids Res. 1987 May 26;15(10):4179–4191. doi: 10.1093/nar/15.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa-Sehara A., Yamane M., Fujii-Kuriyama Y. A DNA-binding factor specific for xenobiotic responsive elements of P-450c gene exists as a cryptic form in cytoplasm: its possible translocation to nucleus. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5859–5863. doi: 10.1073/pnas.85.16.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelboin H. V. Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev. 1980 Oct;60(4):1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- Gidoni D., Dynan W. S., Tjian R. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. 1984 Nov 29-Dec 5Nature. 312(5993):409–413. doi: 10.1038/312409a0. [DOI] [PubMed] [Google Scholar]
- Gonzalez F. J., Nebert D. W. Autoregulation plus upstream positive and negative control regions associated with transcriptional activation of the mouse P1(450) gene. Nucleic Acids Res. 1985 Oct 25;13(20):7269–7288. doi: 10.1093/nar/13.20.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapgood J., Cuthill S., Söderkvist P., Wilhelmsson A., Pongratz I., Tukey R. H., Johnson E. F., Gustafsson J. A., Poellinger L. Liver cells contain constitutive DNase I-hypersensitive sites at the xenobiotic response elements 1 and 2 (XRE1 and -2) of the rat cytochrome P-450IA1 gene and a constitutive, nuclear XRE-binding factor that is distinct from the dioxin receptor. Mol Cell Biol. 1991 Sep;11(9):4314–4323. doi: 10.1128/mcb.11.9.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Hoffman E. C., Reyes H., Chu F. F., Sander F., Conley L. H., Brooks B. A., Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991 May 17;252(5008):954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- Jones K. W., Whitlock J. P., Jr Functional analysis of the transcriptional promoter for the CYP1A1 gene. Mol Cell Biol. 1990 Oct;10(10):5098–5105. doi: 10.1128/mcb.10.10.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. B., Durrin L. K., Fisher J. M., Whitlock J. P., Jr Control of gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Multiple dioxin-responsive domains 5'-ward of the cytochrome P1-450 gene. J Biol Chem. 1986 May 25;261(15):6647–6650. [PubMed] [Google Scholar]
- Kageyama R., Merlino G. T., Pastan I. Nuclear factor ETF specifically stimulates transcription from promoters without a TATA box. J Biol Chem. 1989 Sep 15;264(26):15508–15514. [PubMed] [Google Scholar]
- Kim C. H., Heath C., Bertuch A., Hansen U. Specific stimulation of simian virus 40 late transcription in vitro by a cellular factor binding the simian virus 40 21-base-pair repeat promoter element. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6025–6029. doi: 10.1073/pnas.84.17.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Martin K. J. The interactions of transcription factors and their adaptors, coactivators and accessory proteins. Bioessays. 1991 Oct;13(10):499–503. doi: 10.1002/bies.950131003. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Reaction of nucleosome DNA with dimethyl sulfate. Proc Natl Acad Sci U S A. 1979 May;76(5):2133–2137. doi: 10.1073/pnas.76.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Moyer R., Mariën K., van Holde K., Bailey G. Site-specific aflatoxin B1 adduction of sequence-positioned nucleosome core particles. J Biol Chem. 1989 Jul 25;264(21):12226–12231. [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Kamataki T., Waxman D. J., Guengerich F. P., Estabrook R. W., Feyereisen R., Gonzalez F. J., Coon M. J., Gunsalus I. C., Gotoh O. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993 Jan-Feb;12(1):1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- Neuhold L. A., Shirayoshi Y., Ozato K., Jones J. E., Nebert D. W. Regulation of mouse CYP1A1 gene expression by dioxin: requirement of two cis-acting elements during induction. Mol Cell Biol. 1989 Jun;9(6):2378–2386. doi: 10.1128/mcb.9.6.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasco D. S., Boyum K. W., Merchant S. N., Chalberg S. C., Fagan J. B. Transcriptional and post-transcriptional regulation of the genes encoding cytochromes P-450c and P-450d in vivo and in primary hepatocyte cultures. J Biol Chem. 1988 Jun 25;263(18):8671–8676. [PubMed] [Google Scholar]
- Pasco D. S., Fagan J. B. Efficient DNA-mediated gene transfer into primary cultures of adult rat hepatocytes. DNA. 1989 Sep;8(7):535–541. doi: 10.1089/dna.1.1989.8.535. [DOI] [PubMed] [Google Scholar]
- Pelkonen O., Nebert D. W. Metabolism of polycyclic aromatic hydrocarbons: etiologic role in carcinogenesis. Pharmacol Rev. 1982 Jun;34(2):189–222. [PubMed] [Google Scholar]
- Poland A., Knutson J. C. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Reyes H., Reisz-Porszasz S., Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992 May 22;256(5060):1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- Rigaud G., Roux J., Pictet R., Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991 Nov 29;67(5):977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- Saatcioglu F., Perry D. J., Pasco D. S., Fagan J. B. Aryl hydrocarbon (Ah) receptor DNA-binding activity. Sequence specificity and Zn2+ requirement. J Biol Chem. 1990 Jun 5;265(16):9251–9258. [PubMed] [Google Scholar]
- Saatcioglu F., Perry D. J., Pasco D. S., Fagan J. B. Multiple DNA-binding factors interact with overlapping specificities at the aryl hydrocarbon response element of the cytochrome P450IA1 gene. Mol Cell Biol. 1990 Dec;10(12):6408–6416. doi: 10.1128/mcb.10.12.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S. H. Comparative toxicology and mechanism of action of polychlorinated dibenzo-p-dioxins and dibenzofurans. Annu Rev Pharmacol Toxicol. 1986;26:371–399. doi: 10.1146/annurev.pa.26.040186.002103. [DOI] [PubMed] [Google Scholar]
- Shen E. S., Whitlock J. P., Jr Protein-DNA interactions at a dioxin-responsive enhancer. Mutational analysis of the DNA-binding site for the liganded Ah receptor. J Biol Chem. 1992 Apr 5;267(10):6815–6819. [PubMed] [Google Scholar]
- Shen E. S., Whitlock J. P., Jr The potential role of DNA methylation in the response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1989 Oct 25;264(30):17754–17758. [PubMed] [Google Scholar]
- Watson A. J., Hankinson O. Dioxin- and Ah receptor-dependent protein binding to xenobiotic responsive elements and G-rich DNA studied by in vivo footprinting. J Biol Chem. 1992 Apr 5;267(10):6874–6878. [PubMed] [Google Scholar]
- Watson A. J., Weir-Brown K. I., Bannister R. M., Chu F. F., Reisz-Porszasz S., Fujii-Kuriyama Y., Sogawa K., Hankinson O. Mechanism of action of a repressor of dioxin-dependent induction of Cyp1a1 gene transcription. Mol Cell Biol. 1992 May;12(5):2115–2123. doi: 10.1128/mcb.12.5.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L. P., Koeiman N., Whitlock J. P., Jr Dioxin-inducible, Ah receptor-dependent transcription in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8545–8549. doi: 10.1073/pnas.87.21.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr Genetic and molecular aspects of 2,3,7,8-tetrachlorodibenzo-p-dioxin action. Annu Rev Pharmacol Toxicol. 1990;30:251–277. doi: 10.1146/annurev.pa.30.040190.001343. [DOI] [PubMed] [Google Scholar]
- Wu L., Whitlock J. P., Jr Mechanism of dioxin action: Ah receptor-mediated increase in promoter accessibility in vivo. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4811–4815. doi: 10.1073/pnas.89.11.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Whitlock J. P., Jr Mechanism of dioxin action: receptor-enhancer interactions in intact cells. Nucleic Acids Res. 1993 Jan 11;21(1):119–125. doi: 10.1093/nar/21.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]