Abstract
Smoking is a risk factor for most of the diseases leading in mortality1. We conducted genome-wide association (GWA) meta-analyses of smoking data within the ENGAGE consortium to search for common alleles associating with the number of cigarettes smoked per day (CPD) in smokers (N=31,266) and smoking initiation (N=46,481). We tested selected SNPs in a second stage (N=45,691 smokers), and assessed some in a third sample (N=9,040). Variants in three genomic regions associated with CPD (P< 5·10−8), including previously identified SNPs at 15q25 represented by rs1051730-A (0.80 CPD,P=2.4·10−69), and SNPs at 19q13 and 8p11, represented by rs4105144-C (0.39 CPD, P=2.2·10−12) and rs6474412-T (0.29 CPD,P= 1.4·10−8), respectively. Among the genes at the two novel loci, are genes encoding nicotine-metabolizing enzymes (CYP2A6 and CYP2B6), and nicotinic acetylcholine receptor subunits (CHRNB3 and CHRNA6) highlighted in previous studies of nicotine dependence2-3. Nominal associations with lung cancer were observed at both 8p11 (rs6474412-T,OR=1.09,P=0.04) and 19q13 (rs4105144-C,OR=1.12,P=0.0006).
Smoking behavior and nicotine dependence (ND) are under genetic influence4. While environmental influences play a strong role in the initiation of smoking5, the heritability of smoking persistence, smoking quantity and ND is high in most twin studies5,6. Sequence variants within a cluster of nicotinic acetylcholine receptor (nAChR) genes on chromosome 15q25 have recently been shown to associate with the number of cigarettes smoked per day (CPD)7-8, ND3,7, and smoking-related diseases such as lung cancer (LC)7,9-10, peripheral arterial disease (PAD)7, and chronic obstructive pulmonary disease (COPD)11
To search for additional common variants associating with smoking behavior we performed meta-analyses of GWA studies, mainly using samples of European ancestry from the ENGAGE consortium (See URL section), focusing on two smoking phenotypes: CPD and smoking initiation. The smoking initiation analysis was performed with a total of 30,431 ever-smokers and 16,050 never-smokers, using data from 12 GWA studies, Corogene, deCODE, EGPUT, ERF, NFBC, KORA, NTR-NESDA, Rotterdam, SUSOD, TwinUK and WTCCC-CAD. For CPD we combined data from the same studies and the NL-BLC with a total of 31,266 subjects. Information on the meta-analysis studies for CPD and smoking initiation is provided in Supplementary Table 1, the Supplementary Note, and in the Methods online. After genomic control correction of each component study, we combined association data for ~2,500,000 imputed and genotyped autosomal SNPs with a fixed-effects additive meta-analysis using the inverse-variance method for CPD and smoking initiation. QQ-plots for CPD, excluding markers in the 15q25 region, displayed only modest inflation of the χ2-test statistic (λGC=1.02) (Supplementary Figure 1A). In addition to the 15q25 locus, SNPs at two loci, 19q13 and 7p14, were genome-wide significanct (GWS) for CPD (P<5·10−8) in the meta-analysis data. The QQ-plot for smoking initiation displayed weak inflation of the χ2-test statistic (λGC=1.03) and no GWS associations (Supplementary Figure 1B).
We selected 15 regions for smoking initiation totaling 277 SNPs, and 14 regions totaling 443 SNPs for CPD, for in silico replication in samples from the Tobacco and Genetics (TAG) and the Oxford-Glaxo Smith Kline (Ox-GSK) consortia (see accompanying papers12-13) (Supplementary Table 2). For CPD we included a region on chromosome 8p11 based on large number of SNPs exhibiting suggestive associations with CPD, strong candidacy of region genes (encoding nAChR subunits α6 and β3 (CHRNA6 and CHRNB3)), and prior suggestive evidence for association between SNPs within this region and ND3-4.
In addition to the 15q25 locus, three novel loci, 7p14, 8p11 and 19q13, were GWS for CPD after combining the results from the ENGAGE meta-analysis set with those of TAG and OX-GSK (Table 1, Fig 1, and Supplementary Table 2). No GWS associations for the selected smoking initiation regions were observed in the combined analysis of the meta-analysis and the in silico data (Supplementary Table 2).
Table 1.
Association of markers in 4 chromosomal regions with CPD. Results are given for the ENGAGE analysis (ENGAGE), the in-silico replication obtained by combining results from TAG and OX-GSK (in-silico), and the results of single-SNP assay replications in samples from Iceland, Australia, Denmark, Germany, and Spain (ISL-AUST-DEN-GER-SPA). Samples that were both in ENGAGE and either TAG or OX-GSK were removed before obtaining the combined in-silico results. Shown are the number of smokers (N), the effect allele and the other allele, the allele frequencies (Freq), the chromosome number and position, the estimated allelic effects on CPD and their standard errors in CPD (Effect and SE), the P value for the test of association (P), the P value for the test for heterogeneity in effect size, and an estimate of the proportion of total variation in study estimates that is due to heterogeneity (I2)
| Allele | ENGAGE (meta-analysis) (N=31,266) |
TAG and OX-GSK (In silico replication) (N=45,691) |
ISL-AUS-DEN-GER- SPA (direct genotyping) (N=9,040) |
Combined (N=85,997) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| SNP | Effect | Other | Freq | Chr | Position | Effect±SE | P | Effect±SE | P | Effect±SE | P | N | Effect±SE | P | Phet | I2 |
| rs1051730 | A | G | 0.339 | 15q25 | 76,681,394 | 0.84±0.07 | 2.1·10−33 | 0.78±0.06 | 5.6·10−38 | 76,972 | 0.80±0.05 | 2.4·10−69 | 0.035 | 32 | ||
| rs6474412 | T | C | 0.784 | 8p11 | 42,669,655 | 0.31±0.08 | 1.7·10−4 | 0.30±0.07 | 2.6·10−5 | 0.19±0.18 | 0.30 | 84,956 | 0.29±0.05 | 1.4·10−8 | 0.24 | 13 |
| rs13280604 | A | G | 0.784 | 8p11 | 42,678,743 | 0.31±0.08 | 1.2·10−4 | 0.30±0.07 | 2.7·10−5 | 76,670 | 0.31±0.05 | 1.3·10−8 | 0.24 | 14 | ||
| rs215614 | G | A | 0.356 | 7p14 | 32,313,860 | 0.38±0.07 | 2.4·10−8 | 0.17±0.06 | 3.6·10−3 | −0.15±0.16 | 0.35 | 86,259 | 0.22±0.04 | 2.1·10−7 | 0.018 | 34 |
| rs215605 | G | T | 0.357 | 7p14 | 32,303,490 | 0.39±0.07 | 1.7·10−8 | 0.17±0.06 | 3.5·10−3 | 77,012 | 0.26±0.04 | 5.4·10−9 | 0.12 | 22 | ||
| rs7937 | T | C | 0.560 | 19q13 | 45,994,546 | 0.34±0.07 | 2.2·10−7 | 0.19±0.06 | 1.1·10−3 | 0.19±0.14 | 0.17 | 86,319 | 0.24±0.04 | 2.4·10−9 | 0.45 | 1 |
| rs1801272 | A | T | 0.961 | 19q13 | 46,046,373 | 1.08±0.27 | 7.0·10−5 | 0.41±0.24 | 8.4·10−2 | 66,380 | 0.68±0.18 | 1.1·10−4 | 0.50 | 0 | ||
| rs4105144 | C | T | 0.704 | 19q13 | 46,050,464 | 0.59±0.10 | 1.2·10−9 | 0.31±0.08 | 5.8·10−5 | 0.27±0.15 | 0.069 | 83,317 | 0.39±0.06 | 2.2·10−12 | 0.51 | 0 |
| rs7260329 | G | A | 0.687 | 19q13 | 46,213,478 | 0.43±0.07 | 1.1·10−9 | 0.06±0.06 | 0.36 | 0.08±0.16 | 0.65 | 86,092 | 0.20±0.04 | 5.5·10−6 | 0.12 | 21 |
Figure 1.
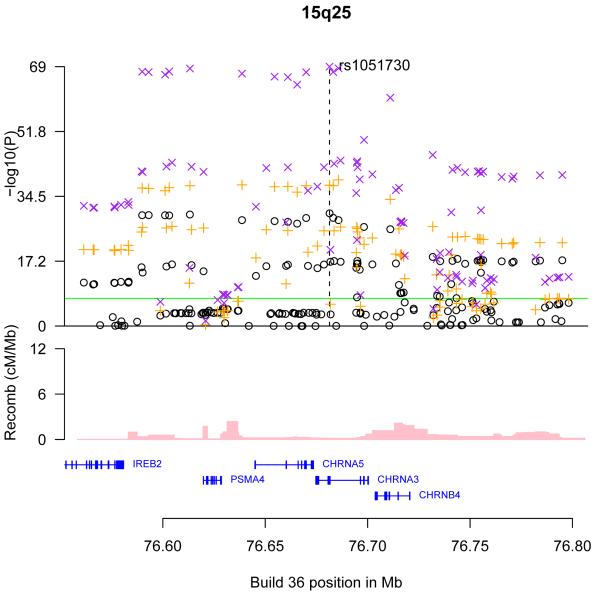
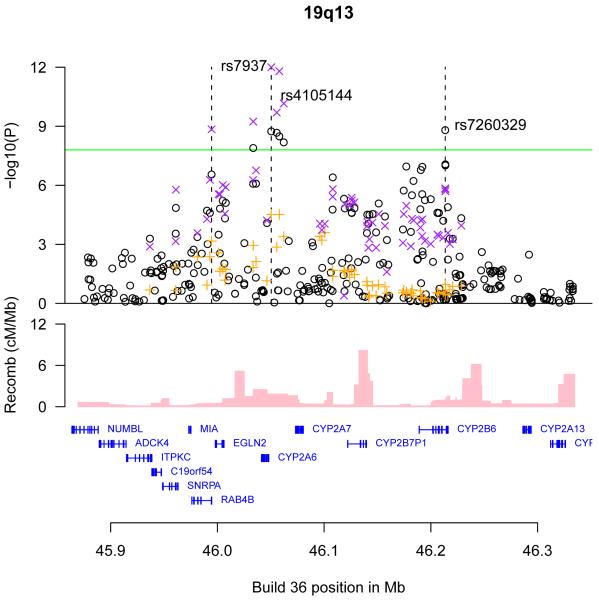
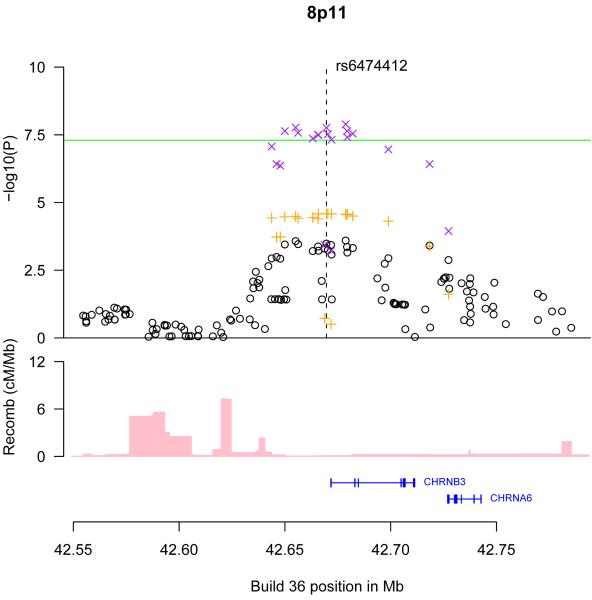
The genomic regions of association with CPD on chromosomes 15q25 (A), 19q13 (B), and 8p11 (C) and 7p14 (D). Shown are the −log10 association P values of SNPs in the region with CPD from the ENGAGE meta analysis (black circles), the in silico replication studies (orange crosses), and joint analysis of ENGAGE, TAG, and OX-GSK GWA data (magenta crosses), the SNP build 36 coordinates, the genes in the region and their exons (in blue) and recombination rates in centimorgans (cM) per megabase (Mb) (pink histogram).
For further confirmation of the CPD association signals at the 7p14, 8p11 and 19q13 loci, selected markers from these regions were genotyped in additional samples (n=9,040) from Iceland, Australia, Denmark, Germany and Spain (Table 1). Markers at 8p11 and the 19q13 loci had effects in the same directions, but not the marker on 7p14 (Table 1). After combining these data with ENGAGE results and the in silico replication the 8p11 and the 19q13 loci remained GWS but not the 7p14 locus (Table 1). The CPD association results for the SNPs in Table 1 for each study are presented in Supplementary Table 3 and Supplementary Figure 2.
Nominally significant heterogeneity in the strength of association with CPD was observed for rs1051730 at 15q25 (P=0.035, I2=32%) and rs215614 at 7p14 (P=0.018, I2=34%). There is little doubt that the 15q25 association with CPD is real. Therefore its heterogeneity must be due to differences between the study population such as different CPD information ascertainment, different types of cigarettes being used, different phenotypic and demographic ascertainment strategies or different genetic structures. The heterogeneity observed at 7p14 could be caused by a false positive finding or, if it is indeed a true positive, some combination of the “winner’s curse” and the same sort of differences driving the heterogeneity at 15q25.
Not surprisingly, the strongest associations observed with CPD in the combined analysis were with SNPs within the previously identified region on chromosome 15q25 (rs1051730-A, P= 2.4 10−69, effect size = 0.80 CPD) (Table 1, Figure 1). We searched for additional association signals in the 15q25 region, not accounted for by rs1051730, by performing linear regression using the rs1051730 allele count as a covariate in a subset of the ENGAGE samples (N=23,089). The residual signals were mostly tagged by two SNPs in relatively low LD, rs2869046-T (P=4.8·10−5, Effect=0.5 CPD) and rs2036534-T (P=9.1·10−5, Effect=0.5 CPD) (r2=0.080 and D’=0.65 in the HapMap CEU samples) (Supplementary Figure 3). These two SNPs are also in fairly weak LD with rs1051730 (r2=0.12, D’=0.49 and r2=0.18, D’=1.05 in the HapMap CEU samples for rs2869046 and rs2036534, respectively). These data suggest that, either, the three variants, rs2869046-T, rs2036534-T and rs1051730-A, represent independent association signals, or that a variant(s) captured by a combination of these variants remains to be identified. A SNP, rs578776, in LD with rs2036534 (r2=0.74, D’=0.95 in the HapMap CEU samples) was previously reported to associate with ND to an extent that could not be accounted for by rs105173014. As with rs2036534, rs578776 is in weak LD with rs1051730 (r2=0.21, D’=1.0 in the HapMap CEU samples). However, rs578776 is not correlated with rs2869046 (r2=0.0.038, D’=0.41 in the HapMap CEU samples) and thus does not explain the signal described here for rs2869046 at the 15q25 locus (Supplementary Figure 3).
The SNPs on chromosome 8p11 that reached genome–wide significance, rs6474412 (effect=0.29 CPD, P=1.4 10−8) and rs13280604 (effect=0.31 CPD, P=1.310−8), are in perfect LD in the HapMap CEU samples with a variant (rs13277254-T) that was previously highlighted as suggestively associated in a GWA study of ND using controls who had been exposed to smoking but had not developed ND (rs13277254-T, OR=1.19, P=6·10−5)2-3. The other SNPs in the region showing association with CPD (Figure 1) are all in strong LD with these SNPs. Rs6474412 is located about 2.1 kb from the 5′ end of the β3 nAChR subunit gene (CHRNB3), and belongs to a group of highly correlated SNPs, that includes two SNPs in exons of CHRNB3: a synonymous SNP (rs4593), and a non-synonymous SNP in CHRNB3 (rs4952)3. Although the CHRNB3 gene is implicated by the location of the associating SNPs, these markers could be tagging variation elsewhere within the LD block that also contains the α6 nAChR subunit gene (CHRNA6) (Figure 1).
Nine different nicotinic cholinergic receptor subunits (α2-α7, β2-β4) are expressed in the human brain, and they combine with each other in diverse patterns to form various types of functional pentameric receptors distinguished by subunit composition and sensitivity to nicotine15. Rodent studies have implicated CHRNA6 and CHRNB3 receptor subunits in nicotine-induced dopamine release16. Neither CHRNA6 nor CHRNB3 are expressed in lung tissue17.
The CPD associated markers on chromosome 19q13 are located in a region harboring CYP2A6, which encodes CYP2A6, an enzyme that plays a major role in the oxidation of nicotine in human liver microsomes, as well as several other genes and pseudogenes belonging to the CYP gene family (Figure 1). A number of sequence variants in or near CYP2A6 that reduce CYP2A6’s enzymatic activity have been identified18. For some of these variants, effects on smoking behavior have been suggested18. In the present study, the most significant association in the region was observed with rs4105144. This SNP is in LD with CYP2A6*2 (rs1801272) (r2=0.13 and D’=1.0 in the HapMap CEU samples) and the CYP2A6*2 reduced function allele is only found on the background of rs4105144-C which associates with reduced smoking quantity. Although the effect of rs4105144 (0.39±0.06) is smaller than that of rs1801272 (0.68±0.18)(Table 1), its association is more significant (lower P value) because of higher minor allele frequency. This suggests that rs4105144-C may be tagging many reduced function variants. The second most significant association in the region was with rs7937, in the untranslated 3′ end of the RAB4B gene, which is in LD with rs4105144 (r2=0.32, D’=0.82 in the HapMap CEU samples). The third most significant association in the region was with rs7260329 that is almost independent of rs4105144 (r2=0.0064, D’=0.091 in the HapMap CEU samples). Rs7260329 is an intronic SNP in CYP2B6, but its product converts nicotine to cotinine with about 10% of the catalytic activity of the CYP2A6 enzyme, and also metabolizes several drugs of abuse, and buproprion, an atypical antidepressant also used as a smoking cessation aid18. The CYP2B6 levels in the human brain are higher than those of CYP2A6 and are altered in smokers and alcoholics18-19.
One of the ENGAGE studies (GenMetS) had information on immune-reactive serum cotinine levels for a set of samples (N=485)20, that reflects the catalytic activity of the CYP2A and CYP2B gene products18. One of the SNPs associating with CPD at 19q13 (rs7937-G) showed nominally significant association with cotinine levels (effect=1.16,P=0.0031), while the markers at 8p11 and 7p14 did not associate with cotinine levels in this sample (P>0.26). However, two SNPs at 19q13 showed a stronger association with cotinine levels than rs7937, rs2233152-A and rs2287692-A (effect=1.92, P=0.00021), but did not associate strongly with CPD in the ENGAGE samples (P=0.013) and most of the markers showing the strongest association with CPD did not associate with cotinine levels.
We next assessed the SNPs from the novel regions associating with CPD for association with ND, defined as a score of four or higher on the Fagerstrom Test for Nicotine Dependence (FTND), or endorsement of at least three of the seven Diagnostic and Statistical Manual of Mental Disorders 4th edition (DSM-IV) criteria. Allele frequencies for 1,979 Icelandic (deCODE) and 835 Dutch (NTR-NESDA) ND cases were compared to 36,202 Icelandic and 611 Dutch population controls. SNPs on chromosome 8p11, and chromosome 7p14 associated nominally with ND, but none of the SNPs on chromosome 19q13 (Supplementary Table 4).
As we had previously found association of the 15q25 region with LC and PAD7, we directly genotyped selected markers from the 7p14, 8p11 and 19q13 regions for association with LC (2,019 cases and 40,509 controls) and PAD (2,855 cases, 40,424 controls) in samples of European ancestry. The LC data were also combined with summary-level data from the publicly available GWA dataset on LC (2,518 case and 1,921 controls) from the International Agency for Research on Cancer (IARC)10 (Table 2). Nominally significant associations with LC were observed for rs6474412-T on 8p11 (OR =1.12, 95%CI: 1.05-1.20, P=0.00060), rs215614-G on 7p14 (OR =1.07, 95%CI: 1.02-1.13, P=0.011), and rs7260329-G and rs4105144-C on 19q13 (OR =1.06, 95%CI: 1.00-1.12, P=0.041 and (OR =1.09, 95%CI: 1.00-1.18, P=0.040) (Table 2). As for the effect on CPD (Table 1) the effects of these variants on LC is substantially weaker than that of the 15q25 variants (OR =1.31, P=1.5 10−8)7,9-10(Table 2), warranting further analysis in additional sample sets. No significant associations with PAD were observed for the markers tested (Supplementary Table 5). The potential effect of rs7260329-G and rs4105144-C on LC, is interesting in light of the fact that CYP2A6 gene product activates procarcinogenic nitrosamines18.
Table 2.
Association of SNPs in 4 chromosomal regions with lung cancer in four populations. Shown are the number of cases and controls (N), the frequencies of the effect allele (see Table 1) in cases and controls, the odds ratio and 95% confidence intervals (OR and 95%CI), the P value for the test of association (P). The results for Peripheral Arterial Disease are shown in Supplementary Table 4.
| N | Freq | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Population | case | control | case | control | OR (95% CI) | P |
| rs6474412-T, chromosome 8p11 | ||||||
| Iceland | 839 | 36,606 | 0.784 | 0.770 | 1.08 (0.96, 1.22) | 0.19 |
| Denver | 192 | 856 | 0.805 | 0.790 | 1.09 (0.83, 1.44) | 0.53 |
| Spain | 351 | 1,195 | 0.819 | 0.764 | 1.40 (1.13, 1.72) | 0.0019 |
| Netherlands | 515 | 769 | 0.828 | 0.809 | 1.13 (0.92, 1.39) | 0.23 |
| IARCa | 2,506 | 1,914 | 0.763 | 0.778 | 1.10 (0.99, 1.22) | 0.072 |
| Combined | 4,403 | 41,340 | - | - | 1.12 (1.05, 1.20) | 0.00060 |
| rs215614-G, chromosome 7p14 | ||||||
| Iceland | 839 | 36,606 | 0.366 | 0.355 | 1.05 (0.95, 1.16) | 0.37 |
| Denver | 195 | 864 | 0.403 | 0.376 | 1.12 (0.89, 1.40) | 0.33 |
| Spain | 450 | 1,281 | 0.370 | 0.335 | 1.17 (1.00, 1.37) | 0.055 |
| Netherlands | 502 | 1,709 | 0.366 | 0.367 | 0.99 (0.86, 1.15) | 0.92 |
| IARCa | 2,513 | 1,917 | 0.344 | 0.365 | 1.09 (1.00, 1.19) | 0.057 |
| Combined | 4,499 | 42,377 | - | - | 1.07 (1.02, 1.13) | 0.011 |
| rs7937-T, chromosome 19q13 | ||||||
| Iceland | 836 | 36,552 | 0.555 | 0.549 | 1.03 (0.93, 1.13) | 0.60 |
| Denver | 193 | 864 | 0.567 | 0.595 | 0.89 (0.71, 1.12) | 0.32 |
| Spain | 453 | 1,330 | 0.532 | 0.512 | 1.08 (0.93, 1.26) | 0.31 |
| Netherlands | 528 | 1,629 | 0.552 | 0.548 | 1.02 (0.89, 1.17) | 0.80 |
| IARC | 2,518 | 1,921 | 0.559 | 0.580 | 1.09 (1.00, 1.18) | 0.048 |
| Combined | 4,528 | 42,296 | - | - | 1.05 (0.99, 1.10) | 0.080 |
| rs4105144-C, chromosome 19q13 | ||||||
| Iceland | 839 | 36,606 | 0.713 | 0.705 | 1.04 (0.90, 1.20) | 0.61 |
| Denver | 193 | 848 | 0.725 | 0.688 | 1.20 (0.94, 1.53) | 0.14 |
| Spain | 437 | 1,288 | 0.669 | 0.620 | 1.24 (1.06, 1.46) | 0.0085 |
| Netherlands | 513 | 1,665 | 0.638 | 0.640 | 0.99 (0.86, 1.15) | 0.93 |
| Combined | 1,982 | 40,407 | - | - | 1.09 (1.00, 1.18) | 0.040 |
| rs7260329-G, chromosome 19q13 | ||||||
| Iceland | 831 | 36,454 | 0.688 | 0.669 | 1.09 (0.98, 1.21) | 0.11 |
| Denver | 189 | 808 | 0.728 | 0.694 | 1.18 (0.92, 1.51) | 0.20 |
| Spain | 457 | 1,305 | 0.702 | 0.674 | 1.14 (0.97, 1.35) | 0.11 |
| Netherlands | 519 | 1,660 | 0.701 | 0.678 | 1.12 (0.96, 1.30) | 0.15 |
| IARC | 2,481 | 1,899 | 0.662 | 0.670 | 1.02 (0.95, 1.10) | 0.61 |
| Combined | 4,477 | 42,126 | - | - | 1.06 (1.00, 1.12) | 0.041 |
For IARC, results for rs6474412 and rs215614 were not available and here we report results for rs6474414 and rs215605, respectively, both of which are perfect surrogates in the HapMap CEU samples (r2=1).
The 13 regions that were selected from the ENGAGE meta-analysis of CPD but were not GWS, nominate a number of interesting functional candidate genes, including genes encoding for γ-aminobutiric acid receptor subunits (GABRA1 and GABRG2), and PDE1C, CDH13, and A2BP1, that were all highlighted in a GWA study of ND and smoking cessation21-22, and some of these genes may play a role in smoking behavior.
In conclusion, we have discovered sequence variants associating with smoking behavior within regions harboring nAChR genes (CHRNB3-CHRNA6, 8p11) and nicotine-metabolizing enzymes (CYP2A6 -CYP2B6, 19q13). The 8p11 association is reminiscent of that with chromosome 15q25; both regions contain nAChR genes, and the key variants associate with ND and LC, bringing up the question of whether the risk for LC is through the effect on smoking behavior, or involves increased vulnerability to the harmful effects of smoking as well7,23-27. However, the dissection of the causal pathway will be even more difficult in case of these new variants, as their effects on both CPD and LC are smaller, and further studies are warranted.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the participants in the genetic studies whose contributions made this work possible. This work was supported in part by NIH (R01-DA017932) and the European Commission’s Sixth Framework Program, Integrated Project GENADDICT (LSHM-CT-2004-005166). The ENGAGE smoking consortium was formed through a component of the Integrated Project ENGAGE, supported by the European Commission’s Seventh Framework Program, grant agreement HEALTH-F4-2007- 201413. ENGAGE projects have benefited from the SIMBioMS platform28 that has greatly facilitated data exchange and annotation. Additional acknowledgements are listed in the Supplementary Note online.
Footnotes
COMPETING INTERESTS STATEMENT Authors whose affiliations are listed as deCODE genetics are employees of deCODE genetics, a biotechnology company.
URLs. ENGAGE Consortium (www.euengage.org).
References
- 1.WHO Report on the Global Tobacco Epidemic, 2008. 2008.
- 2.Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rose R, Broms U, Korhonen T, Dick D, Kaprio J. Genetics of Smoking Behavior. In: Kim Y, editor. Handbook of Behavior Genetics. Springer Science+Business Media; 2009. [Google Scholar]
- 5.Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 6.Koopmans JR, Slutske WS, Heath AC, Neale MC, Boomsma DI. The genetics of smoking initiation and quantity smoked in Dutch adolescent and young adult twins. Behav Genet. 1999;29:383–93. doi: 10.1023/a:1021618719735. [DOI] [PubMed] [Google Scholar]
- 7.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berrettini W, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 11.Pillai SG, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furberg H. TAG study. Nat Genet. 2010 [Google Scholar]
- 13.Marchini J. OX-GSK Study. Nat Genet. 2010 [Google Scholar]
- 14.Stevens VL, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17:3517–25. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins AC, Salminen O, Marks MJ, Whiteaker P, Grady SR. The road to discovery of neuronal nicotinic cholinergic receptor subtypes. Handb Exp Pharmacol. 2009:85–112. doi: 10.1007/978-3-540-69248-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mineur YS, Picciotto MR. Genetics of nicotinic acetylcholine receptors: Relevance to nicotine addiction. Biochem Pharmacol. 2008;75:323–33. doi: 10.1016/j.bcp.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West KA, et al. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest. 2003;111:81–90. doi: 10.1172/JCI16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray R, Tyndale RF, Lerman C. Nicotine dependence pharmacogenetics: role of genetic variation in nicotine-metabolizing enzymes. J Neurogenet. 2009;23:252–61. doi: 10.1080/01677060802572887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miksys S, Lerman C, Shields PG, Mash DC, Tyndale RF. Smoking, alcoholism and genetic polymorphisms alter CYP2B6 levels in human brain. Neuropharmacology. 2003;45:122–32. doi: 10.1016/s0028-3908(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 20.Keskitalo K, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet. 2009;18:4007–12. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhl GR, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65:683–93. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhl GR, et al. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chanock SJ, Hunter DJ. Genomics: when the smoke clears. Nature. 2008;452:537–8. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 24.Thorgeirsson TE, Stefansson K. Genetics of smoking behavior and its consequences: the role of nicotinic acetylcholine receptors. Biol Psychiatry. 2008;64:919–21. doi: 10.1016/j.biopsych.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–6. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lips EH, et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int J Epidemiol. 2009 doi: 10.1093/ije/dyp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorgeirsson TE, Stefansson K. Commentary: Gene-environment interactions and smoking-related cancers. Int J Epidemiol. 2010 doi: 10.1093/ije/dyp385. [DOI] [PubMed] [Google Scholar]
- 28.Krestyaninova M, et al. A System for Information Management in BioMedical Studies--SIMBioMS. Bioinformatics. 2009;25:2768–9. doi: 10.1093/bioinformatics/btp420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutyavin IV, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice JA. Mathematical Statistics and Data Analysis. Wadsworth Inc.; Belmont, CA: 1995. [Google Scholar]
- 34.Higgins J, Thompson S. Quantifying heterogeneity in a meta-analysis. Statistics in Medinine. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 35.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.