Abstract
AIM: To evaluate the biological and clinical characteristics of miR-622 in gastric cancer.
METHODS: We analyzed the expression of miR-622 in 57 pair matched gastric neoplastic and adjacent non-neoplastic tissues by quantitative real-time polymerase chain reaction. Functional analysis of miR-622 expression was assessed in vitro in gastric cancer cell lines with miR-622 precursor and inhibitor. The roles of miR-622 in tumorigenesis and tumor metastasis were analyzed using a stable miR-622 expression plasmid in nude mice. A luciferase reporter assay was used to assess the effect of miR-622 on inhibitor of growth family, member 1 (ING1) expression.
RESULTS: Expression of miR-622 was down-regulated in gastric cancer. MiR-622 was found involved in differentiation and lymphatic metastasis in human gastric cancer. Ectopic expression of miR-622 promoted invasion, tumorigenesis and metastasis of gastric cancer cells both in vitro and in vivo. ING1 is a direct target of miR-622.
CONCLUSION: These findings help clarify the molecular mechanisms involved in gastric cancer metastasis and indicate that miR-622 modulation may be a bona fide treatment of gastric cancer.
Keywords: MicroRNA, MiR-622, Gastric cancer, Metastasis, Inhibitor of growth family member 1
INTRODUCTION
Gastric cancer is the fourth most common cancer and is the second most common cause of death from cancer in the world[1-3]. For most solid malignancies, metastasis is the predominant cause of cancer death[4-6]. Elucidation of the molecular mechanisms that regulate the sequential steps of metastasis formation is critical for the reduction of cancer mortality. The regulatory mechanism involved in the development of gastric cancer is not well understood; the discovery of critical carcinogenic pathways and the identification of new therapeutic targets for gastric cancer are crucial for local and global public health.
Recently, researchers discovered a novel class of short, endogenous non-coding RNAs, called microRNAs (miRNAs) described in not only animals and plants, but also in humans as well[7-9]. It is clear that miRNAs play pivotal roles in a wide array of biological processes, including cell proliferation, differentiation and apoptosis[10,11]. MiRNAs regulate the expression of protein-coding genes by degrading target mRNAs or by inhibiting gene translation[12]. Emerging evidence strongly suggests that abnormal miRNA expression is a common and important feature of human malignancies[13,14], and some studies show that some miRNAs are associated with metastasis from gastric cancer[15-18].
In this study, we investigated the biological effects and potential mechanisms of miR-622 in gastric carcinoma. We examined the expression of miR-622 in gastric cancer and found that miR-622 was associated with differentiation and lymphatic metastasis of human gastric cancer. Exogenous expression of miR-622 promotes invasion, tumorigenesis and metastasis formation both in vitro and in vivo. Using bioinformatics analysis, we identified inhibitor of growth family, member 1 (ING1) as a putative miR-622 target. Subsequent experiments confirmed that up-regulation of miR-622 repressed the expression of ING1 at translational level.
MATERIALS AND METHODS
Cell lines and tissue samples
MKN-45, AGS, MKN-28, SGC-7901 and human embryonic kidney cell lines 293T (HEK 293T) were maintained in our central laboratory. Gastric cancer cell lines SNU-1 and NCI-N87 were obtained from American Type Culture Collection (Manassas, VA, USA). Cells were grown in RPMI-1640 or Dulbecco’s modified Eagle’s medium (DMEM; Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum. Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2. Primary gastric tumor tissues and adjacent non-tumor gastric tissues were collected from either routine therapeutic surgery or gastrointestinal endoscopy at our department. All samples were obtained with informed consent from the patients and approval by the hospital institutional review board.
Analysis of miRNA expression using TaqMan real-time polymerase chain reaction
Total RNA from tissue samples and cell lines was isolated using the mirVana miRNA Isolation Kit (Ambion, USA). Expression of mature miRNAs was detected using the Taqman MicroRNA Assay (Applied Biosystems, Carlsbad, CA, USA) specific for hsa-miR-622. Briefly, 5 ng small RNA or total RNA was reversely transcribed using specific stem-loop RT primers. The reverse transcription products were then amplified and detected using PCR with specific primers and TaqMan probes. The PCR was run in a 7900 HT Fast Real-Time polymerase chain reaction (RT-PCR) System (Applied Biosystems) and SDS2.2.2 software (Applied Biosystems) was used for comparative ΔCt analysis. U6 snRNA (RNU6B, Applied Biosystems) was used as an endogenous control.
MiRNA target prediction
The miR-622 sequence was obtained from miRBase (http://www.microrna.sanger.ac.uk). Predicted miRNA targets were determined using the TargetScan[19], PicTar[20], and Miranda[7] algorithms.
Construction of miR-622 expression plasmids
For miR-622 expression, human miR-622 precursor (622 bp) was cloned into pSilencer 4.1 (Ambion, Austin, TX, USA) using the following primers: Forward: 5’-ATCCCAGGGAGACAGAGATCGAGG-3’, Reverse: 5’-AAGCTTGGTGGTGGACTTTTGGTTGT-3’. The plasmid was named pS-miR-622, and the control plasmid, consisting of a scrambled sequence (Ambion), was named pS-control. Transfected cells were selected with puromycin 48 h after transfection and then diluted for clonal selection. Mature miRNA expression in selected clones was assessed by RT-PCR as described above. All constructs were verified by sequencing. Stability-enhanced miR-622 precursor and negative control 1 ribo-oligonucleotides were obtained from Ambion. To inhibit miR-622 function, an Ambion miRNA inhibitor for miR-622 (AS-miR-622) was used, along with the negative control (AS-control).
Transfection
For transfection, a complex of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and 200 nmol/L RNAs described above was prepared according to the manufacturer’s instructions and directly mixed with cells in 24-well cell culture plates at a density of 4 × 104 cells per well. The level of miR-622 expression in transfected cell lines was assayed by real-time RT-PCR 72 h after transfection.
Invasion and scratch healing assays
Cell invasion assays were performed using insert membranes coated with diluted Matrigel (BD Biosciences, San Jose, CA, USA). Cells (1 × 105) were added to the upper chamber and cultured for 48 h. Finally, the insert membranes were cut and stained with Crystal violet (0.04% in water, 100 mL) and permeable cells were counted under an inverted microscope and photographed. For the scratch assay, cells were treated with 10 mg/mL mitomycin C (Sigma) for 3 h and then wounded with a pipette tip. Fresh, full medium was added, and wound healing was observed for 48 h. Photographs were taken every 6 h.
Tumorigenesis and metastasis assay in nude mice
Cells (1 × 106), transfected with pS-miR-622 or pS-control, were collected and inoculated subcutaneously into right flank regions of 4-wk-old male BALB/c nude mice (Institute of Zoology, Chinese Academy of Sciences, Shanghai). In another group, male nude mice were injected through the tail vein. Tumor nodules were measured every 4 d with a caliper. Mice were sacrificed at the end of one month and the number of metastatic tumors in the liver; tumor growth rate and rate of inhibition were calculated. Two independent experiments were performed for each experimental group.
Immunohistochemistry
Mouse tissues were fixed in 10% neutralized formalin and embedded in paraffin blocks. Sections were then prepared for immunohistochemical examination. After deparaffinization and rehydration, antigen retrieval was performed by boiling samples in 10 mmol/L citrate buffer (pH 6.0) for 10 min. After inhibition of endogenous peroxidase activity for 30 min with methanol containing 0.3% H2O2, sections were blocked with 2% bovine serum albumin in phosphate-buffered saline (PBS) for 30 min and then incubated with human anti-rabbit ING1 monoclonal antibody (Abcam, dilution 1:500). The immune complex was visualized using the Dako REA™ Envision™ Detection System and Peroxidase/DAB (Dako) according to the manufacturer’s instructions. Nuclei were counterstained with hematoxylin.
Western blotting
ING1 protein levels were quantified by standard Western blotting procedures, using the anti-rabbit ING1 monoclonal antibody (Abcam, dilution 1:500). Protein levels were normalized to the total glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using a rabbit monoclonal anti-GAPDH antibody (Sigma).
Luciferase miRNA target reporter assay
Total cDNA from SGC-7901 cells was used to amplify the 3’-UTR of ING1 by PCR, using the forward primer: 5’-TAGTCGAGTGGTTCCACTTCTCGTGC-3’ and the reverse primer: 5’-TTACAGCTCACATACAGCAGGAAG-3’. After digestion of the PCR product by SpeI and Hind III, the ING1 3’UTR was cloned into the SpeI and Hind III sites of pMir-Report (Ambion), yielding pMir-Report-ING1. Mutations were introduced at potential miR-622 binding sites using the QuikChange site-directed mutagenesis kit (Stratagene). HEK 293T cells were transfected with the pMir-Report vectors containing the 3’UTR variants, and 5 h after transfection, the cells were transfected again with 100 nmol/L of miR-622 precursor, miR-622-inhibitor or control plasmids. Five ng of the phRL-TK vector (Promega) harboring the Renilla luciferase gene was co-transfected as an internal control to determine the transfection efficiency. Cells were harvested 48 h after transfection and analyzed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) and the GloMaxTM 20/20 detection system (E5331, Promega).
Statistical analysis
Statistical analysis was performed using SPSS 15.0 software (SPSS, USA). Data were expressed as the mean ± SD from at least three separate experiments. Differences between groups were analyzed using Student’s t test and the χ2 test. P < 0.05 was considered statistically significant.
RESULTS
Relationship between miR-622 expression levels and clinicopathological factors in patients with gastric cancer
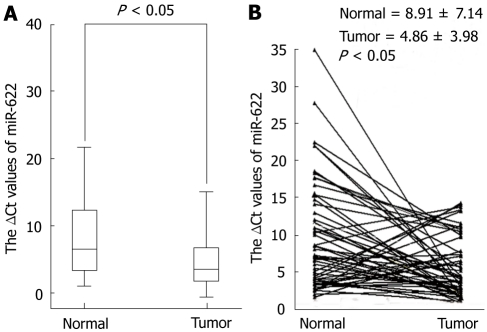
To determine whether miR-622 expression is associated with gastric cancer, we examined and compared miR-622 expression in primary gastric cancer tissues and in pair-matched adjacent non-tumor tissues using quantitative RT-PCR. A decrease of miR-622 expression was found in 70.18% of the 57 patients with gastric cancer with a median change by about 1.83-fold (P < 0.05, Figure 1). We then investigated the relationship between miR-622 expression levels and clinicopathological factors in the 57 patients. We found that the expression level of miR-622 was associated with the differentiation and lymphatic metastasis (P < 0.05, Table 1). As shown in Table 1, miR-622 expression levels were not associated with tumor size and distal metastasis, TNM stage or invasion.
Figure 1.
Expression of miR-622 was down-regulated in gastric cancer. A: Level of miR-622 expression in gastric cancer tissues (n = 57) was lower than that in non-tumor tissues (n = 57). B: The values (ΔCt) are relative to those of U6 small RNA.
Table 1.
Clinicopathological features and miR-622 expression in gastric cancer patients
| Factors | No. of patients | Mean expression of miR-622 | P value |
| Age (yr) | |||
| < 65 | 42 | 4.84 ± 3.78 | 0.9731 |
| ≥ 65 | 15 | 4.89 ± 4.65 | |
| Gender | |||
| Male | 30 | 5.51 ± 3.89 | 0.1911 |
| Female | 27 | 4.12 ± 4.03 | |
| Cell differentiation | |||
| Poor | 19 | 5.71 ± 4.14 | 0.021a |
| Moderate | 38 | 3.14 ± 3.07 | |
| Tumor size (cm) | |||
| < 5 | 48 | 4.60 ± 3.79 | 0.2591 |
| ≥ 5 | 9 | 6.24 ± 4.91 | |
| Gross appearance | |||
| Borrmann I + II type | 10 | 3.72 ± 3.69 | 0.3251 |
| Borrmann III + IV type | 47 | 5.10 ± 4.04 | |
| Site of tumor | |||
| Cardia | 9 | 3.60 ± 4.00 | 0.371 |
| Body | 8 | 3.84 ± 5.57 | |
| Antrum | 40 | 5.34 ± 3.682 | |
| Lymphatic metastasis | |||
| Positive | 31 | 6.03 ± 4.16 | 0.041a |
| Negative | 26 | 3.87 ± 3.60 | |
| Depth of cancer invasion | |||
| T2 | 11 | 4.71 ± 5.03 | 0.5861 |
| T3 | 44 | 5.02 ± 3.78 | |
| T4 | 2 | 2.02 ± 1.37 | |
| TNM Stage | |||
| I | 2 | 0.82 ± 0.72 | 0.3851 |
| II | 12 | 4.02 ± 3.31 | |
| III | 35 | 5.34 ± 4.12 | |
| IV | 8 | 5.00 ± 4.49 | |
| Distal metastasis | |||
| Positive | 6 | 3.67 ± 3.22 | 0.4441 |
| Negative | 51 | 5.00 ± 4.07 |
Chi-square test.
P < 0.05; TNM: Tumor-node-metastasis.
Expression of miR-622 and ING1 in gastric cancer cell lines
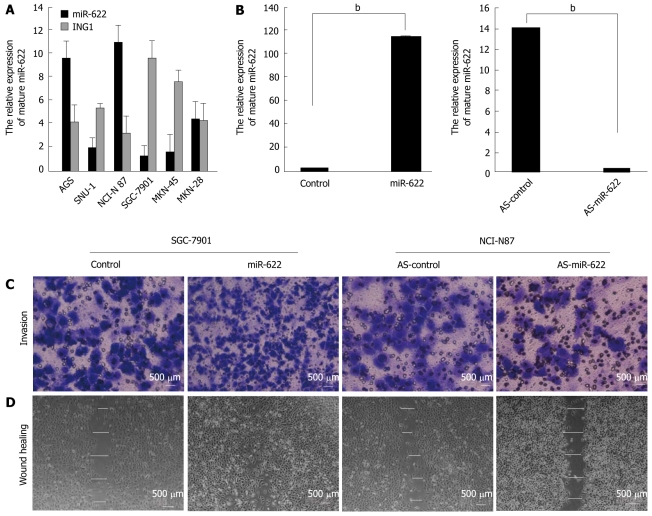
We detected miR-622 and ING1 expression using qRT-PCR in six different gastric cancer cell lines (AGS, SNU-1, NCI-N87, SGC-7901, MKN-45 and MKN-28). Based on the miR-622 expression levels in these cell lines, we chose to further analyze SGC-7901 and NCI-N87 cells for miR-622 gain-of-function and loss-of-function studies, respectively (Figure 2A). After transfection, we examined the expression of miR-622 in cancer cells by real-time PCR. The expression level of miR-622 was significantly up-regulated in SGC-7901 cells transfected with miR-622 precursor (P < 0.01, Figure 2B) and the expression level of miR-622 was significantly down-regulated in NCI-N87 cells transfected with miR-622 inhibitor, compared with the negative controls (P < 0.01, Figure 2B). We also analyzed the expression of ING1 in the same gastric cancer cells. The expression of miR-622 was inversely correlated with the expression of ING1 (Figure 2A).
Figure 2.
Restoration of miR-622 promotes gastric cancer cell invasion and migration. A: q-real time polymerase chain reaction for the expression of miR-622 and ING1 was performed using six gastric cancer cell lines. The lowest and highest levels of miR-622 expression among the six gastric cancer cell lines were found in SGC-7901 and NCI-N87 cells, respectively, and the expression of miR-622 was inversely correlated with the expression of ING1; B: Expression of miR-622 was restored or suppressed in gastric cancer cells after miR-622 precursor or miR-622 inhibitor transfection, compared with the controls (P < 0.01); C and D: Over-expression of miR-622 promotes increased cell invasion and migration compared with the miR-control transfectants. However, down-regulation of miR-622 inhibited cell invasion and migration compared with AS-control transfectants. bP < 0.01, compared with miR-control transfectants. Scale bars: 500 μm. AS-miR-622: An ambion miRNA inhibitor for miR-622.
Ectopic expression of miR-622 promotes invasion and migration of gastric cancer cells in vitro
We examined cell invasion and migration ability using the transwell invasion (Figure 2C) and scratch healing assays (Figure 2D). The number of invasive cells in the miR-622 precursor-transfected sample was significantly increased compared with the control (731.51 ± 3.16 vs 362.24 ± 5.18, P < 0.01). MiR-622 precursor-transfected cells healed the wound 48 h after scratching, whereas control-transfected cells were unable to heal the wound. The mean wound distances of the experimental sample and the control 48 h after scratching were significantly different (11.23 ± 7.19 μm vs 176.31 ± 7.24 μm, P < 0.01). Conversely, miR-622 inhibitor-transfected cells showed inhibited invasion (127.31 ± 11.62 vs 329.18 ± 5.18, P < 0.05) and migration ability (384.06 ± 9.35 μm vs 176.31 ± 7.24 μm, P < 0.01) compared with the control group (Figure 2C and D).
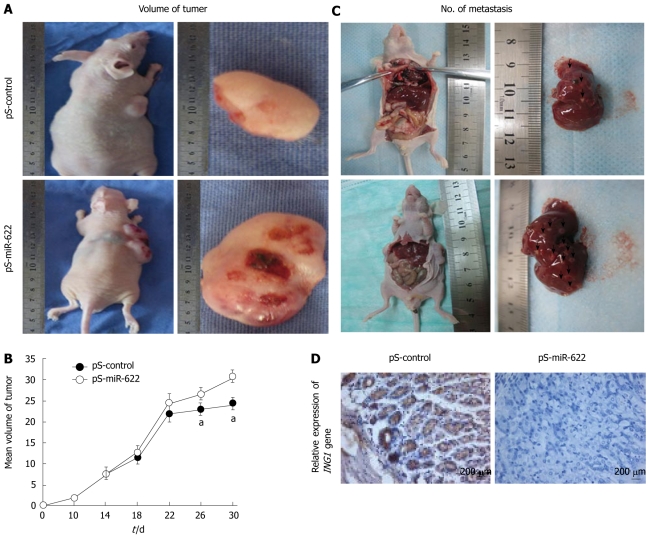
Ectopic expression of miR-622 promotes tumorigenesis and metastasis in vivo
We then tested whether ectopic expression of miR-622 could promote tumor growth in vivo. We constructed a miR-622 expression vector (pS-miR-622) and selected SGC-7901 cells stably transfected with pS-miR-622. Subsequently, pS-miR-622-transfected SGC-7901 cells and pS-control-transfected SGC-7901 cells were injected subcutaneously into nude mice, and tumor formation was monitored. After 30 d, the animals were euthanized, and the tumor volume was measured. Tumor growth was significantly promoted in the mice injected with pS-miR-622-transfected SGC-7901 cells compared with pS-control-transfected SGC-7901 cells (Figure 3A and B). The average tumor volume of mice inoculated with pS-miR-622-transfected SGC-7901 cells at day 30 was 29.91 ± 2.13 cm3, which was significantly larger than that of mice inoculated with pS-control-transfected SGC-7901 cells (23.81 ± 1.95 cm3, P < 0.05, Figure 3A and B). Thus, miR-622 can promote tumorigenesis in vivo. We found that ectopic expression of miR-622 promoted migration and invasion of gastric cancer cells in vitro; therefore, we further tested whether ectopic expression of miR-622 could affect tumor metastasis in vivo. SGC-7901 cells stably transfected with pS-miR-622 or pS-control vector were injected into 4 -wk-old male nude mice through the tail vein. One month after the injection, the mice were euthanized. The average number of hepatic metastatic nodes per mouse was much larger in mice injected with SGC-7901 cells stably transfected with pS-miR-622 vector than that in the control group (3 ± 0.68 vs 12.32 ± 0.21, P < 0.01) (Figure 3C). In addition, immunohistochemical analysis confirmed that the ING1 protein was down-regulated in pS-miR-622-transfected SGC-7901 cells compared with pS-control-transfected SGC-7901 cells (Figure 3D). Thus, these results indicate that miR-622 has the ability to promote metastasis of gastric cancer cells in vivo, which is consistent with the data obtained from the in vitro migration and invasion assays.
Figure 3.
MiR-622 promotes tumorigenesis and metastasis in vivo. A: Photographs of tumors in nude mice resulting from injection of miR-622 expression vector (pS-miR-622) or control plasmid (pS-control) stably transfected SGC-7901 cells; B: Tumorigenesis curves. Rapid tumor growth was observed in the pS-miR-622 group (data expressed as mean ± SD; P < 0.05); C: Photographs of the number of liver metastatic nodes in nude mice injected with pS-miR-622 or pS-control vector stably transfected SGC-7901 cells. The arrows indicate liver metastatic nodes; D: Histological examination found that the expression of ING1 was markedly reduced in the pS-miR-622-transfection group compared with the control groups. aP < 0.05, compared with miR-control transfectants. Scale bars: 200 μm. pS-control: Control plasmid; pS-miR-622: miR-622 expression vector.
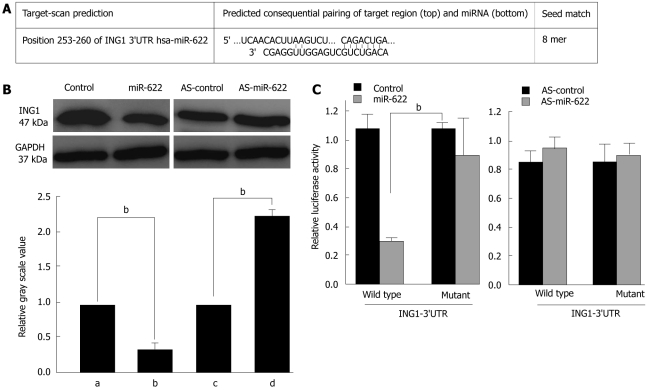
MiR-622 directly inhibits the expression of ING1 through ING1 3’UTR
Bioinformatics analysis identified several candidate miR-622 target genes. As shown in Figure 4A, a target sequence for miR-622 was found in the 3’UTR of ING1 at position 253-260 nt. To test whether the 3’UTR of ING1 is a functional target of miR-622, we engineered a reporter plasmid containing the wild-type 3’UTR of ING1 in the 3’ position of the firefly luciferase reporter gene. In parallel, we engineered another reporter plasmid in which the conserved target sequence within nt253-260 was specifically mutated, a modification that was predicted to inhibit miR-622-nt253-260 interactions (Figure 4A). To investigate the influence of miR-622 on ING1 expression, we searched for changes in ING1 protein and mRNA levels in HEK 293T cells transfected with the miR-622 precursor or the miR-622 inhibitor or in control-transfected cells. As shown in Figure 4B, miR-622 precursor transfection led to a significant decrease in ING1 protein levels (P < 0.05, Figure 4B). In contrast, miR-622 inhibitor transfection did not significantly alter ING1 protein levels compared with control-transfected cells.
Figure 4.
MiR-622 binds to the 3’UTR of Inhibitor of growth family, member 1 to post-transcriptionally repress the protein levels. A: Representative nucleotide sequence matches between possible target genes and miRNAs. Putative binding site of miR-622 in the ING1 3’UTR region (detected by TargetScan). Only matched nucleotides with miRNA seed sequences are indicated with vertical lines; B: Effects of miR-622 on ING1 protein levels. HEK 293T cells were transfected with 100 nmol/L of miR-622 precursor or miR-622 inhibitor. Whole-cell lysates were prepared 72 h after transfection and subjected to immunoblot analysis using antibodies against ING1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Relative gray scale values were compared with GAPDH; C: Luciferase reporter assay. A total of 1 × 105 HEK 293T cells were transfected with 100 ng wild-type-UTR-reporter or mutant-UTR-reporter constructs together with 100 nmol/L of miR-622 precursor or miR-622 inhibitor as indicated. bP < 0.01. Each bar represents mean values ± SE from three independent experiments. ING1: Inhibitor of growth family, member 1; AS-miR-622: An ambion miRNA inhibitor for miR-622.
These findings suggest an interaction between miR-622 and ING1 that we further investigated by luciferase assay analysis. To show a direct interaction between miR-622 and the 3’UTR of ING1, we cloned the 3’UTR region that is predicted to interact with miR-622 into a luciferase reporter vector. We then assessed luciferase activity by co-transfection the luciferase reporter vector bearing the 3’UTR of ING1 with the miR-622 precursor or the miR-622 inhibitor or with control plasmids. Luciferase activity was markedly diminished in the cells transfected with the miR-622 precursor and wild-type 3’UTR reporter plasmid, compared with the cells transfected with the miR-622 precursor and mutant 3’UTR reporter plasmids (Figure 4C, P < 0.05). Conversely, a significant increase in luciferase activity was observed after transfection with miR-622 inhibitor (Figure 4C). Taken together, these data imply that miR-622 may attenuate the expression of ING1 by directly targeting the ING1 3’UTR.
DISCUSSION
Invasion and metastasis, two of the most important hallmarks of cancer, are the leading factors of malignant cancer that lead to lethality, especially for gastric cancer. The long-term survival of patients with gastric cancer after curative resection is confounded by a high recurrence rate, which is mainly due to the spread of lymphatic metastasis[4,21]. Therefore, the identification of metastatic factors and an understanding of the underlying molecular pathways involved in the progression of metastasis are critical issues. Recent studies have shown that miRNAs play a fundamental role in the invasion and metastasis of gastric cancer[15,16,18], thereby opening a novel avenue to investigate the molecular mechanism of gastric cancer progression and to develop potential therapeutics against gastric cancer.
Prior to this study, very little was known about miR-622 expression in gastric carcinoma and its correlation with the clinicopathologic features of these patients. To address these issues, miR-622 expression levels and the clinicopathologic characteristics of 57 patients with gastric cancer were examined. There was a significant association between miR-622 expression and differentiation as well as lymphatic metastasis. However, no relationship was found between miR-622 expression and gender, tumor size, differentiation grade, distant metastasis or TNM stage. Given that the expression levels of miR-622 were significantly associated with lymphatic metastasis and differentiation, we speculated that up-regulation of miR-622 might promote the malignant phenotypes of gastric cancer cells. Therefore, we used miR-622 precursor and inhibitor constructs to perform miR-622 gain-of-function or loss of-function studies in human gastric cells. Invasion and migration ability assays revealed that miR-622 over-expression induced invasion and migration of gastric cancer cells in vitro. Conversely, miR-622 inhibitor can inhibit invasion and migration. These findings suggest that miR-622 functions as a key mediator of cell invasion and migration in gastric cancers. Cell migration plays an important role in many diverse biological processes[22]. Aberrant activation of cell migration in neoplastic cells results in tumor metastasis, which is the principal event leading to death in the majority of cancer patients[5].
We observed that miR-622 promotes cell invasion and migration ability in gastric cancers; however it was unclear whether up-regulation of miR-622 also affects gastric cancer cell tumorigenesis and metastasis. Therefore, we constructed specific miR-622 plasmids and established permanent transfected cell lines to investigate the potential role of miR-622 in tumorigenesis and metastasis of gastric cancer. We showed, for the first time, that miR-622 over-expression significantly induced tumorigenesis and metastasis of gastric cancer cell lines in nude mice. Cellular tumorigenesis assays and metastasis assays both in vitro and in vivo, revealed that miR-622 over-expression resulted in the promotion of tumorigenesis and metastasis of SGC7901 cells. These findings lend evidence that miR-622 may indeed function as a key mediator of cell tumorigenesis and metastasis and that it will be a promising target for gastric cancer treatment.
The fundamental function of miRNAs is to regulate target genes by direct cleavage of the mRNA and/or by inhibition of protein synthesis, according to the degree of complementarity with the target mRNA 3’UTR[23]. Computational algorithms have been the major driving force in predicting miRNA targets, which are based mainly on base pairing of miRNAs and target gene 3’UTRs[24]. To explore the molecular mechanism underlying miR-622 function, we searched for its direct target genes using bioinformatics analysis of miRNA-mRNA 3’UTR matching. Among the putative targets for miR-622, ING1 was detected by the TargetScan program. This gene encodes a tumor suppressor protein that can induce cell growth arrest and apoptosis; the encoded protein is a nuclear protein that physically interacts with the tumor suppressor protein TP53 and is a component of the p53 signaling pathway[25-28]. The bioinformatics analysis revealed that the conserved binding sites on ING1 that can be recognized by miR-622 are located in the 3’UTR. To test this assumption, we investigated whether miR-622 affects ING1 protein levels. We found that miR-622 leads to a significant decrease in ING1 protein levels, suggesting that ING1 is a functional target of miR-622. Lastly, results from our dual-luciferase reporter assays suggest that ING1 is a functional downstream target of miR-622.
In summary, we have shown that expression levels of miR-622 in gastric cancer tissues correlates with the differentiation and lymphatic metastasis. Using gain-of-function and loss-of-function studies, we demonstrated that up-regulation of miR-622 expression promotes gastric cancer cell invasion, tumorigenesis and metastasis both in vitro and in vivo. Therefore, miR-622 is likely to play an important role in tumorigenesis and metastasis of human gastric cancer and is a promising molecular target for gastric cancer therapy.
COMMENTS
Background
Gastric cancer is a worldwide cancer with poor prognosis. Identification of diagnostic biomarkers and effective therapeutic targets is important in the treatment and diagnosis of gastric cancer. Recently, researchers discovered a novel class of short, endogenous non-coding RNAs, called microRNAs (miRNAs), in plants, animals, and humans. It is clear that miRNAs play pivotal roles in a wide array of biological processes, including cell proliferation, differentiation and apoptosis.
Research frontiers
MiRNAs are 18-22 nucleotides, non-coding RNAs that regulate gene expression in a post-transcriptional manner. The fact that miRNAs are widely expressed in various species and tissues indicates that miRNAs may play an essential role in cell growth, differentiation, apoptosis as well as carcinogenesis. MiRNAs regulate the expression of protein-coding genes by degrading target mRNAs or by inhibiting gene translation. Emerging evidence strongly suggests that abnormal miRNA expression is a common and important feature of human gastric cancer.
Innovations and breakthroughs
The present study demonstrated that miR-622 is associated with differentiation and lymphatic metastasis in human gastric cancer. Exogenous expression of miR-622 promotes invasion, tumorigenesis and metastasis, both in vitro and in vivo. Using bioinformatics analysis, the authors identified the inhibitor of growth family, member 1 (ING1) as a putative miR-622 target. Subsequent experiments confirmed that up-regulation of miR-622 represses the expression of ING1 at the translational level.
Applications
In this study, the expression of miR-622 was found down-regulated in gastric cancer. MiR-622 was involved in differentiation and lymphatic metastasis in human gastric cancer. The ectopic expression of miR-622 promoted invasion, tumorigenesis and metastasis of gastric cancer cells both in vitro and in vivo. ING1 was a direct target of miR-622. These findings help clarify the molecular mechanisms involved in gastric cancer metastasis and indicate that miR-622 modulation may be a bona fide treatment of gastric cancer.
Terminology
MiRNAs are 18-22 nucleotides, non-coding RNAs that regulate gene expression in a post-transcriptional manner. ING1 is an inhibitor of growth family, member 1. This gene encodes a tumor suppressor protein that can induce cell growth arrest and apoptosis. The encoded protein is a nuclear protein that physically interacts with the tumor suppressor protein TP53 and is a component of the p53 signaling pathway.
Peer review
The authors evaluated the possible pathogenetic role of miR-622 in metastasis formation and invasiveness of gastric cancer using not only human samples and miRNA expression arrays, but also functional assays on cell lines and mouse animal model. The topic is of significant clinical importance and their results may be used in clinical research of gastric cancer, and it may give some evidences for new therapeutic strategies.
Acknowledgments
We thank the colleagues from our central laboratory of Shandong Provincial Hospital affiliated to Shandong University for their excellent technical support.
Footnotes
Supported by Grants from Science Foundation of Shandong Province of China (2003-23), Key Research Project from Shandong Science and Technology Commission, No. 2005GG3202066
Peer reviewer: Ferenc Sipos, MD, PhD, Cell Analysis Laboratory, 2nd Department of Internal Medicine, Semmelweis University, Szentkirályi u. 46., Budapest 1088, Hungary
S- Editor Tian L L- Editor Ma JY E- Editor Ma WH
References
- 1.Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010;14:302–308. [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Coburn NG. Lymph nodes and gastric cancer. J Surg Oncol. 2009;99:199–206. doi: 10.1002/jso.21224. [DOI] [PubMed] [Google Scholar]
- 5.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 10.Bueno MJ, Pérez de Castro I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143–3148. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- 11.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 16.Li Z, Zhan W, Wang Z, Zhu B, He Y, Peng J, Cai S, Ma J. Inhibition of PRL-3 gene expression in gastric cancer cell line SGC7901 via microRNA suppressed reduces peritoneal metastasis. Biochem Biophys Res Commun. 2006;348:229–237. doi: 10.1016/j.bbrc.2006.07.043. [DOI] [PubMed] [Google Scholar]
- 17.Saito Y, Suzuki H, Hibi T. The role of microRNAs in gastrointestinal cancers. J Gastroenterol. 2009;44 Suppl 19:18–22. doi: 10.1007/s00535-008-2285-3. [DOI] [PubMed] [Google Scholar]
- 18.Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y, et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6:e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 20.Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- 21.D’Ugo D, Persiani R, Zoccali M, Cananzi F, Vigorita V, Mazzeo P, Tufo A, Biondi A. Surgical issues after neoadjuvant treatment for gastric cancer. Eur Rev Med Pharmacol Sci. 2010;14:315–319. [PubMed] [Google Scholar]
- 22.Vicente-Manzanares M, Sancho D, Yáñez-Mó M, Sánchez-Madrid F. The leukocyte cytoskeleton in cell migration and immune interactions. Int Rev Cytol. 2002;216:233–289. doi: 10.1016/s0074-7696(02)16007-4. [DOI] [PubMed] [Google Scholar]
- 23.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 25.Gunduz M, Demircan K, Gunduz E, Katase N, Tamamura R, Nagatsuka H. Potential usage of ING family members in cancer diagnostics and molecular therapy. Curr Drug Targets. 2009;10:465–476. doi: 10.2174/138945009788185086. [DOI] [PubMed] [Google Scholar]
- 26.Gunduz M, Gunduz E, Rivera RS, Nagatsuka H. The inhibitor of growth (ING) gene family: potential role in cancer therapy. Curr Cancer Drug Targets. 2008;8:275–284. doi: 10.2174/156800908784533454. [DOI] [PubMed] [Google Scholar]
- 27.Shah S, Smith H, Feng X, Rancourt DE, Riabowol K. ING function in apoptosis in diverse model systems. Biochem Cell Biol. 2009;87:117–125. doi: 10.1139/O08-107. [DOI] [PubMed] [Google Scholar]
- 28.Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R. The new tumor suppressor genes ING: genomic structure and status in cancer. Int J Cancer. 2008;123:1483–1490. doi: 10.1002/ijc.23790. [DOI] [PubMed] [Google Scholar]