Abstract
Huntington's disease (HD) is caused by an expanded CAG tract in the Interesting transcript 15 (IT15) gene encoding the 350 kDa huntingtin protein. Cellular stresses can trigger the release of huntingtin from the endoplasmic reticulum, allowing huntingtin nuclear entry. Here, we show that endogenous, full-length huntingtin localizes to nuclear cofilin–actin rods during stress and is required for the proper stress response involving actin remodeling. Mutant huntingtin induces a dominant, persistent nuclear rod phenotype similar to that described in Alzheimer's disease for cytoplasmic cofilin–actin rods. Using live cell temporal studies, we show that this stress response is similarly impaired when mutant huntingtin is present, or when normal huntingtin levels are reduced. In clinical lymphocyte samples from HD patients, we have quantitatively detected cross-linked complexes of actin and cofilin with complex formation varying in correlation with disease progression. By live cell fluorescence lifetime imaging measurement–Förster resonant energy transfer studies and western blot assays, we quantitatively observed that stress-activated tissue transglutaminase 2 (TG2) is responsible for the actin–cofilin covalent cross-linking observed in HD. These data support a direct role for huntingtin in nuclear actin re-organization, and describe a new pathogenic mechanism for aberrant TG2 enzymatic hyperactivity in neurodegenerative diseases.
INTRODUCTION
In 1993, an expanded CAG tract in the interesting transcript 15 (IT15) gene encoding the polyglutamine-expanded huntingtin protein was found to be the cause of Huntington's disease (HD): a progressive, neurodegenerative disorder with typical late-age onset affecting as many as 1 in 4000 individuals (1,2). This 350 kDa protein contains little homology to other known proteins, but does share a repetitive peptide structure termed HEAT-repeats, found in many large scaffolding proteins (3). HEATs are seen to allosterically regulate the shape of proteins and allow multiple conformations and protein–protein interactions controlled by elastic and tensor forces, mediated by tethers to cell structural components or organelles (4). Normal huntingtin functions have been described in transcriptional regulation (5), epigenetic control (6,7), vesicle trafficking (8), endoplasmic reticulum (ER)–stress response (9) and neurogenesis during development (10). Previously, we defined the amino-terminal 17 amino acids of huntingtin as a critical ER tether that mediates its release from the ER upon stress (9). This 17-amino-acid signal in huntingtin, or N17, is a modulator of mutant huntingtin toxicity in many systems (9,11) and the target of post-translational modifications (12–14). This signal is also found in all mouse models of HD, and is protective to the effects of polyglutamine expansion if phospho-mimicked at serines 13 and 16 in the bacterial artificial chromosome HD mouse model (13). From this work, we focused our cell biological studies on the effects of stress on the localization of the normal, full-length, huntingtin protein that is endogenous to mouse striatal-derived cells (15), without any over-expression of huntingtin fragments. Our goal was to observe normal huntingtin cell biology under transient stresses, similar to those types of stresses associated with normal human aging.
Heat shock stress recapitulates a strong transient stress event that causes a global unfolding of proteins, or proteotoxic stress, a temporary drop in adenosine triphosphate (ATP) levels and spike in calcium signaling via the ER without having to use small molecules (16,17). The heat shock stress response is well characterized, leading to the transient activation of several chaperones, transcription factors and components of the unfolded protein response (UPR) at the ER (18). Some of these responses involve classic transcriptional activation, but other responses are at the post-translational level only, to rapidly respond to a potentially toxic cell stress event. The cell stress response has been of intense interest to the neurodegeneration community. Neuronal cells have a decreased ability to deal with oxidative and cellular stress during the aging process which has been tightly linked to neurodegeneration in HD, Alzheimer's disease (AD) and Parkinson's disease (PD) (19–21). Upon induction of heat shock stress, we observed the nuclear localization of endogenous huntingtin, similar to previous studies using cold shock or UPR stresses with huntingtin fragments containing N17 (9). However, we also observed the localization of huntingtin within the nucleus to numerous straight rod-like structures of 3–5 µm, which disappeared upon the relief of stress. Similar structures were reported as rods of cofilin and super-twisted F-actin in neurites, observed in models of AD (22,23). In the nucleus, during the heat shock response, cofilin, an actin binding protein normally required for actin treadmilling, saturates actin causing bundles of actin filaments. These bundles are referred to as ‘cofilin rods’ (24). Cofilin rods can form in the cytoplasm during certain cell stresses and stop a proportion of actin from treadmilling. This liberates ATP normally involved in this process so it can be used elsewhere in the cell during stress (25). We observed a dominant loss of function in cells expressing full-length mutant huntingtin's ability to respond to and recover from stress through the cofilin–actin pathway.
In order to determine the relevance of these observations to HD, we performed western blot analysis of cofilin on the lymphocytes of HD patient blood samples. Patients ranged from premanifest through the four clinically defined Shoulson–Fahn stages of disease. From this cohort, we detected the presence of a cross-linked cofilin–actin band by western blot in which higher degrees of cross-linking quantifiably correlated to disease stage. These correlations were apparent both across the population of each disease stage and also longitudinally within individuals.
Tissue transglutaminase 2 (TG2) is a calcium sensitive multifunctional enzyme. TG2 has guanosine triphosphate signaling activity as well as transamidating activity. TG2 can catalyze the formation of a covalent bond between polyamine, or the ɛ-amino group of polypeptide-bound lysine, and the γ-carboxamide group of polypeptide bound glutamine to form polyamine or isopeptide bonds, respectively (26). TG2 activity is known to be elevated in HD brains, and in HD lymphocytes (27,28). Others have shown that transglutaminase can cross-link cofilin and actin in vitro (29). We therefore hypothesized that the higher order cross-link of cofilin and actin observed in the lymphocyte samples may be due to elevated TG2 activity. Using western blot assays, we show that cofilin–actin cross-linking, similar to what is observed in HD lymphocytes, was dependent not only on TG2 protein levels, but also stress, consistent with the requirement for activated TG2.
At the single cell level, we used biophotonic fluorescence lifetime imaging measurement (FLIM) technology (30) to measure Förster resonant energy transfer (FRET) between enhanced yellow fluorescent protein (eYFP)-TG2 and mCerulean–cofilin, at rods in live cells undergoing stress, to determine whether the proteins directly (<8 nm) interact in vivo. We show that TG2 interacts in a stress-dependent manner with both nuclear and cytoplasmic cofilin rods, potentially mediating the cofilin–actin cross-link and the rod persistent phenotype observed in AD and HD. Thus, our data lead us to hypothesize a new model for TG2 hyperactivity in HD. This model describes defective actin turnover during stress due to the polyglutamine expansion in huntingtin causing ER disruption, increased calcium levels and activation of TG2, resulting in aberrant cofilin–actin covalent cross-links. This model predicts that actin turnover may be universally affected in all cell types in HD, but would be most critical to highly dendritic neurons (31), such as the medium-sized spiny neurons (MSNs) which are predominantly affected in HD. Our data also show that there may be quantifiable HD phenotypes in peripheral cells that may be developed into biomarkers for HD.
RESULTS
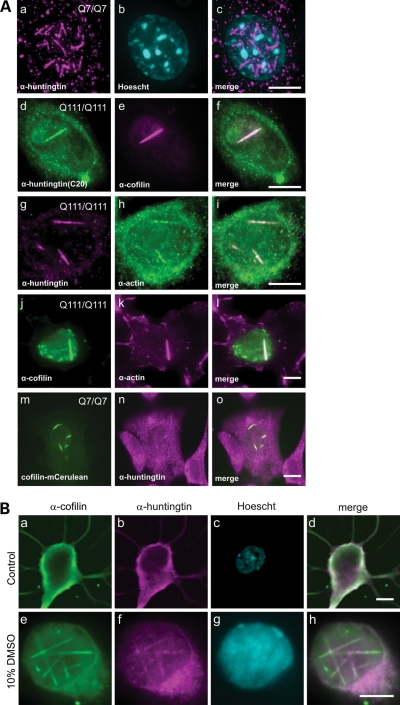
We have previously shown that huntingtin localizes to the ER under steady state growth conditions, and that it can be released from the ER and enter the nucleus under conditions of temperature or UPR stress (9). In order to determine whether the huntingtin protein had any direct role in cellular stress response, we observed a mouse striatal neuron-derived cell line (STHdh) (32) expressing full-length endogenous levels of either wild-type (STHdhQ7/Q7), or mutant (STHdhQ111/Q111) huntingtin, under typical heat shock conditions of 42°C for 60 min. Huntingtin was then visualized by a fully validated huntingtin monoclonal antibody, MAb2166 (33), by immunofluorescence (Fig. 1). Within the nucleus, we could visualize huntingtin localized to numerous 3–5 µm rod structures in STHdhQ7/Q7 (Fig. 1Aa–c, m–o), and to phenotypically different, fewer and longer rod structures in STHdhQ111/Q111 (Fig. 1Ad–l). These nuclear rods could be visualized with the amino-terminal epitope antibody MAb2166, as well as the carboxy-terminal epitope antibody C20 (Fig. 1Ad–f), suggesting that full-length huntingtin localizes to these structures. Nuclear cofilin–actin rods have been observed by others in the past in response to various cell stress agents, including heat shock (33–35). To test if these huntingtin-positive nuclear structures may be cofilin–actin rods, we performed co-immunofluorescence with antibodies against huntingtin and cofilin (Fig. 1Ad–f), huntingtin and actin (Fig. 1Ag–i) and cofilin and actin (Fig. 1Aj–l). From these data, we concluded that the huntingtin-positive nuclear rods induced by heat shock are cofilin–actin rods. This was confirmed directly by co-imaging and co-localization of the mCerulean–cofilin fusion protein over-expressed in STHdh cells and immunofluorescence to visualize endogenous huntingtin using MAb2166 (Fig. 1Am–o). To verify that these observations were not artifactual to immortalized tissue culture cells, or to heat shock, we imaged endogenous cofilin and huntingtin in primary mouse hippocampal neurons, with and without 10% dimethyl sulfoxide (DMSO) treatment (35) to induce cofilin rods (Fig. 1Be–h), with similar localization of endogenous wild-type mouse huntingtin to cofilin rods. In summary, upon heat shock or DMSO induced cell stress, huntingtin protein entered the nucleus and localized to cofilin–actin rods. We did not observe huntingtin at cytoplasmic cofilin–actin rods under any cell stress conditions tested in any cell type, indicating that this is likely a nuclear function of huntingtin during stress.
Figure 1.
Full-length, endogenous huntingtin protein is a component of nuclear cofilin–actin stress rods. (A) Immunofluorescence in mouse STHdhQ7/Q7 or STHdh Q111/Q111 striatal-derived cell lines, in cells heat-shocked at 42°C for 60 min. Secondary antibodies were either Alexa 488 (green) or Alexa 595 (magenta) labeled. (a–c) Huntingtin monoclonal antibody MAb2166, co-stained with Hoechst DNA dye (cyan). (d–f) Huntingtin monoclonal antibody C20, co-stained with cofilin monoclonal MAb22. (g–i) Huntingtin monoclonal MAb2166 co-stained with an antibody against actin. (j–l) Co-staining with antibodies against cofilin and actin. (m–o) Huntingtin monoclonal antibody MAb2166, in cells expressing an mCerulean–cofilin fusion protein. (B) Immunofluorescence on 6-day-old primary hippocampal neurons treated with 10% DMSO for 90 min to induce rod formation. Untreated cell (a–d) stained with affinity purified rabbit IgG to chick ADF (a), huntingtin monoclonal antibody MAb2166 (b) and co-stained with Hoechst DNA dye. Secondary antibodies were either Alexa 488 (green) or Alexa 595 (magenta) labeled. DMSO treated cell (e–h). Merged overlapping signal is pseudo-colored white. Scale bar is 10 µm.
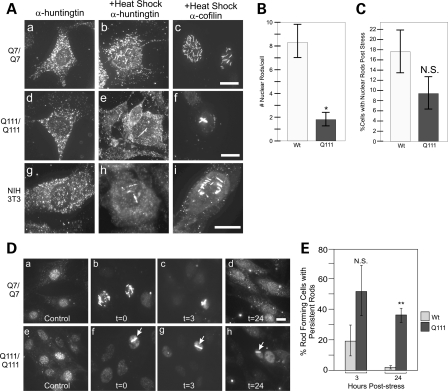
In order to quantify the effect of the polyglutamine expansion on nuclear cofilin–actin rods, we quantitatively compared the rod formation in STHdhQ7/Q7 and STHdhQ111/Q111 cell lines during stress. STHdhQ7/Q7 cells formed numerous (8 ± 5) (Fig. 2Aa–c and B), short 3–5 µm rods upon heat shock (33), whereas STHdhQ111/Q111 cells formed fewer (2 ± 1) and longer rods that often spanned the nucleus to 10 µm (Fig. 2Ad–f and B), phenotypically similar to persistent rods previously defined by others in AD models (23). The percentage of cells that formed rods during stress was not statistically different at the 60 min time point (Fig. 2C). We also observed endogenous huntingtin protein in a heat-shocked mouse fibroblast cell line, NIH 3T3 (Fig. 2Ag–i), and found that huntingtin localized to cofilin rods phenotypically similar to STHdhQ7/Q7 cells. Thus, huntingtin localization to nuclear cofilin rods upon stress is not limited to neuron-derived cells. This is consistent with endogenous huntingtin protein expression in all cell types outside of the brain (1).
Figure 2.
Mutant huntingtin protein affects nuclear cofilin rod formation and induces persistence of cofilin rods. Immunofluorescence with either huntingtin or cofilin antibodies in either STHdhQ7/Q7 or STHdhQ111/Q111 cell lines. (A) Mutant huntingtin protein affects the number and size of nuclear cofilin stress rods upon 60 min heat shock. (a–c) Huntingtin or cofilin immunofluorescence before (a), or after heat shock (b–c) in STHdhQ7/Q7 cells. (d–f) Huntingtin or cofilin immunofluorescence before (d), or after heat shock (e–f) in STHdhQ111/Q111 cells. (g–i) Huntingtin or cofilin immunofluorescence before (g), or after heat shock (h–i) in NIH 3T3 mouse fibroblast cell line. (B). Comparison of number of nuclear rods per cell immediately after 60 min heat shock in STHdhQ111/Q111 or STHdhQ7/Q7 lines. Rod forming cells were imaged as a z-stack and the mass projection was used to count number of rods per nucleus in each cell type. N = 3, n= 10 per replicate, total n= 30. *P-value of n< 0.001. (C) Comparison of number of cells with nuclear rods immediately after 60 min heat shock. Cells were fixed immediately after 60 min heat shock and 20 sequential images at a random area in the dish were taken at 63×. Number of cells with nuclear rods were counted and expressed as a percentage. N = 3, P-value of 0.210. N.S., not statistically significant. (D) Immunofluorescence staining against cofilin in either STHdhQ7/Q7 (a–d) or STHdhQ111/Q111 (e–h) cell lines. Images shown directly after heat shock stress (b, f) or after 3 h (c, g) or 24 h (d, h) recovery at optimal conditions. Control, no stress, images shown (a, e). (E) Quantification of percent rod forming cells with persistent nuclear rods after 3 and 24 h at 33°C was performed as described for (C). Percent cells with persistent rods expressed as a percentage of initial rod forming cells immediately after heat shock stress (t = 0) for each trial. N = 3; *P-value of 0.163; N.S., not statistically significant. **P-value of 0.003. Persistent rod phenotype highlighted with white arrows. All scale bars are 10 µm.
In AD, cytoplasmic cofilin–actin rods are shown to persist beyond the stress when cells are returned to normal conditions (23). In order to assay whether mutant huntingtin rods were persistent or not, we heat-shocked cells, and fixed them immediately following heat shock (t= 0), or placed them back at 33°C and allowed them to recover for 3 or 24 h (t= 3 or t= 24). This was followed by fixation and immunofluorescence with anti-cofilin MAb22 (Fig. 2D). A minimum of 20 sequential frames at ×63 magnification were imaged and the number of rod forming cells were counted. Recovery values were normalized to the number of rod forming cells observed at t= 0. After 24 h recovery, the wild-type cell line recovered almost completely with only 2% of rods persisting, whereas mutant cells still had a 36% persistence rate (Fig. 2E). Strikingly, 100% of nuclear rods that persist beyond 3 h (in any cell type) have a persistent rod phenotype (Fig. 2Dc, g, h, see arrows).
For subsequent experiments, based on our quantitative phenotyping of the different cell lines, we generated a phenotypic threshold defining a cell with the wild-type rod phenotype as having >4 rods/nucleus (rod length spanning 1–6 µm) and a cell that has a persistent rod phenotype to have between 1 and 4 rods/nucleus with at least 1 rod being >4 µm. Using these parameters, we quantified the percentage of rod forming cells with persistent rod phenotypes immediately following heat shock. We found that only 27.6 ± 6.1% of rod forming STHdhQ7/Q7 cells form nuclear rods with a persistent phenotype compared with 83 ± 5.6% of rod forming STHdhQ111/Q111 cells. Collectively, these results indicate that huntingtin has a normal universal role in the nucleus with respect to nuclear rod formation and that this role is disrupted by a polyglutamine expansion. In addition, the presence of mutant huntingtin causes an increase in the persistence of nuclear cofilin–actin rods once the cell has been allowed to recover from stress, thus directly implying huntingtin dysfunction in this stress response.
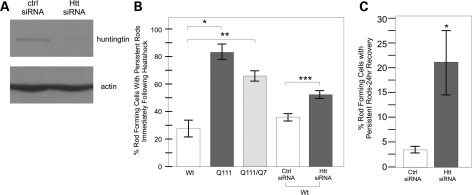
In order to consistently assay cofilin rod formation, we made a stable STHdhQ7/Q7 cell line expressing the mCerulean–cofilin fusion protein. To assess any essential role of wild-type huntingtin in this cell stress response, we treated STHdh mCerulean–cofilin cell lines with specific small interfering ribonucleic acid (siRNA) to knockdown levels of huntingtin. After siRNA knockdown of huntingtin was confirmed by western blot (Fig. 3A), we compared rod formation in STHdh mCerulean–cofilin cells treated with huntingtin siRNA or a control siRNA during and after heat shock stress by visualizing mCerulean–cofilin (Fig. 3B and C). We did not observe any difference in percentage of cells forming rods upon heat shock at 60 min whether huntingtin levels were reduced or not (Fig. 3B). After 1h stress, 35.5 ± 2.3% of rod forming cells had a persistent rod phenotype when scramble siRNA was transfected versus 51.6 ± 4.3% when huntingtin siRNA was transfected (Fig. 3B). After 24 h recovery, we observed a significant increase in persistent rods after huntingtin knockdown; 3.5 ± 0.38% of rods persisted in cells transfected with scrambled siRNA versus 21.4 ± 6.5% when huntingtin siRNA was transfected (Fig. 3C). These results show that when wild-type huntingtin is knocked down, the cofilin–actin rod phenotype and defects are similar to cells expressing mutant huntingtin (Fig. 3B), indicating a loss of function with polyglutamine expansion in huntingtin.
Figure 3.
Huntingtin protein is required for proper cofilin nuclear rod formation and clearance following cell stress. (A) Western blot against huntingtin protein after control siRNA or huntingtin-specific siRNA was expressed in wild-type huntingtin STHdhQ7/Q7 cells for 72 h. Anti-actin shown as a loading control. (B) Comparison of percent nuclear rod forming cells with persistent rod phenotype in heat-shocked cells. Comparison between STHdhQ7/Q7, STHdhQ111/Q111 and STHdhQ7/Q111 cell lines as well as in STHdhQ7/Q7 cells following siRNA treatment. Experiment performed as described previously. N = 3, *P-value of <0.003, **P-value of 0.028. (C) Comparison of percent of rod forming cells with nuclear persistent rods after 72 h siRNA treatment, heat shock and 24 h recovery. Quantification performed as previously described. N = 3, *P-value of 0.046.
We then wanted to ascertain whether the effect of mutant huntingtin presence was dominant, an important factor when considering the genetics of HD. Immortalized striatal neurons from a heterozygous (STHdhQ111/Q7) mouse were heat-shocked, and we performed similar immunofluorescence for cofilin. At the 1h time point, the heterozygous cells formed rods with a similar phenotype to mutant cells: 69 ± 3.3% (SE) of rod forming cells had a persistent rod phenotype (Fig. 3B). This indicates that the changes in rod formation can happen even when wild-type huntingtin is present, and thus the persistent rod phenotype is dominant.
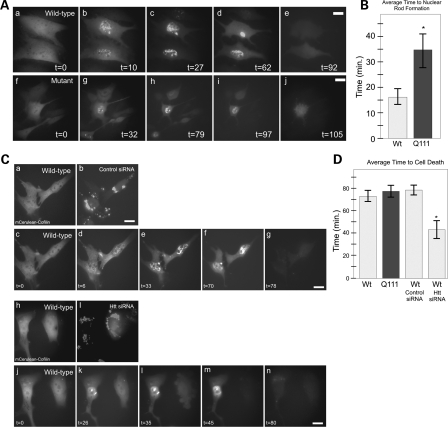
In order to visualize the dynamics of cofilin rod formation during cell stress, we used STHdhQ7/Q7 and STHdhQ111/Q111 cell lines stably expressing mCerulean–cofilin fusion protein, and observed live cells over time during heat shock stress by fluorescence microscopy using a heated stage (Supplementary Material, Videos S1 and S2). Cells were imaged every 60 s during heat shock. These temporal experiments were repeated multiple times and parameters of rod formation, rod persistence time, rod length and time to cell death were quantified and compared (Supplementary Material, Table S1A). Typical results are presented in Figure 4. These experiments revealed that the average time before nuclear cofilin rods was observed during cell stress were significantly longer in STHdhQ111/Q111 (35 min) versus STHdhQ7/Q7 (16 min) cell lines (Fig. 4A and B) (Supplementary Material, Videos S1 and S2). A detailed quantitative comparison of the rod phenotypes between the two cell lines demonstrated that mutant huntingtin cell lines were significantly delayed in rod formation during stress, and that rods were typically fewer and longer, with a persistent phenotype (Supplementary Material, Table S1A). This is consistent with our earlier results in fixed cell lines expressing only endogenous cofilin. In contrast to fixed cells, live cell temporal observations demonstrate that fewer STHdhQ111/Q111 cells form rods during stress in the presence of mutant huntingtin. These data highlight the importance of using live cell imaging for these observations.
Figure 4.
Huntingtin is required for normal cell heat shock stress response and mutant huntingtin affects the rate of the nuclear cofilin rod stress response. Temporal imaging in live STHdhQ7/Q7 or STHdhQ111/Q111 cells stably expressing mCerulean–cofilin fusion protein. (A) mCerulean–cofilin imaged over time showing nuclear cofilin rod formation in both wild-type and mutant cell lines at 10 and 32 min, respectively (b versus g), following panels show length of times rods exist and time of clearance during maintained heat shock at 42°C (c–e and h–j). (B) Comparison graph of average time to nuclear rod formation for stable mCerulean–cofilin wild-type (N = 15) and mutant (N = 9) STHdh cells during live cell imaging experiments. *P-value of 0.007. (C) Temporal imaging in live STHdhQ7/Q7 cells stably expressing mCerulean–cofilin. Cells were co-transfected with control siRNA or huntingtin-specific siRNA and Block-iT™ Alexa Fluor® red for 72 h prior to experiments. Single-cell visualization of control (a–b) or huntingtin siRNA (h–i) transfection with labeled Block-iT™Alexa Fluor® red. Visualization of STHdhQ7/Q7 cells with mCerulean–cofilin during heat shock, treated with control siRNA (c–g). STHdhQ7/Q7 cells with cofilin–mCerulean during heat shock, treated with siRNA to huntingtin (j–n). (D) Comparison graph of average time to cell death in mCerulean–cofilin stable STHdhQ7/Q7 (N = 13) or STHdhQ111/Q111 (N = 24) cells imaged live and STHdhQ7/Q7 mCerulean–cofilin cells treated with control siRNA (N = 5) or huntingtin siRNA (N = 6) imaged live during heat shock. *P-value of 0.003. Scale bars are 10 µm.
We used the same assay to track huntingtin siRNA effects on a cell-by-cell basis using a siRNA transfection fluorescent marker indicator (Block-it™, Alexa Fluor Red) in our mCerulean–cofilin stable STHdhQ7/Q7 cell line (Fig. 4Ca–b and h–i). When cells were transfected with huntingtin siRNA, the cells had a delayed rod formation response upon stress, and a persistent rod phenotype (Fig. 4Ck–m) mimicking what occurred in our STHdhQ111/Q111 cofilin stable cells. There was also a significantly faster cell death due to stress after siRNA treatment (44 min versus 79 min for the scrambled control) (Fig. 4D; Supplementary Material, Table S1B and Videos 3 and 4). Thus, huntingtin has a critical normal function in cell heat shock stress response, and the mutant huntingtin protein or knockdown of normal huntingtin protein appears to delay the onset of this stress response and affect the normal recovery from stress.
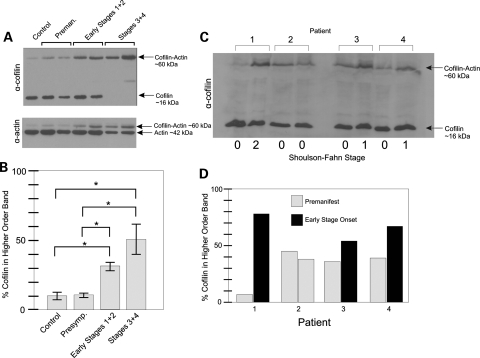
Under normal growth conditions, cofilin has an essential function in actin treadmilling, or turnover from the monomeric soluble state to the filamentous fibers. This function is controlled by the reversible phosphorylation of cofilin by LIM kinase (36). During stress, cofilin becomes hyper-dephosphorylated. Dephosphorylated cofilin has a strong affinity for adenosine diphosphate actin and saturates F-actin, forming rods or bundles that sequester most of the cofilin and thus slow turnover of the remaining F-actin sparing ATP for other cellular processes (24,25). LIM is targeted by Parkin in PD (37), and cofilin levels are highly variant in PD lymphocytes (38). Therefore, we western blotted cofilin in protein samples from HD patient lymphocyte samples obtained from gradient density centrifugation-separated pellets from blood. These samples were collected from patients at the four clinically defined (Shoulson–Fahn) stages of HD, including pre-manifest patients and age-matched controls, for a total of 39 individuals. We performed western blots using monoclonal antibodies raised against cofilin protein (Fig. 5A). We observed a higher order cofilin complex that was enhanced with advancing stages of HD. The size of the higher order band was consistent with the additive molecular weights of actin and cofilin proteins, which have been shown by others to be capable of stably cross-linking in vitro (39). Blotting for actin revealed a band migrating at the same size in these samples (Fig. 5A). Therefore, this complex contained cofilin and actin, and was stable despite sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), boiling and 1% dithiothreitol treatment, indicating a covalently linked complex. When quantitatively comparing the ratios of cofilin trapped in the cofilin–actin complex to the free cofilin (higher order band to lower order band) by pixel density analysis, we noted that more cofilin was present in the higher order complex as the stages of HD advanced. This effect was statistically significant between control or pre-manifest individuals and early or late stages of HD (Fig. 5B). Thus, the degree of cross-linked cofilin–actin complex from patient lymphocytes could be tracked from pre-manifest patients to patients with advancing stages of HD.
Figure 5.
White blood cell populations from HD patients show a cross linked cofilin–actin complex on a western blot which increases with clinical onset and severity of disease. Western blots on the protein extracted from blood buffy coat histopaque-treated pellets from HD patients at different clinically defined stages of HD (the Shoulson–Fahn method). Western blot was done using MAb22 cofilin or anti-actin antibodies. (A) Typical comparison of samples between age-matched controls and either pre-symptomatic, early stage 1 and 2 patients and late stage 3 and 4 patients. Mobility of either cofilin or actin is indicated by black arrows. (B) Quantification of percent cofilin signal in higher order cofilin–actin band from western blots performed on control and HD patient blood samples. Analysis done using pixel intensity analysis using NIH Image J. Control (N = 7), pre-symptomatic (N = 11), early stages 1 + 2 (N = 9), late stages 3 + 4 (N = 12). *P-value < 0.05. (C) Western blot of cofilin in longitudinal blood samples (prepared as described previously) from four HD patients, three of whom had progressed in HD from pre-symptomatic to early HD between samplings. (D) Graph shows percent cofilin signal in higher order cofilin–actin band in western blot determined by pixel intensity analysis using NIH Image J.
We additionally obtained longitudinal primary lymphocyte samples from four HD patients, three of whom had progressed from pre-manifest to early HD, while one patient remained pre-manifest on both samplings. For all three individuals who had progressed to early HD, we observed an increase in trapped cofilin–actin complexes (Fig. 5C and D). No increase in cofilin–actin complex was observed in the pre-manifest individual (Fig. 5D, patient #2) consistent with the population studies, and the greatest increase in ratio of cofilin–actin complex to free cofilin was observed in a patient who had progressed from pre-manifest to a later stage of HD (Fig. 5D, patient #1). The two sets of data from different patient populations, and from individual patients over time similarly demonstrated that trapped cofilin–actin complexes increase with disease onset and increasing severity of HD. This raises the question as to what is actually mediating this cross-linkage and what connection there may be between cofilin and actin cross-links in lymphocytes and cofilin–actin rods in model systems.
In the past, others have observed the ability of cofilin and actin to be covalently cross-linked in vitro upon addition of transglutaminase (29). Tissue transglutaminase (TG2) is an intracellular protein with many functions, but is known to catalyze the formation of a covalent bond between polypeptide-bound lysine and polypeptide-bound glutamine (40). Several lines of evidence prompted us to examine TG2 activity in our system. First, TG2 activity and expression are highly elevated in neurodegenerative diseases, including HD in the brain (27), cerebrospinal fluid and plasma (41), as well as in AD (42). For example, TG2 has been shown to co-localize with pathological lesions in AD brains (43) and isopeptide cross-links have been detected in paired helical Tau filaments (44). Of relevance to this study, TG2 activity has been shown to be increased in HD patient lymphocytes, in a CAG length-dependent manner (28). Furthermore, a TG2 knockout mouse crossed with an HD model mouse results in reduced neuronal death and increased lifespan (45). Regular polyglutamine (46) as well as expanded polyglutamine (47) have been shown to be TG2 substrates and TG2 is mis-regulated in HD; however, there is evidence showing that TG2, although involved in HD progression, does not cross-link or co-localize to inclusions of mutant huntingtin (48). Consistent with this, there is evidence showing that in an HD mouse model TG2 is involved in HD progression in an aggregate-independent manner (49). Recently, TG2 has been shown to modulate transcriptional changes in HD indicating aberrant nuclear activity apart from aggregation (50). Given these data, we tested whether TG2 could be responsible for the cofilin–actin cross-links observed in clinical HD lymphocyte samples.
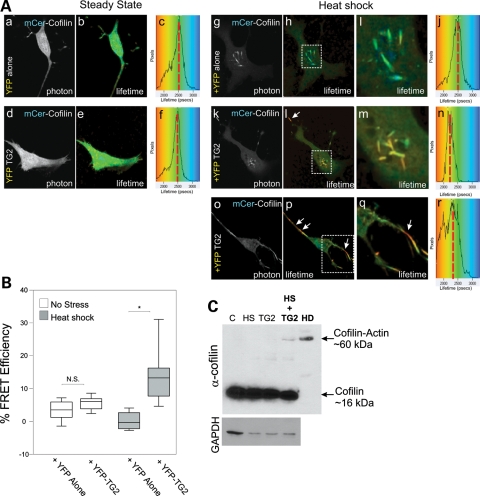
We wanted to ascertain whether TG2 could have activity at rods in our live cell STHdh system. FLIM measures the change in fluorescence lifetime of the donor fluorophore, in this case mCerulean blue cyan fluorescent protein (CFP), which inversely correlates to FRET efficiency in the presence of an acceptor, in this case eYFP. This is not affected by spectral bleed-through or protein concentration, and thus is the best standard of FRET measurement between two individual proteins observed in live cells (30). This method is used to determine whether two proteins are directly interacting within 8 nm of 3D space in vivo, as FRET efficiency drops off to the sixth power with distance (30). FRET efficiency, E, is calculated as 1 minus the lifetime of the donor in the presence of acceptor/lifetime of donor without acceptor. mCerulean–cofilin was used as our donor and eYFP (Venus yellow variant) or eYFP-TG2 as our acceptor. Under steady-state conditions, mCerulean–cofilin could be visualized in both the nucleus and cytoplasm, with mCerulean–cofilin having a fluorescent lifetime of 2600 ps, either with eYFP or eYFP-TG2 co-expressed (Fig. 6Aa–c versus d–f shown on the color scale map as green and red dashed line). This indicated that there is no FRET under steady-state conditions, hence no interaction between the proteins. Under heat shock conditions in mCerulean–cofilin expressing cells, the lifetime of mCerulean–cofilin did not change from 2600 picoseconds when only eYFP was co-expressed (Fig. 6Ag–j and B). The lifetime of mCerulean–cofilin was significantly shorter due to FRET in the presence of eYFP-TG2 (Fig. 6Ak–r and B), with the strongest FRET efficiency at the cofilin–actin rods, from 2600 to under 2000 ps (indicated by yellow–orange in Fig. 6Bl, m, p, q, see arrows). The interaction between mCerulean–cofilin and eYFP-TG2 gave an average of 15.6% FRET efficiency over the whole cell area (with the maximum possible being ∼30% for this donor–acceptor FRET pair) versus the average of the eYFP control interacting with mCerulean–cofilin of 0.7% (Fig. 6Aj versus n, quantified in 6B). These data show that eYFP-TG2 can directly interact with mCerulean–cofilin on both nuclear and cytoplasmic cofilin rods in vivo (see arrows in Fig. 6Bl, p, q) which may lead to a cross-link of cofilin and actin, especially if elevated calcium levels are present, as seen in HD (51).
Figure 6.
TG2 directly interacts with cofilin–actin rods during stress and TG2 over-expression induces a cofilin–actin complex in stressed cells. (A) STHdh cells were transiently transfected with mCerulean–cofilin and either eYFP alone (a–c, g–j) or eYFP-TG2 (d–f, k–r), and FLIM analysis was performed either before (a–c, d–f) or following a 45 min heat shock at 42.5°C (g–j, k–n, o–r). Fluorescence lifetimes for mCerulean blue are presented with a continuous pseudocolor rainbow scale representing time values ranging from 1750 to 3250 ps. The lifetime distribution curve of the mCerulean–cofilin is shown as a histogram on the right representing the number of pixels at each lifetime. The red vertical broken line marks the median lifetime distribution for the cell. Red arrows connect histogram value position with lifetime image value. (B) Box and whisker plot representing FRET efficiency, with FRET occurring at distances <8 nm for these FRET pairs. All imaging and FRET analysis were done in Hank's HEPES buffer pH 7.3. N = 3, n > 10 for each donor–acceptor pair. *P< 0.001. Line = mean; box = 1 standard deviation from the mean; whiskers = 2 standard deviations from the mean. (C) STHdhQ7/Q7 cells were heat-shocked (at 42.5°C for 45 min) and/or transfected with eYFP-TG2 (for ∼36 h). Western blot using MAb22 anti-cofilin was performed on cell pellets to determine whether a higher order cofilin band would be observed. Higher order band observed only in stressed cells over-expressing TG2. Patient HD stage 4 lymphocyte sample was used as a size control for cofilin–actin band. Glyceraldehyde 3-phosphate dehydrogenase loading control was used. Cells were verified for YFP-TG2 expression by fluorescent microscopy.
To ask whether TG2 could catalyze a cross-linked cofilin–actin band as observed in our HD lymphocyte sample western blots, we assayed for the presence of the cofilin–actin complexes in STHdh cells by western blotting for endogenous cofilin under conditions of steady state, heat shock, with TG2 over-expressed, and under heat shock with TG2 over-expressed (Fig. 6C). When over-expressing eYFP-TG2, we had ∼25% transfection rate by direct visualization of YFP (data not shown). A cofilin–actin band co-migrating with a similar higher order cofilin–actin band in clinical HD patient lymphocytes was observed when TG2 was over-expressed coupled with stress conditions (Fig. 6C). The effect seen in the HD patient sample was more robust than that in our model system, which may indicate that lymphocyte population in HD may be undergoing a chronic stress, as opposed to the transient stress of our assay. The data we provide indicate that a disrupted mechanism of TG2 in HD may be aberrant cross-linking of cytoskeletal proteins.
DISCUSSION
Previous work has described the first 17 amino acids of huntingtin as being an amphipathic alpha helix with an affinity for the ER. Huntingtin has the ability to come off the ER and enter the nucleus during stress (9), following the classic model of ER stress-sensor proteins (52). Here, we demonstrate that huntingtin directly affects nuclear actin remodeling during the heat shock stress response. In the past, huntingtin has been shown to bind F-actin (53) and localize to tubulin-rich structures at the mitotic spindle (10). Our data show that huntingtin localizes to an additional form of the dynamic cytoskeleton, nuclear cofilin–actin rods, during stress. The cofilin rod stress response occurs to halt a small proportion of actin from treadmilling, so the ATP that would be used in this dynamic turnover can be used elsewhere for more crucial cell functions (25). Other than freeing up ATP (25), the exact role of nuclear actin rods is not known. Rods may influence chromatin structure and dynamics as well as transcription in response to stress, all of which are affected in HD (7,54). All of our data studying nuclear cofilin rods were acquired by observing full-length endogenous huntingtin expressed at normal levels. While data suggest that the amino terminus of huntingtin is involved in cytoskeletal association (9,53), optimal huntingtin interactions with the cytoskeleton may require additional domains at the carboxyl-terminus of the protein (55). In live cells, FRET with cofilin was only observed with full-length huntingtin and not with smaller fragments (Supplementary Material, Fig. S1), implying that this huntingtin function is a loss of function of fragments of huntingtin. Our data show that in the presence of mutant huntingtin this stress response is impaired: the stress response is slower, fewer cells respond and a persistent nuclear rod phenotype is induced. Defective actin turnover is quickly establishing itself as a common theme among other neurodegenerative diseases, including PD (37) and AD (22,23) indicating that defective actin remodeling through cofilin may be universally contributing to the age-onset progression of these neurodegenerative diseases. Specifically in AD, cofilin–actin rods have been shown to persist in neurites leading to axonal dystrophy (23).
Cofilin rods can be induced by stresses that cause protein misfolding, increased calcium and decreased available ATP. These include temperature stress, 10% DMSO, ATP depletion with sodium azide and 2-deoxyglucose to inhibit metabolism, heavy metals and hydrogen peroxide (23,33). Chronic low ATP levels have been described in HD patient brains and HD animal models (56,57), presumably from defective mitochondrial metabolism (58). A defect in the actin stress response could further contribute to the aberrant ATP regulation in HD. Our findings suggest that defective actin remodeling during stress may lead to persistent rods causing a decline in the available cofilin in the cell, or that the altered actin dynamics during stress may be toxic. Either of these outcomes would predict that a population of neurons requiring higher levels of active actin turnover and having high ATP demands would be greatly affected in HD. Notably, the highly dendritic MSNs are predominantly affected in HD, whereas the smaller interneurons in the striatum, with fewer projections, are spared (59). Dynamic actin turnover is critical to the health of neurons due to its role in maintaining the plasticity of dendritic spines, which is critical in projection neurons (31). Furthermore, active actin remodeling to maintain synaptic and dendritic structures is a huge energy burden, using up to half of the total ATP in a steady-state cell (60). Therefore, the reduced ability of these cells to free up ATP during stress or defective actin turnover, even subtly, would conceivably lead to the changes in spine density, axonal atrophy and eventual neurodegeneration, all observed in MSNs in HD.
Although this is the first report of defective actin turnover via cofilin in HD, several previous studies have reported beneficial effects of the Rho-associated kinase (ROCK) inhibitor, Y-27632, in HD model systems (61). ROCK is an upstream modulator of cofilin. ROCK inhibition effectively leads to reduced phosphorylation of cofilin and therefore increased actin binding cofilin activity (62). ROCK inhibitors have shown in vivo beneficial effects in both mouse (63) and Drosophila models (64) of HD. Thus, our findings provide a potential mechanistic link between the role of huntingtin in the cell stress response and the independent observations that ROCK inhibition has protective benefit in several HD models. This suggests that modulating the activity of cofilin and actin may have therapeutic potential in HD.
Given our findings that the polyglutamine expansion affects cofilin dynamics in neuronal populations, we sought to test whether changes in cofilin properties could be observed in peripheral cells from HD patients. We performed western blotting against cofilin and actin from HD patient lymphocytes and observed a higher order protein band corresponding with the size of a putative cofilin–actin cross-link which increased with disease progression. In our attempt to determine what may cause the SDS resistant, cofilin–actin cross-link, we noted that transglutaminase has the ability to cross-link cofilin and actin in vitro (29) and that tissue transglutaminase (TG2) is thought to be involved in multiple neurodegenerative disorders, including HD (40). TG2 is a transamidating enzyme that can cross-link the ɛ-amino group of polypeptide bound lysine and the γ-carboxamide group of polypeptide bound glutamine through an acyl transfer reaction forming a stable covalent cross-link. This reaction may be reversible and is highly dependent upon calcium levels for activation (40). TG2 binds calcium which activates its enzymatic activity in a concentration-dependent manner (65). Disrupted calcium signaling is also noted in HD (66) and an increase in calcium levels is a hallmark of most cellular stress events. TG2 activity and expression are up-regulated in HD (27), and notably have been shown to have aberrant activity in HD lymphocyte populations in a CAG length-dependent manner (28). To test whether TG2 could cause a similar cofilin–actin cross-link in vivo, we over-expressed TG2 in STHdh cells and activated it using heat shock stress. In this manner, we can recapitulate the higher order cofilin protein band from patient lymphocytes in STHdh cells. Furthermore, using FLIM–FRET, we show that TG2 can directly localize to cofilin rods during stress suggesting that TG2 may not only cross-link free cofilin and actin, but that its activity may be involved in the aberrant nuclear cofilin–actin stress rod response in HD. Consistent with our data highlighting the importance of full-length huntingtin, disrupted calcium signaling in an HD mouse model requires the polyglutamine expansion in the context of the full huntingtin protein (66). Calcium levels can also be elevated upon massive over-expression of a small fragment of polyglutamine-expanded huntingtin that requires the presence of N17 (11). This indicates that aberrant TG2 activation may be present in many HD models, but for different reasons. It is especially notable that the genetic cross between a TG2 knockout mouse and an HD model mouse can rescue HD toxicity supporting the dysfunctional role of TG2 as a strong contributor to the HD phenotype in this model (45).
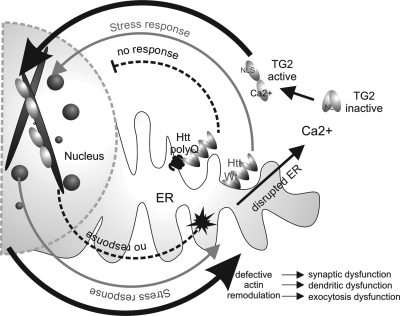
Based on the findings presented in this paper, we propose a model in which huntingtin protein acts as a cell stress response protein at the ER (Fig. 7). Under cell stress conditions, which lead to ER stress and increased cellular calcium, full-length huntingtin translocates to the nucleus and plays a role in actin remodeling in response to stress. Once the cell has recovered from stress, actin remodeling should return to normal. However, when polyglutamine-expanded huntingtin is present, this nuclear actin stress response is impaired; there is a prolonged aberrant regulation of calcium-causing activation of TG2 where it can act on cofilin–actin. This defective actin remodeling would have multiple undesirable effects in a neuron, such as synaptic and dendritic dysfunction, leading to the initial synaptic dysfunction and changes in spine density observed in the early stages of HD. This would likely lead to additional cell stress, further decreases in cellular ATP and additional intracellular calcium increases, progressively augmenting the cycle.
Figure 7.
A model for defective huntingtin-mediated cofilin rod stress response leading to activation of TG2. The grey arrow pathway highlights normal huntingtin stress response by releasing from the ER, entering the nucleus and binding cofilin–actin rods, then exiting the nucleus upon stress relief. With mutant huntingtin present, the dashed arrow pathways show a defect in huntingtin stress response, resulting in less nuclear activity and persistent rods. Back in the cytoplasm, the black arrow pathways highlight elevated calcium due to a defective ER, which results in aberrant TG2 activation and cross-linking of cofilin–actin in both the nucleus and cytoplasm. Defective actin remodeling critically affects neurons at the level of dendritic and synaptic dysfunction, as well as exocytosis activity in peripheral cells.
The implications of our findings may lead to new therapeutic targets for HD, including: alteration of mutant huntingtin nuclear localization in response to stress; inhibition of TG2 enzymatic activity (67) or activation of alternative stress–response pathways that override or circumvent the function of the huntingtin protein in actin-mediated cell stress response. The ratio of free cofilin to cross-linked cofilin can be quantifiably measured in human blood pellets, suggesting that this assay may be developed into a potential biomarker for HD.
MATERIALS AND METHODS
Tissue culture and generation of stable cell lines
Mouse striatal STHdhQ7/Q7, STHdhQ111/Q111 and STHdh7/Q111 (a kind gift of M. E. MacDonald, MGH) cell lines derived from the mouse striatum of wild-type mice and knock-in HD mice, were grown in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (FBS) (Invitrogen) at 33°C with 5% CO2. Striatal cells were clonally selected and grown under G418 drug selection at 33°C to ensure temperature sensitive selection. NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) with 10% FBS (Invitrogen) at 37°C with 5% CO2. Stable STHdhQ7/Q7 and STHdhQ111/Q111 cell lines were generated by co-transfecting pmCerulean–Cofilin and pPuro using turbofect (Fermentas). Cells were grown under puromycin drug selection (Sigma) and positive colonies selected manually using colony selection rings. Stable cell lines were then maintained normally under only G418 selection.
Primary cell growth and immunofluorescence
Sterile 0.1 mg/ml Poly-d-Lysine (Sigma-Aldrich) was diluted in Borate Buffer (50 mm boric acid, Sigma-Aldrich; 12 mm sodium tetraborate, Fischer Scientific) and applied to a LAB-TEK™ eight-well chamber slide (Nalge Nunc International) and incubated at room temperature for 30 min. Each well was washed five times with NANO pure water and washed once with Neurobasal medium (Invitrogen). Cultures were prepared as described previously (38). A total of 15000 cells per well were plated in 400 µl of Neurobasal medium supplemented with B27 (Gibco) and GlutaMAX (Gibco) in a humidified 95% air/5% CO2 incubator at 37°C. Medium was changed on days 2 and 4. On day 6, the medium was replaced with 10% DMSO (Sigma) in Neurobasal with B27 and GlutaMAX and incubated for 90 min in a humidified 95% air/5% CO2 incubator at 37°C. Cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) pH 7.2 for 50 min at room temperature. Cells were permeabilized in 0.05% Triton (Sigma), 2% FBS in PBS and this was applied to cells for 3 min at room temperature and then washed three times with 2% FBS (Atlas, Fort Collins, CO, USA) in PBS over the course of 1h. Primary antibodies include: affinity-purified rabbit IgG to chick actin depolymerizing factor (ADF) (2 ng/l rabbit 1439), which cross-reacts with mammalian ADF and cofilin, and MAB2166 ant huntingtin (Millipore; 1:50 dilution), which were both diluted in 2% FBS in PBS plus 0.05% Tween-20 (PBST) and incubated with the cells overnight at 4°C. Cells were washed two times with 2% FBS in PBS. Secondary antibodies were diluted in 2% FBS in PBST at 1:1000 and include Alexa 594 goat anti-mouse (Invitrogen) and Alexa 488 goat and rabbit (Invitrogen). 4′,6-diamidino-2-phenylindole (Invitrogen) was also added to the secondary antibody solution at 1:1000. Secondary antibodies were incubated for 30 min and then washed four times with PBS. Antifade Gold (Invitrogen) was applied after final wash.
Plasmid construct and expression
Primers to express human cofilin1 were made (McMaster Mobix facility) with 5′ BspEI and 3′ Acc651 overhangs and cloning was performed, using PCR product from human cofilin1 cDNA (OriGene), between BspEI/Acc651 sites of pemCerCI (BD Biosciences/Clontech) to create mCer-cofilin plasmid.
Primers—forward 5′-GATCTCCGGAATGGCCTCCGGTGTGGC-3′;
reverse 5′-GATCGGTACCCAAAGGCTTGCCCTCCAG-3′.
TG2 YFP was a kind gift from Dr Gail Johnson. We used PCR primers to express TG2 with 5′ BglI and 3′ ACC651 overhangs. Cloning was performed using PCR product from TG2 PCR between BglI/ACC651 sites of YFPCI (BD Biosciences/Clontech) to create YFP-TG2.
Primers—forward 5′-GATCAGATCTGGTGGCGGAGG GATGGCCGAGGAGCTGGTCTTAG-3′;
reverse 5′CTATGGTACCCCCTCCGCCACC GGCGGGGCCAATGATGACATTC-3′.
Huntingtin expression constructs were previously described (8).
Plasmids were transfected using the transfection reagent Turbofect™ (Fermentas) according to the manufacturer's instructions. For the full-length huntingtin FLIM-FRET experiments, Lipofectamine 2000 was used as a transfection reagent according to the manufacturer's protocol (Invitrogen). All co-transfections were performed using a 1:1 molar ratio of plasmids.
Heat shock, immunofluorescence and recovery
Cells were cultured in 25 mm live-cell culture dishes at ∼75–85% confluence. Once settled, media were exchanged for HEPES-buffered (20 mm pH 7.4) media. Plates were wrapped with parafilm and placed in a pre-warmed water bath at 42.5°C for 60 min. Cells were fixed in 4% paraformaldehyde for 45 min at room temperature. Cells were permeabilized using ice-cold methanol at −20°C for 5 min and blocked in 1% FBS in PBS for 45 min. Primary antibody α-cofilin (mAB22 a kind gift from J. Bamburg 1/250), anti-actin (20-33 sigma 1/100) or anti-huntingtin C20 (Santa Cruz) were applied in antibody solution (Blocker + 0.02% TWEEN-20) for 2 h. Secondary antibodies conjugated to Alexa probes (Invitrogen) or Cy5 and FITC (Jackson laboratories) were used for 30 min at room temperature in antibody solution. For recovery experiments, three dishes of each cell type were heat-shocked as described. One dish from each cell type was fixed immediately. Recovery dishes had media exchanged for non-buffered media and were placed back in the incubator and allowed to recover for either 3 or 24 h before fixation. After fixation of all dishes, immunofluorescence was performed as described. When immunostaining using the primary antibody mAB2166 anti-huntingtin (1/250 Chemicon) and anti-actin (20-33 Sigma 1/100) cells were permeabilized using 0.5% Triton X in PBS and 2% FBS for 12 min at 4°C with the rest of the immunofluorescence procedure being performed as described. All double immunofluorescence was done in series with non-cross reactive secondary antibodies.
Stealth RNAi duplexes and transfection
Huntingtin Stealth™ Select RNAi was purchased from Invitrogen as a set of three (MSS205082, MSS205083, MSS205084). Stealth™ RNAi Negative Control Medium GC (Invitrogen) was purchased as a negative control. 120 pmol of RNAi was transfected into 25 mm dishes plated at 50% confluency using the lipid-based transfection reagent, Lipofectamine™ 2000 (Invitrogen) according to the manufacturers protocols. Cells were treated for biochemical analysis or processed for fluorescence microscopy 72 h after transfection. For delta T dish live cell experiments, Block-iT™ Alexa Fluor® red (Invitrogen) was co-transfected with huntingtin Stealth™ Select RNAi or Stealth™ RNAi Negative Control Medium GC, at a 1:3 ratio, as a control for transfected cells during live cell imaging experiments.
Protein extraction and immunoblot assay
For STHdh cell lines, cells were grown in 10 cm dishes to confluency, washed with PBS and collected using a rubber scraper. Cells were pelleted, incubated and resuspended in NP-40 lysis buffer with protease inhibitor cocktail. Cells were spun at 14000g for 10 min and the supernatant (protein fraction) was collected.
Or when protein from insoluble fraction was collected, protein was extracted as described above and pellet was re-suspended in NP-40 lysis buffer with protease inhibitors. Slurry was sonicated for three cycles of 15 pulses. The settings were set with a duty cycle of 10% and output control power of 3.
Lymphocyte samples were obtained by extracting the buffy coat from 3 ml total human blood using Accuspin Histopaque 1077 tubes from Sigma (Product code A6929, 3–6 ml). Protein was extracted from lymphocyte samples as described for the cell lines.
Equal amounts of proteins were loaded on 12% or 7% SDS–polyacrylamide gel and electroblotted to a polyvinylidene fluoride membrane. Membranes were blocked with 5% non-fat dry milk in TBST for 1 h followed by 1 h incubation at room temperature with anti-cofilin mAB22 (1:5000, a kind gift from Dr J. Bamburg), anti-cofilin H12 (1:1000 Santa Cruz), anti-htt-N18 (1:10000) or anti-actin 20-33 (1:2500, Sigma). After incubation with appropriate horseradish peroxidase-conjugated secondary antibody (Sigma), bands were visualized by enhanced chemiluminescence. Quantification of western blot bands was performed using the Image J software and densitometry, normalized to actin. For lymphocyte samples, densitometry was performed individually on each lane and values are expressed as percent of total area in the higher order band.
Microscopy
All wide-field fluorescence microscope images were captured on a Nikon TE200 epifluorescence inverted microscope equipped with a 63× oil immersion plan apochromat NA1.4 objective and a Hamamatsu Orca ER digital camera (Hamamatsu Photonics, Japan). Qualitative images of nuclear rods (Figs 1 and 3) were produced by obtaining a multichannel Z-stack and performing blind 3D non-iterative deconvolution (Autodeblur, Media Cybernetics). Images shown are mass-projections from deconvolved Z-stacks with gamma alterations.
Live cell imaging
Live cell visualization was done using the Delta T4 heated stage, lid and objective system (Bioptechs). Cells were seeded and treated in 0.17 mm delta T dishes (Bioptechs). Cells were heated to 42.5°C using the heated stage and objective and visualized at 100× plan apochromat oil N.A1.3. As soon as dish reached temperature, fluorescent, images were recorded once every 60 s for the duration of the session.
Statistical analysis
Statistical analysis was performed using the SigmaPlot Software 11.0 (Systat Software Inc.) For single comparisons, Student's t-tests were performed if data passed normality assumptions. If data did not pass normality assumptions, it was analyzed by the Mann–Whitney method. For multiple comparisons, analysis of variance by the ranks was performed and multiple comparisons were performed using the Dunn method.
Fluorescence lifetime imaging measurement (FLIM)
FLIM was conducted using an inverted confocal laser-scanning microscope (Leica TCS SP5) with a 63× glycerol immersion NA 1.4 Plan apochromat objective. The SP5 is run using the LAS Advanced Fluorescence software from Leica. Two-photon excitation of samples was done using a tunable Chameleon laser, mode-locked to deliver femtosecond pulses at a rate of 80 MHz with an output power of 1.8 W for a peak wavelength of 820 nm.
mCerulean and YFP fluorophores were used as FRET pairs. Excitation of the mCerulean donor using the two-photon laser was found to be optimal at 820 nm. Collection of mCerulean fluorescence emission was limited to 480 ± 20 nm using a bandpass filter. All live cell imaging and FLIM were done in Hank's saline HEPES buffer pH 7.3.
Photons from the donor fluorophores were collected and counted using TCSPC software from Becker & Hickl (SPC-830). The laser power was adjusted in order to give a photon collection count of ∼105 photons/s, where all FLIM measurements were conducted over a 60 s collection time. The lifetimes of all the pixels in the field of view (256 × 256) were calculated by the SPC image analysis software (Becker & Hickl GmbH) to generate exponential decay curves. Binning and thresholding values (bin= 3, threshold= 10) were kept constant to ensure consistency of lifetime measurements over multiple trials.
Förster resonant energy transfer (FRET) analysis
FRET analysis was performed using the Becker and Hickl FLIM plug in for the ImageJ software (www.macbiophotonics.ca). The lifetime of every pixel in the image was calculated to give a mean lifetime value for each cell. Pixels with lifetimes outside the range of 1750–3250 were excluded from further analysis. FRET efficiency for each image was determined using the equation E FRET= 1 − (average lifetime D.A/average lifetime D), where average lifetime D.A indicates the average lifetime of mCerulean–cofilin in the presence of the indicated acceptor and average lifetime D indicates the overall average lifetime of mCerulean–cofilin alone (no acceptor present).
SUPPLEMENTARY MATERIAL
FUNDING
This work is supported by operating grants from the Canadian Institutes of Health Research (MOP-165174), the Krembil Foundation and CHDI Inc. (to R.T.). L.M. has a CIHR Doctoral Research Award. S.J.T. is funded by the MRC, Wellcome Trust, CHDI, and Brain Research Trust. The clinical evaluation, genetic testing and patient blood collections were undertaken at UCLH/UCL who received a proportion of funding from the UK Department of Health's NIHR Biomedical Research Centers funding scheme. NIH grant NS40371 to J.R.B. Funding to pay the Open Access publication charges for this article was provided by CIHR.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the technical assistance of Ms L. S. Minamide, Dr Nayana Lahiri and valuable discussions with Drs O. Wiggan and B.W. Bernstein.
Conflict of Interests statement. None declared.
REFERENCES
- 1.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Rawlins M. Huntington's disease out of the closet? Lancet. 2010;23:1372–1373. doi: 10.1016/S0140-6736(10)60974-9. [DOI] [PubMed] [Google Scholar]
- 3.Andrade M.A., Petosa C., O'Donoghue S.I., Muller C.W., Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 4.Grinthal A., Adamovic I., Weiner B., Karplus M., Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc. Natl Acad. Sci. USA. 2010;107:2467–2472. doi: 10.1073/pnas.0914073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuccato C., Ciammola A., Rigamonti D., Leavitt B.R., Goffredo D., Conti L., MacDonald M.E., Friedlander R.M., Silani V., Hayden M.R., et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 6.Seong I.S., Woda J.M., Song J.J., Lloret A., Abeyrathne P.D., Woo C.J., Gregory G., Lee J.M., Wheeler V.C., Walz T., et al. Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 2010;19:573–583. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benn C.L., Sun T., Sadri-Vakili G., McFarland K.N., DiRocco D.P., Yohrling G.J., Clark T.W., Bouzou B., Cha J.H. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J. Neurosci. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauthier L.R., Charrin B.C., Borrell-Pages M., Dompierre J.P., Rangone H., Cordelieres F.P., De Mey J., MacDonald M.E., Lessmann V., Humbert S., Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 10.Godin J.D., Colombo K., Molina-Calavita M., Keryer G., Zala D., Charrin B.C., Dietrich P., Volvert M.L., Guillemot F., Dragatsis I., et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Rockabrand E., Slepko N., Pantalone A., Nukala V.N., Kazantsev A., Marsh J.L., Sullivan P.G., Steffan J.S., Sensi S.L., Thompson L.M. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum. Mol. Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 12.Aiken C.T., Steffan J.S., Guerrero C.M., Khashwji H., Lukacsovich T., Simmons D., Purcell J.M., Menhaji K., Zhu Y.Z., Green K., et al. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J. Biol. Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu X., Greiner E.R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J.S., Thompson L.M., Wetzel R., Yang X.W. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 15.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., MacDonald M.E. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 16.Morimoto R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummond I.A., McClure S.A., Poenie M., Tsien R.Y., Steinhardt R.A. Large changes in intracellular pH and calcium observed during heat shock are not responsible for the induction of heat shock proteins in Drosophila melanogaster. Mol. Cell Biol. 1986;6:1767–1775. doi: 10.1128/mcb.6.5.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder M., Kaufman R.J. ER stress and the unfolded protein response. Mutat. Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 19.Gibson G.E., Starkov A., Blass J.P., Ratan R.R., Beal M.F. Cause and consequence: mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochim. Biophys. Acta. 2010;1802:122–134. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosoi T., Ozawa K. Endoplasmic reticulum stress in disease: mechanisms and therapeutic opportunities. Clin. Sci. (Lond.) 2010;118:19–29. doi: 10.1042/CS20080680. [DOI] [PubMed] [Google Scholar]
- 21.Keller J.N. Age-related neuropathology, cognitive decline, and Alzheimer's disease. Ageing Res. Rev. 2006;5:1–13. doi: 10.1016/j.arr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Maloney M.T., Bamburg J.R. Cofilin-mediated neurodegeneration in Alzheimer's disease and other amyloidopathies. Mol. Neurobiol. 2007;35:21–44. doi: 10.1007/BF02700622. [DOI] [PubMed] [Google Scholar]
- 23.Minamide L.S., Striegl A.M., Boyle J.A., Meberg P.J., Bamburg J.R. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat. Cell Biol. 2000;2:628–636. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- 24.Bamburg J.R., Wiggan O.P. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein B.W., Chen H., Boyle J.A., Bamburg J.R. Formation of actin-ADF/cofilin rods transiently retards decline of mitochondrial potential and ATP in stressed neurons. Am. J. Physiol. Cell Physiol. 2006;291:C828–C839. doi: 10.1152/ajpcell.00066.2006. [DOI] [PubMed] [Google Scholar]
- 26.Lesort M., Chun W., Tucholski J., Johnson G.V. Does tissue transglutaminase play a role in Huntington's disease? Neurochem. Int. 2002;40:37–52. doi: 10.1016/s0197-0186(01)00059-6. [DOI] [PubMed] [Google Scholar]
- 27.Lesort M., Chun W., Johnson G.V., Ferrante R.J. Tissue transglutaminase is increased in Huntington's disease brain. J. Neurochem. 1999;73:2018–2027. [PubMed] [Google Scholar]
- 28.Cariello L., de Cristofaro T., Zanetti L., Cuomo T., Di Maio L., Campanella G., Rinaldi S., Zanetti P., Di Lauro R., Varrone S. Transglutaminase activity is related to CAG repeat length in patients with Huntington's disease. Hum. Genet. 1996;98:633–635. doi: 10.1007/s004390050273. [DOI] [PubMed] [Google Scholar]
- 29.Benchaar S.A., Xie Y., Phillips M., Loo R.R., Galkin V.E., Orlova A., Thevis M., Muhlrad A., Almo S.C., Loo J.A., et al. Mapping the interaction of cofilin with subdomain 2 on actin. Biochemistry. 2007;46:225–233. doi: 10.1021/bi0610754. [DOI] [PubMed] [Google Scholar]
- 30.Wallrabe H., Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Hotulainen P., Hoogenraad C.C. Actin in dendritic spines: connecting dynamics to function. J. Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cattaneo E., Conti L. Generation and characterization of embryonic striatal conditionally immortalized ST14A cells. J. Neurosci. Res. 1998;53:223–234. doi: 10.1002/(SICI)1097-4547(19980715)53:2<223::AID-JNR11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 33.Nishida E., Iida K., Yonezawa N., Koyasu S., Yahara I., Sakai H. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc. Natl Acad. Sci. USA. 1987;84:5262–5266. doi: 10.1073/pnas.84.15.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iida K., Matsumoto S., Yahara I. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct. Funct. 1992;17:39–46. doi: 10.1247/csf.17.39. [DOI] [PubMed] [Google Scholar]
- 35.Ohta Y., Nishida E., Sakai H., Miyamoto E. Dephosphorylation of cofilin accompanies heat shock-induced nuclear accumulation of cofilin. J. Biol. Chem. 1989;264:16143–16148. [PubMed] [Google Scholar]
- 36.Endo M., Ohashi K., Mizuno K. LIM kinase and slingshot are critical for neurite extension. J. Biol. Chem. 2007;282:13692–13702. doi: 10.1074/jbc.M610873200. [DOI] [PubMed] [Google Scholar]
- 37.Lim M.K., Kawamura T., Ohsawa Y., Ohtsubo M., Asakawa S., Takayanagi A., Shimizu N. Parkin interacts with LIM Kinase 1 and reduces its cofilin-phosphorylation activity via ubiquitination. Exp. Cell Res. 2007;313:2858–2874. doi: 10.1016/j.yexcr.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Mila S., Albo A.G., Corpillo D., Giraudo S., Zibetti M., Bucci E.M., Lopiano L., Fasano M. Lymphocyte proteomics of Parkinson's disease patients reveals cytoskeletal protein dysregulation and oxidative stress. Biomark. Med. 2009;3:117–128. doi: 10.2217/bmm.09.4. [DOI] [PubMed] [Google Scholar]
- 39.Mannherz H.G., Ballweber E., Galla M., Villard S., Granier C., Steegborn C., Schmidtmann A., Jaquet K., Pope B., Weeds A.G. Mapping the ADF/cofilin binding site on monomeric actin by competitive cross-linking and peptide array: evidence for a second binding site on monomeric actin. J. Mol. Biol. 2007;366:745–755. doi: 10.1016/j.jmb.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 40.Ruan Q., Johnson G.V. Transglutaminase 2 in neurodegenerative disorders. Front Biosci. 2007;12:891–904. doi: 10.2741/2111. [DOI] [PubMed] [Google Scholar]
- 41.Jeitner T.M., Matson W.R., Folk J.E., Blass J.P., Cooper A.J. Increased levels of gamma-glutamylamines in Huntington disease CSF. J. Neurochem. 2008;106:37–44. doi: 10.1111/j.1471-4159.2008.05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson G.V., Cox T.M., Lockhart J.P., Zinnerman M.D., Miller M.L., Powers R.E. Transglutaminase activity is increased in Alzheimer's disease brain. Brain Res. 1997;751:323–329. doi: 10.1016/s0006-8993(96)01431-x. [DOI] [PubMed] [Google Scholar]
- 43.Wilhelmus M.M., Grunberg S.C., Bol J.G., van Dam A.M., Hoozemans J.J., Rozemuller A.J., Drukarch B. Transglutaminases and transglutaminase-catalyzed cross-links colocalize with the pathological lesions in Alzheimer's disease brain. Brain Pathol. 2009;19:612–622. doi: 10.1111/j.1750-3639.2008.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norlund M.A., Lee J.M., Zainelli G.M., Muma N.A. Elevated transglutaminase-induced bonds in PHF tau in Alzheimer's disease. Brain Res. 1999;851:154–163. doi: 10.1016/s0006-8993(99)02179-4. [DOI] [PubMed] [Google Scholar]
- 45.Mastroberardino P.G., Iannicola C., Nardacci R., Bernassola F., De Laurenzi V., Melino G., Moreno S., Pavone F., Oliverio S., Fesus L., Piacentini M. ‘Tissue’ transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington's disease. Cell Death Differ. 2002;9:873–880. doi: 10.1038/sj.cdd.4401093. [DOI] [PubMed] [Google Scholar]
- 46.Kahlem P., Terre C., Green H., Djian P. Peptides containing glutamine repeats as substrates for transglutaminase-catalyzed cross-linking: relevance to diseases of the nervous system. Proc. Natl Acad. Sci. USA. 1996;93:14580–14585. doi: 10.1073/pnas.93.25.14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper A.J., Sheu K.F., Burke J.R., Onodera O., Strittmatter W.J., Roses A.D., Blass J.P. Polyglutamine domains are substrates of tissue transglutaminase: does transglutaminase play a role in expanded CAG/poly-Q neurodegenerative diseases? J. Neurochem. 1997;69:431–434. doi: 10.1046/j.1471-4159.1997.69010431.x. [DOI] [PubMed] [Google Scholar]
- 48.Chun W., Lesort M., Tucholski J., Ross C.A., Johnson G.V. Tissue transglutaminase does not contribute to the formation of mutant huntingtin aggregates. J. Cell Biol. 2001;153:25–34. doi: 10.1083/jcb.153.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey C.D., Johnson G.V. Tissue transglutaminase contributes to disease progression in the R6/2 Huntington's disease mouse model via aggregate-independent mechanisms. J. Neurochem. 2005;92:83–92. doi: 10.1111/j.1471-4159.2004.02839.x. [DOI] [PubMed] [Google Scholar]
- 50.McConoughey S.J., Basso M., Niatsetskaya Z.V., Sleiman S.F., Smirnova N.A., Langley B.C., Mahishi L., Cooper A.J., Antonyak M.A., Cerione R.A., et al. Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol. Med. 2010;2:335–337. doi: 10.1002/emmm.201000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang T.S., Slow E., Lupu V., Stavrovskaya I.G., Sugimori M., Llinas R., Kristal B.S., Hayden M.R., Bezprozvanny I. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington's disease. Proc. Natl Acad. Sci. USA. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao L., Ackerman S.L. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Angeli S., Shao J., Diamond M.I. F-actin binding regions on the androgen receptor and huntingtin increase aggregation and alter aggregate characteristics. PLoS ONE. 2010;5:e9053. doi: 10.1371/journal.pone.0009053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luthi-Carter R., Cha J.H. Mechanisms of transcriptional dysregulation in Huntington's disease. Clin. Neurosci. Res. 2003;3:165–177. [Google Scholar]
- 55.Pal A., Severin F., Lommer B., Shevchenko A., Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington's disease. J. Cell Biol. 2006;172:605–618. doi: 10.1083/jcb.200509091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gines S., Seong I.S., Fossale E., Ivanova E., Trettel F., Gusella J.F., Wheeler V.C., Persichetti F., MacDonald M.E. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington's disease knock-in mice. Hum. Mol. Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 57.Koroshetz W.J., Jenkins B.G., Rosen B.R., Beal M.F. Energy metabolism defects in Huntington's disease and effects of coenzyme Q10. Ann. Neurol. 1997;41:160–165. doi: 10.1002/ana.410410206. [DOI] [PubMed] [Google Scholar]
- 58.Seong I.S., Ivanova E., Lee J.M., Choo Y.S., Fossale E., Anderson M., Gusella J.F., Laramie J.M., Myers R.H., Lesort M., MacDonald M.E. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 59.Richfield E.K., Maguire-Zeiss K.A., Vonkeman H.E., Voorn P. Preferential loss of preproenkephalin versus preprotachykinin neurons from the striatum of Huntington's disease patients. Ann. Neurol. 1995;38:852–861. doi: 10.1002/ana.410380605. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein B.W., Bamburg J.R. Actin-ATP hydrolysis is a major energy drain for neurons. J. Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao J., Welch W.J., Diprospero N.A., Diamond M.I. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell Biol. 2008;28:5196–5208. doi: 10.1128/MCB.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maekawa M., Ishizaki T., Boku S., Watanabe N., Fujita A., Iwamatsu A., Obinata T., Ohashi K., Mizuno K., Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 63.Li M., Huang Y., Ma A.A., Lin E., Diamond M.I. Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol. Dis. 2009;36:413–420. doi: 10.1016/j.nbd.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 64.Agrawal N., Pallos J., Slepko N., Apostol B.L., Bodai L., Chang L.W., Chiang A.S., Thompson L.M., Marsh J.L. Identification of combinatorial drug regimens for treatment of Huntington's disease using Drosophila. Proc. Natl Acad. Sci. USA. 2005;102:3777–3781. doi: 10.1073/pnas.0500055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kiraly R., Csosz E., Kurtan T., Antus S., Szigeti K., Simon-Vecsei Z., Korponay-Szabo I.R., Keresztessy Z., Fesus L. Functional significance of five noncanonical Ca2+-binding sites of human transglutaminase 2 characterized by site-directed mutagenesis. FEBS J. 2009;276:7083–7096. doi: 10.1111/j.1742-4658.2009.07420.x. [DOI] [PubMed] [Google Scholar]
- 66.Zhang H., Li Q., Graham R.K., Slow E., Hayden M.R., Bezprozvanny I. Full length mutant huntingtin is required for altered Ca2+ signaling and apoptosis of striatal neurons in the YAC mouse model of Huntington's disease. Neurobiol. Dis. 2008;31:80–88. doi: 10.1016/j.nbd.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaertl S., Prime M., Wityak J., Dominguez C., Munoz-Sanjuan I., Pacifici R.E., Courtney S., Scheel A., Macdonald D. A profiling platform for the characterization of transglutaminase 2 (TG2) inhibitors. J. Biomol. Screen. 2010;15:478–487. doi: 10.1177/1087057110366035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.