Abstract
The dynamic chromatin activities of Mi-2/Nucleosome Remodeling and Histone deacetylation (Mi-2/NuRD) complexes in mammals are at the basis of current research on stemness, longevity/ageing, and cancer (4-2-1/SLAC), and have been widely studied over the past decade in mammals and the elegant model organism, Caenorhabditis elegans. Interestingly, a common emergent theme from these studies is that of distinct coregulator-recruited Mi-2/NuRD complexes largely orchestrating the 4-2-1/SLAC within a unique paradigm by maintaining genome stability via DNA repair and controlling three types of transcriptional programs in concert in a number of cellular, tissue, and organism contexts. Thus, the core Mi-2/NuRD complex plays a central role in 4-2-1/SLAC. The plasticity and robustness of 4-2-1/SLAC can be interpreted as modulation of specific coregulator(s) within cell-specific, tissue-specific, stage-specific, or organism-specific niches during stress induction, ie, a functional module and its networking, thereby conferring differential responses to different environmental cues. According to “Occam’s razor”, a simple theory is preferable to a complex one, so this simplified notion might be useful for exploring 4-2-1/SLAC with a holistic view. This thought could also be valuable in forming strategies for future research, and could open up avenues for cancer prevention and antiageing strategies.
Keywords: stemness, longevity, ageing, cancer, 4-2-1/SLAC, Mi-2/NuRD, mammals, Caenorhabditis elegans
Introduction
The average human lifespan has increased dramatically in the past century, and many people are now living long enough to suffer from a range of predominantly age-related diseases, for example, certain cancers, neurodegenerative conditions, and Type 2 diabetes. In addition to genetic mutations, a number of environmental factors, such as chemicals and radiation, may contribute to the development of these disorders.1–4 However, the human dream of finding the “fountain of youth” is now becoming more achievable in light of recent advances in a number of areas, in particular, stemness, longevity/ageing, and cancer (4-2-1/SLAC).
How can we live healthier, longer, and happier lives? A better understanding of the underlying mechanisms of the Mi-2/nucleosome remodeling and histone deacetylation (Mi-2/NuRD) complex in 4-2-1/SLAC could shed some light on this question. The spatiotemporal chromatin remodeling activities of Mi-2/NuRD, a family of protein complexes, form the basis of both normal and abnormal pathways of 4-2-1/SLAC during development and differentiation, although it is likely that they work alongside other remodeling complexes, including esBAF, PcG, and HP1/Rb.5,6
The family of Mi-2/NuRD complexes has a number of core polypeptides, each of which has a distinct context-dependent role. Some components of the Mi-2/NuRD complexes are important in gene regulation and DNA repair, and are uniquely characterized by their adenosine triphosphate (ATP)-dependent chromatin remodeling, histone deacetylase, and demethylase activities, and higher order chromatin organization. These complexes simultaneously modulate all three types of transcriptional programs, ie, regulation of transcription factors in subsets of genes, epigenetic regulation via DNA methylation and histone modification, and regulation involving higher order chromatin organization.6–9
In eukaryotic cells, the nucleus is vital for many cellular functions. The chromatin nucleosomes that pack DNA generally inhibit processes that require access to the DNA template, such as DNA transcription and repair. The structure of chromatin changes dynamically during development and carcinogenesis. 5 Chromatin remodelers are highly specialized enzymes that are responsible for a number of decisions regarding the fate of cells, based on specific inheritance and cell-cell or cell-environment interactions during development of the organism.10,11 The core Mi-2/NuRD complex, which was biochemically isolated by a number of laboratories more than a decade ago, has been shown to use ATP hydrolysis to alter the positions of nucleosomes within DNA. Two highly homologous proteins, Mi-2α/CHD3 and Mi-2β/CHD4, represent the catalytic ATP-hydrolyzing subunits in the complex. In addition, the core of the Mi-2/NuRD complex is so far known to contain histone deacetylases (HDAC1 and HDAC2), Rb-associated proteins (RBBP4/RbAp48 and RBBP7/RbAp46), metastasis-associated proteins 1–3 (MTA1–3), p66α/GATAD2a and β/GATAD2b, methyl-CpG-binding proteins (MBD2 and MBD3), and histone 3 lysine-specific demethylase 1 (LSD-1).6,12 The epigenetic regulation activities of Mi-2/NuRD complexes systematically reprogram and coordinate differential biological networking in response to subtle changes in environmental and microenvironmental cues.
Both stem cells and cancer cells have the capacity for long-term proliferation. The self-renewing adult stem cell population and the germline stem cell population afford an opportunity to elucidate the mechanisms that inhibit cellular senescence and promote continuous growth and division. Systematic ectopic expression of stem-like genes and their products might contribute to longevity via the chromatin remodeling activities of Mi-2/NuRD complexes,13 although a defect in these complexes accelerates ageing, as in the progeria syndrome.14 Cancer stem-like cells have the capacity for additional self-renewal or immortality, and promote prolonged cell longevity. Overexpression of a “cocktail” of stemness factors in induced pluripotent stem cells (iPSCs) systematically reprograms the differentiated cells that inherently have a limited lifespan and gives them stem-like characteristics, including self-renewal and pluripotency.15
The Mi-2/NuRD family complex is a key player in several closely related developmental processes, such as longevity, ageing, stemness (eg, iPSCs, hematopoietic stem cells, embryonic stem cells, germline stem cells, and cancer stem cells) and cancer.8 Distinct Mi-2/NuRD complexes link several transcriptional regulatory processes, including histone deacetylation, histone demethylation, nucleosome mobilization, and higher order chromatin organization, as well as recruiting other transcription factors. Many Mi-2/NuRD complexes are also modulatory corepressors/coactivators comprising multiple protein subunits that are linked directly with SLAC in different organisms.
Caenorhabditis elegans, one of most powerful model organisms in biology, has the number of protein-coding genes that are largely the same as those in mammals, and mostly encode protein functions similar to their mammalian equivalents. This model has provided us with many important discoveries, including programmed cell death,16 RNA interference,17 development of the green fluorescent protein,18 and micro RNAs.19 These discoveries have contributed greatly to our understanding of 4-2-1/SLAC biology. Importantly, C. elegans is among the best models we currently have for deciphering human longevity and ageing,20,21 and it has gained increasing popularity as a genetic model for cancer research.22,23 In fact, micro RNAs were first discovered during genetic screening for regulators of developmental timing using the stem cell-like seam lineage in C. elegans.24 Therefore, C. elegans is a particularly attractive model for the study of mammalian 4-2-1/SLAC, with the additional advantages of simplicity and cost-effectiveness. Finally, use of this model is increasing our understanding of 4-2-1/SLAC, and could provide avenues to achieve healthy longevity as well as potential therapeutic treatments for cancer.
Mammalian Stemness Factors and Mi-2/NuRD Complexes
Many transcriptional regulators interact with the core Mi-2/NuRD complex during development (Fig. 1). The constantly increasing number of known stemness factors has been shown to be physically associated with distinct Mi-2/NuRD complexes, including Oct4, Nanog, c-myc, and Esrrb6,25–27 (Fig. 2). Oct4 is expressed in the germline and its expression is clearly associated with germ cell malignancy.28 Several studies have also identified Oct4 expression in adult somatic stem cells and soma-derived malignancy.29,30 Oct4 regulates the circuitry governing embryonic stem cell pluripotency, particularly the genes encoding Oct4, Sox2, and Nanog.31 These transcription factors mutually enforce expression of each other in a self-sustaining network that helps to maintain a pluripotent state. Like Oct-4, c-myc is one of a cocktail of stemness factors for iPSCs.32 Oct4 can both activate and repress transcriptional targets in mouse and human embryonic stem cells. The MTA1/Mi-2/NuRD complex may have an essential role in pluripotency during normal development.25 Sall4, an Oct4 partner, and other members of the Spalt-like family of transcriptional cofactors have been shown to associate with the Mi-2/NuRD complex33 in mammals (Figure 3).
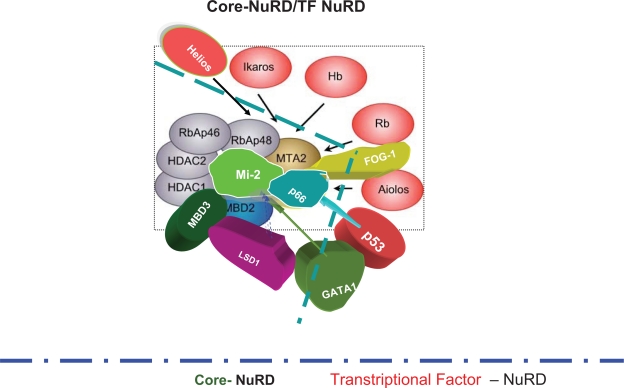
Figure 1.
The core NuRD complex and its transcriptional factors which recruit the core NuRD complex to the promoter of the respective target genes.6
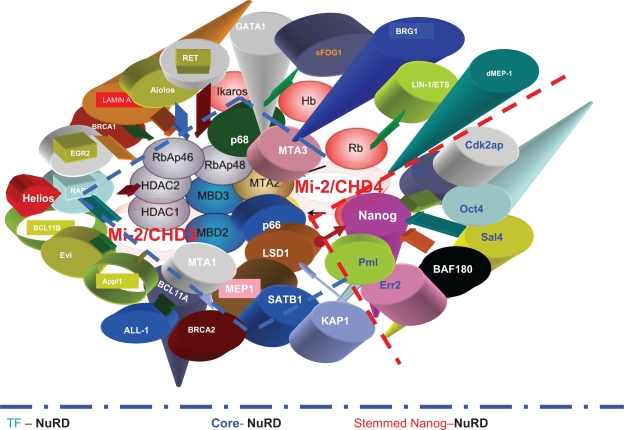
Figure 2.
The core NuRD complex, transcriptional factors NuRD, and stemness factors for Nanog/NuRD. Stemness factors such as Nanog, Oct4, c-myc, and sall4 are associated with the core NuRD complex in some contexts.6
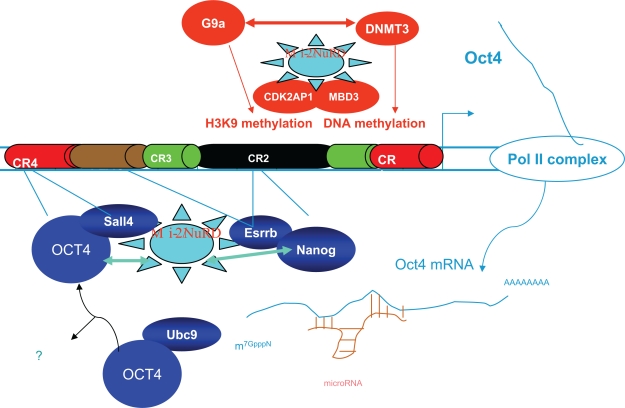
Figure 3.
Interaction and regulation of the Mi-2/NuRD complex on Oct4. Mi-2/NuRD complexes could regulate the activity of Oct4 by mechanisms in transcription at multiple points. Points of regulation by different activities are shown by colored lines. Factors in green and blue (such as Oct4 itself) carry out positive regulatory activities, including H3K4 methylation; those in red act in a negative capacity (eg, DNA and H3K9 methylation). Protein-protein interactions between Mi-2/NuRD with others are shown with arrows or “rays” on a star. CR1–4 are Oct4 promoter and enhancer regions with conservation. Reprinted from Trends Biochem Sci, 2009;34, Kanga J, Shakyaa A, Tantina D., Stem Cells, Stress, Metabolism and Caner: A drama in two Octs, 1–9, copyright 2009, with permission from Elsevier.
Mi-2β/NuRD Complex in SLAC
The Brg1 or Brm ATPases are the catalytic subunit in the Brg/Brahma-associated factor (BAF) complexes that are critical in embryonic stem cell self-renewal, 35 including an embryonic stem cell-specific complex called esBAF.36,37 The esBAF complex appears to play a direct role in mediating the gene regulatory functions of several core embryonic stem cell transcription factors. Like esBAF subunits, the core subunits of the Mi-2/NuRD complex interact directly with the transcription factors Oct4, Sall4, and Nanog.25,37 BRA-1, the C. elegans homolog of BRAM1/BS69 in mammalian cells, has been found to be associated with LET-418/Mi-2β (Brunschwig and Mueller, personal communication, 2006, European Worm meeting), which is the central component of the Mi-2/NuRD complex. Interestingly, a recent study shows that components of the BAF complex could enhance reprogramming.38 However, the core subunits of the Mi-2/NuRD complex, such as Mi-2β/CHD4, MTA3, MBD3, MTA1, and p66, are also among the pluripotent cell-enriched proteins identified using stable isotope labeling with amino acids in cell culture. This raises the possibility of the existence of another form of esBAF, the putative BRG1/BRAM1/Mi-2/NuRD8 or BRAM1/Nanog and Oct4-associated deacetylase (NODE) in mammals that has a similar chromatin remodeling role in reprogramming. Consequently, the stemnessed BRAM/Mi-2/NuRD could function to silence transcription alongside the esBAF complex,37 and maintaining its expression at adequate but not excessive levels.
Mi-2β-Deficient Mi-2/NuRD Complex and Stemness in Mammals
Chromatin remodeling is a leading factor in the differentiation of hematopoietic stem cells into various cells. A hyperdynamic “breathing” chromatin structure keeps hematopoietic stem cells in their pluripotent state,39 and the differentiation-oriented genes of the hematopoietic stem cell are maintained in a “primed” or “poised” state but can be rapidly activated on differentiation. 40 The Mi-2β/NuRD complex is highly expressed in hematopoietic stem cells.41 Targeted disruption of Mi-2β in hematopoietic stem cells has been shown to lead to overproduction of proerythroblasts without proper maturation.41 Importantly, specialized chromatins are necessary for self-renewal and differentiation of somatic stem cells, and this keeps stem cell-specific genes active and key differentiation factors repressed but poised for activation. The Mi-2/NuRD complex could function as a corepressor and coactivator.
Mi-2/NuRD Complex Deficiency and Derepression of Germline Stem Cell Markers in C. elegans
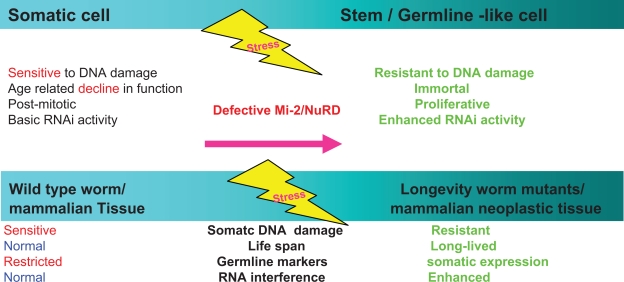
An understanding of how germ cell traits are normally repressed in the soma will improve our understanding of the complex links between stemness and oncogenesis. Components of the Mi-2/NuRD complex, including LET-418/Mi-2β, HDA-1/HDAC-1, and an associated Zn-finger protein, MEP-1/KLF4, are required for maintaining somatic differentiation in C. elegans. In mutant animals that have lost the function of MEP-1 and LET-418, germline-specific genes become derepressed in somatic cells, and the Polycomb group (PcG) and SET domain-related proteins MES-1–4/SET and the human mortality-related gene (MRG)-1, a chromatin-associated protein, are required for this ectopic expression.42–46 This suggests that Mi-2/NuRD complexes normally antagonize the activities of MES-1–4 and MRG-1 in the soma and prevent germline gene expression. However, during embryogenesis, pharyngeal and intestinal in excess Zn-finger (PIE-1) physically associates with members of the MEP-1/KLF4/NuRD complex in primordial germ cells to inhibit their activity42 and to maintain the pluripotency of germ cells. At later times, the MEP-1 and LET-418 complexes remodel chromatin to establish new stage-specific or cell-specific differentiation potential.42
The soma ages during the lifespan and the germline is immortal. Genomic instability in somatic cells increases with age, and this decline in somatic maintenance might be regulated to facilitate resource reallocation towards reproduction at the expense of cellular senescence. C. elegans mutants with increased longevity exhibit a soma-to-germline transformation of gene expression programs that is normally limited to the germ line. Decreased insulin-like signaling causes somatic misexpression of the germline-limited pie-1 and p-granule-like pgl family of genes in intestinal and ectodermal tissues. The fork-head boxO1A (FOXO) transcription factor, DAF-16, in insulin-like46 signaling directly binds to the pie-1 promoter, thereby regulating pie-1 expression. The somatic tissues of insulin-like mutants are more germline-like and protected from genotoxic stress. Gene inactivation of components of the chaperonin complex that induces increased longevity also causes somatic misexpression of P-granule-like 1 (PGL-1) abnormality. These results indicate that the acquisition of germline characteristics by the somatic cells of C. elegans mutants with increased longevity contributes to their increased health and survival.13 Histone demethylase LSD-1, the recently identified subunit of the Mi-2/NuRD complex, regulates neural stem cell proliferation.47
Deficient Mi-2/NuRD Complexes, GLP-1/Notch Signaling, and Germ Cell Stemness in C. elegans
In humans, Notch and epidermal growth factor receptor pathway interaction can regulate the abundance and self-renewal of neural stem cells.48 In C. elegans, Notch and epidermal growth factor receptor pathways typically show “love and hate” cross-talk in vulval development.49,50 In addition, a Notch-like signal from the distal tip cell is both necessary and sufficient for controlling germ stem cell self-renewal and proliferation. The distal tip cell expresses a delta-like Notch ligand, LAG-2, and the mitotic germ cells express a Notch-type receptor, GLP-1. Along with other research laboratories, we have observed that the loss-of-function LET-418/Mi-2β/NuRD derepresses the expression of the LAG-2/delta ligand in the C. elegans intestine.6,51,52 The distal tip cell provides a niche for germline stem cells. The GLP-1/Notch receptor maintains the germ cells in the stem cell state. LAG-2/delta binding triggers cleavage of GLP-1/Notch for transport to the nucleus, where it forms protein complexes with other transcription factors, LAG-1/CSL (CBF-1, Su(H), Lag-1) and LAG-3/Mastermind, which possibly tethers LAG-1/CSL and the cleaved GLP-1/Notch to control target gene expression. Loss of GLP-1/Notch and LAG-2/delta function causes germ stem cell loss and consequently premature entry into meiosis. In contrast, glp-1(ar202gf ) mutation GLP-1/Notch activity causes development of germ cell overproliferation tumors. GLD-1/NOS facilitates meiosis by blocking translation of glp-1 mRNA. However, mutation of the DAF-2/insulin receptor inhibits the tumor growth in the gld-1 mutant, conferring longevity and freedom from cancer.22
MBD-3/Mi-2/NuRD Complex in SLAC
MBD3 knockout embryonic stem cells are viable but unable to form a stable NuRD complex, and also differentiate incorrectly. The Mi-2/NuRD complexes play a crucial role in embryonic stem cell self-renewal and pluripotency. Pluripotency collapses during embryogenesis when cells commit to specific developmental programs. MBD3 knockout embryonic stem cells cannot completely silence genes expressed before implantation of the embryo. MBD3 knockout embryonic stem cells have been shown to self-renew without leukemia-inhibitory factor, and to be able to differentiate into embryoid bodies or chimeric embryos, but fail to become developmental lineages.53 The inner cell mass of MBD3-deficient blastocysts cannot develop into mature epiblasts after implantation. Furthermore, loss of MBD3 affects embryonic stem cell differentiation and alters cell types produced during differentiation. A number of genes show stage-specific expression in inner cell mass cells during preimplantation development, and MBD3 is required for proper gene expression patterns in preimplantation and peri-implantation embryos as well as in embryonic stem cells.54 Knockout or knockdown of MBD3 in embryonic stem cells causes dysregulation of a number of genes, including derepression of the expression of trophectoderm marker genes, ie, Cdx2, eomesodermin, and Hand1 in undifferentiated embryonic stem cells, along with an elevated acetylation level of histone 3 in promoters of the respective genes. Thus, MBD3 helps to restrict embryonic stem cells from differentiating towards the trophectoderm lineage, and is a key epigenetic player in maintaining full pluripotency in mouse embryonic stem cells.55
In particular, one isoform of the NuRD complex, the NODE complex, which lacks MBD3 and RBBP7, represses expression of developmentally regulated genes in embryonic stem cells. MTA1 knockdown showed different expression profile changes from those of MBD3 knockdown or knockout, suggesting that they have some different functions. Unlike MBD3 loss, which inhibits embryonic stem cell differentiation, MTA1 knockdown results in upregulation of differentiation genes of multiple lineages, as well as embryonic stem cell differentiation.25
Lamin A/Mi-2/NuRD Complex in SLAC
Nuclear lamins comprise the nuclear lamina, a scaffold-like structure underneath the inner nuclear membrane. Lamins play a role in adult stem cell differentiation, aging, tumorigenesis, DNA replication, and chromatin organization.56 Lamin A has important roles in transcription, as highlighted by its binding to RNA polymerase II, RNA splicing factors, and a number of known transcription factors. The lamina is involved in transcription.57 Disorganization of the lamina with dominant-negative Lamin A mutants inhibits DNA transcription by RNA polymerase II.58 Lamin A has been shown to interact with specific proteins that affect transcription, such as the retinoblastoma tumor suppressor.59 Lamin A forms a complex with core components of the Mi-2/NuRD complex6 (see below). In C. elegans, LIN-35/Rb was proposed to recruit the LET-418/Mi-2β/NuRD complex during vulval development. Intranuclear lamina foci also colocalize with RNA splicing factors to organize the RNA processing machinery.
Lamin A-Deficient/Mi-2/NuRD Complex and Stemness
Hutchinson-Gilford progeria syndrome is caused by mutations in the gene LMNA, which encodes nuclear Lamins A and C.14 The in vivo physical interaction of Lamin A with RBBP4, RBBP7, and HDAC1 points to Lamin A as being a coregulator for the nuclear lamina and the Mi-2/NuRD complex in normal cells. Multiple Mi-2/NuRD components are lost in Hutchinson-Gilford progeria syndrome. Similar to RBBP4/7 knockdown, silencing of any subunit increased the percentage of cells lacking H3K9me3 and HP1γ heterochromatin foci. Furthermore, knockdown of HDAC1, MTA3, CHD3, or CHD4 in primary human fibroblasts increases the percentage of cells containing phosphor-H2AX-positive foci. Progerin causes the loss of RBB4/7, which is an early event in ageing-associated chromatin defects. Loss of any Mi-2/NuRD component and reduction of HDAC1 activity is sufficient to trigger several ageing-associated chromatin defects. Induction of progerin or knockdown of RBBP4 and RBBP7 results in changes in heterochromatin structure, followed by accumulated DNA damage. Loss of RBBP4/7 compromises the histone modifications and higher order chromatin structure, possibly making chromatin more susceptible to DNA damage. Similar to the effects after silencing RBBP4 and RBBP7, impairment of the H4K20 histone methyltransferase PR-Set7 in heterochromatin interferes with DNA replication and causes increased levels of DNA damage.6 Thus, the Mi-2/NuRD complex is a mediator of ageing-associated chromatin defects.
Unlike Lamin B, which is expressed in embryonic stem cells, Lamin A is expressed predominantly in differentiated cells and maintains the differentiated state. Lamin A is not expressed during mouse development before day 9 (phenotypically similar to no expression of CHD-3/Mi-2α in mouse oocytes and CHD-3/Mi-2α in early C. elegans embryos), nor in undifferentiated mouse embryonic carcinoma cells. Lamin A expression is activated during human embryonic stem cell differentiation before downregulation of Oct-3/4, but not before the downregulation of other pluripotency markers, such as Tra-1-60, Tra-1-81, and SSEA-4. Therefore, Lamin A expression is a marker of both mouse and human embryonic stem cell differentiation. 61 The expression of four pluripotency genes (Oct4, Sox2, c-myc, and Klf4) can reprogram fibroblasts into a pluripotent state.62–65 The expression of all these four pluripotency genes was induced when human somatic 293T cells were reprogrammed with extracts of mouse embryonic stem cells. Lamin A was removed from the nuclei.66 Lamin A-dependent misregulation of adult stem cells is associated with accelerated ageing. Progerin is also expressed in wild-type cells linked with physiological ageing. The expression of progerin activates major downstream effectors of the Notch signaling pathway. Induction of progerin in human mesenchymal stem cells alters their differentiation potential. Adult stem cell exhaustion and progressive dysfunction of tissue functions results in accelerated ageing in patients with Hutchinson-Gilford progeria syndrome, and possibly also physiological ageing.14
Lamin A-Deficient Mi-2/NuRD Complex, Transcription of Satellite III Repeats, and Stress Bodies
Nuclei in Hutchinson-Gilford progeria syndrome cells display a loss of heterochromatin. In cells from a female patient with Hutchinson-Gilford progeria syndrome, the facultative chromatin mark histone H3 trimethylated on lysine 27 (H3K27me3) targeted by EZH2, the pericentric constitutive heterochromatin mark histone H3K9me3, and an altered association of H3K9me3 with heterochromatin protein 1 alpha (HP1α) were shown to be downregulated. Interestingly, this loss of constitutive heterochromatin was accompanied by an upregulation of pericentric satellite III repeat transcripts. In contrast, there was an increase in the histone H4K20me3, an epigenetic mark for constitutive heterochromatin. Expression of LAΔ50 in normal cells induces changes in histone methylation patterns similar to those seen in Hutchinson-Gilford progeria syndrome cells.14
Dermal fibroblasts from Hutchinson-Gilford progeria syndrome expressing a mutant Lamin A have dysmorphic nuclei, hypersensitivity to heat shock, and delayed response to heat stress.67 During upregulation of satellite III transcripts in Hutchinson-Gilford progeria syndrome, in cells without heat shock stress (ie, at 37 °C), stress bodies are observed in nuclei regardless of the extent of their lobulation. However, the normal cell has stress bodies only after heat shock.
Lamin A-Deficient Mi-2/NuRD Complex, Cancer, and Cell Signaling Pathways
Lamina A is overexpressed in gynecological, breast, thyroid, lung, gastrointestinal, skin, colorectal, and genitourinary cancers. A putative Lamin A/Mi-2/NuRD complex regulates gene expression through an interplay with signal transduction pathways, such as the Notch pathway, transforming growth factor beta (TGF-β) pathway, Wnt/β-cantenin pathway, and ERK1/2 signaling pathway,56,68 as well as transcription factors and other chromatin-associated proteins, such as the HP1/Rb complex (Figs. 4 and 5). Premature ageing could prevent cancer69 and some other common age-related diseases, such as brain ageing, cataracts, Type 2 diabetes, and hyperlipidemia.
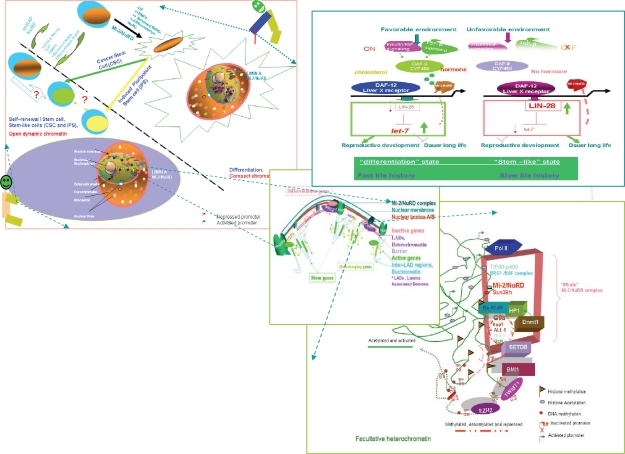
Figure 4.
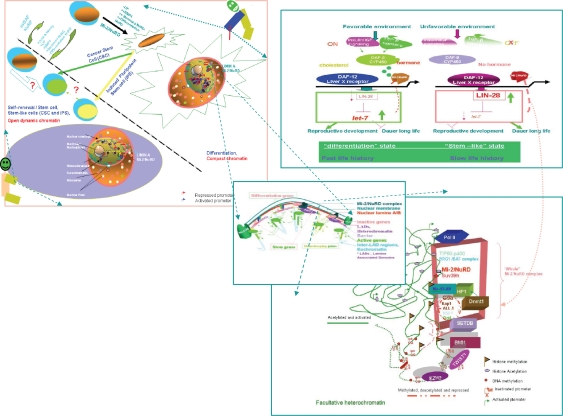
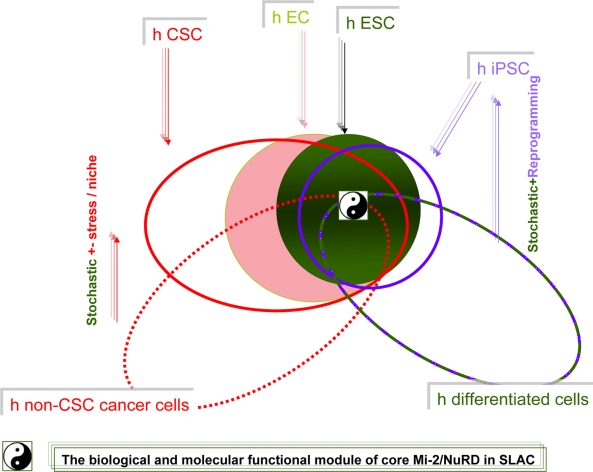
The comprehensive biological and molecular view of the Mi-2/NuRD complexes and SLAC in young mammals and nematode larvae. Upper left: In mammals, the commonality, difference, and mutually induced changes among induced pluripotent stem cells, cancer stem cells, embryonic stem cells and differentiated cells (a high resolution version is presented in Supplementary Fig. 2). Adapted by permission from Macmillan Publishers Ltd: Nature Cell Biology, 2006;8:285–92, copyright 2006.53 Upper right: DAF-12/Liver X receptor and lifespan in Caenorhabditis elegans. In favorable environments, the insulin/IGF-I and transforming growth factor-beta peptide signals converge on the DAF-12/Liver X receptor for the dauer pathways. Cholesterol comes to the DAF-9/cytochrome P450 and a hormone, similar to a sterol-derived dafachronic acid product. The DAF-12/Liver X receptor directs expression of the genes involved in reproductive development, developmental advancements, fat metabolism, and accelerated aging (fast life history traits) mediated by Let-7, LIN-28, and a feedback loop that the Mi-2/NuRD complex is postulated to be involved in. In unfavorable environments, hormonal pathways are suppressed. Unliganded DAF-12 specifies programs for dauer diapause, delayed development, fat storage, and retarded aging (slow life history traits). This process is also mediated by LET-7, LIN-28, and a feedback loop that the Mi-2/NuRD complex is postulated to be involved in (see Supplementary Fig. 3). Middle: Model of the dynamic pop-up and inward gene positioning of nuclear lamina-genome interactions during self-renewal and differentiation in Caenorhabditis elegans. Two major forces drive tissue-specific subnuclear organization of the worm genome, ie, repeat-induced heterochromatin, which associates with the nuclear envelope, and tissue-specific promoters that shift inward in a dominant fashion when they are activated. Tissue-specific promoters shift in a nondominant manner to the nuclear envelope in cells in which they are inactive (Supplementary Figs. 4A and 4B). Bottom: The dynamic interchanges between euchromatin and heterochromatin in stemness and differentiation states. The putative “all-in-one Mi-2/NuRD supercomplex is highlighted” other different “isoforms” of Mi-2/NuRD complexes, include at least the activator-only Mi-2/NuRD complexes and the repressor-only Mi-2/NuRD complexes (see Supplementary Fig. 5).
Figure 5.
The comprehensive biological and molecular view of the Mi-2/NuRD complexes and stemness, longevity/ageing, and cancer in adults in mammals and nematodes.
Lamin A-Deficient/Mi-2/NuRD Complex, Extracellular Matrix Target Genes, Premature Ageing, and Longevity
Lamin A deficiency causes dysregulation of many extracellular matrix genes in humans,70 as occurs in HDA-1 deficiency in C. elegans.71 Moreover, it is known that children with Hutchinson-Gilford progeria syndrome have about seven-fold accelerated premature ageing. Interestingly, however, in C. elegans, removing the reproductive system in daf-2 mutants extends the lifespan by six-fold. These animals look normal and remain active for many months.72 The genetic control of lifespan is conserved, and the long-lived model organisms could provide us with the means to “reverse” Hutchinson-Gilford progeria syndrome in the future. Lifespan and aging in mammals may vary with the efficiency of DNA repair and replacement of damaged tissue by stem cells.73
Mi-2/NuRD Complexes, Heat Shock, and Longevity/Ageing in C. elegans
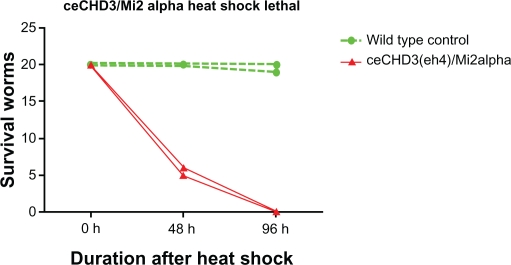
The genome of C. elegans encodes orthologs of major components of the vertebrate Mi-2/NuRD complex. The two Mi-2 homologs in C. elegans are LET-418/CHD4/Mi-2β, and CHD-3/Mi-2α, which share 59% identity with the human Mi-2 protein. In the absence of a maternal contribution, hermaphrodites homozygous for let-418 (loss-of-function) arrest their development at the L1 larval stage. Contrary to let-418, homozygous chd-3 worms have no obvious phenotype. However, a role for chd-3 becomes visible in let-418;chd-3 double mutants, which show a synthetic phenotype. Double mutants with a maternal let-418 contribution arrest at the L4 larval stage, whereas in the absence of a maternal let-418 contribution, they arrest as embryos.74 These organisms are small and unhealthy, and likely represent a progeria-like phenotype. The lin-35/Rb mutants are also dysfunctional and somewhat progeric. Strong loss-of-function alleles of let-418 function in distinct cell signaling pathways, eg, one targeting the promoter of lin-39/Hox via LIN-1/ETS during vulval development.74–76 The C. elegans mutant CHD-3/Mi-2α has an extreme heat stress survival phenotype (Fig. 6). For the knockout chd-3 (eh4) allele, without heat shock treatment, animals have wild-type behavior. However, after heat shock, most, if not all, are dead within two days. Males could comprise the rare survivors, and the exceptionally rare hermaphrodite will have an everted vulva and be sterile. However, probably due to low efficiency, the feeding RNA interference against chd-3 has not completely reproduced this phenotype. In addition, it is a knockout mutation allele and needs further investigation. However, several lines of evidence provide some indirect support for this conclusion. In several mammalian cancerous cell lines and in vitro, during stress, heat shock factor (HSF)-1 could interact with MTA1 and other components of the Mi-2α(CHD3)/NuRD complex.77 Furthermore, HSF-1 inhibitory RNA in C. elegans does not affect the appearance or behavior of young adults, but shortens the lifespan. An hsf-1 inhibitory RNA clone produces a striking progeric phenotype. The animals look old when they are young and they have very short life spans. Thus HSF-1 functions in normal worms to keep them from aging and dying.21
Figure 6.
The survival curve of Caenorhabditis elegans CHD-3 (eh4) after heat stress. Twenty experimental animals were used per group, ie, wild-type (N2) or CHD-3 knockout (FR355). Heat shock treatment is at 33 °C for six hours, and then incubation at 25 °C for the remainder of the experiment. If an animal could not respond to tactile stimulation, it was counted as “dead”. Experiments were repeated many times and a representative figure is shown here.
As aforementioned, human dermal fibroblasts in Hutchinson-Gilford progeria syndrome with the Lamin A G608G mutation are hypersensitive to heat stress.67 It is tempting to hypothesize that this may be functional through the Lamin A/Mi-2/NuRD complex. Finally, the formation of nuclear stress bodies starts soon after the onset of thermal stress with the association of HSF-1, with specific pericentromeric heterochromatic domains of the human genome composed of long arrays of Satellite (Sat III) DNA sequences, which is also seen in the Hutchinson-Gilford progeria syndrome.
Mi-2/NuRD Complex, RNA Silencing, and Stress Bodies in SLAC
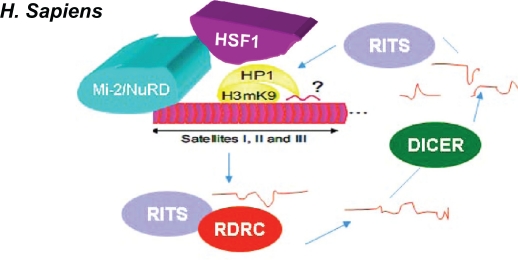
Inhibitory RNA machinery can remain in Sat III RNA to establish a heterochromatin domain78 (Fig. 7). We would like to extend this theme further to a cross-talk between the Mi-2/NuRD complex and RNA silencing. An alteration of the heterochromatin structure may favor an accumulation of Sat III transcripts, suggested by observations made in fibroblasts from patients affected by Hutchinson-Gilford progeria syndrome, where a complete loss of heterochromatic marks is accompanied by the expression of chromosome-9 specific Sat III sequences.42 The expression of pericentric transcripts occurs during replicative senescence at the late stages of both primary fibroblasts and cancer cells.80,81 The expression of Sat III transcripts in embryonic cells82 potentially links the expression of Sat III sequences to developmental programs. Finally, in the formation of nuclear stress bodies (nSBs), the assembly of cytosolic aggregates containing RNA granules contribute to cell survival. In C. elegans, the p-granule genes, cgh-1 and pgl-1, are ectopically expressed in deficient mutants seen in components of the Mi-2/NuRD complex.
Figure 7.
Hypothesized Mi-2/NuRD and DICER silencing. The Mi-2/NuRD complex is possibly enriched in the H3K9me3, heat shock factor-1, and heterochromatin protein-1 regions. RNA molecules are hypothesized to contribute to the structure of the CT and PCT regions. In human cells, all dsRNAs encoded by the PCT regions are hypothetically generated by inhibitory RNA machinery (Dicer, RITS, and RDRC). In humans, CT regions are made of a repetition of AT-rich alpha satellite motifs. The size and structure of the PCT regions are made up of satellite repeats. Such regions might be preferred by stemness factors. Adapted with permission from The International Journal of Developmental Biology (Int. J. Dev. Biol.) 2009;53:259–268.
The transcripts could target human RNP HAP/Saf-B, and possibly other RNA processing factors, to the nuclear stress bodies. With heat shock stress, the snoSNP70 can bind noncoding RNA HSR-1 (unpublished). In yeast, RNA molecules encoded by centromeric repeats have been shown to influence chromatin architecture by the direct formation and maintenance of heterochromatin through RNA interference.83 However, in C. elegans, the relaxed requirement of RRF-1 on somatic cell RNA interference in loss-of-function mutants in components of the Mi-2/NuRD complex has been observed (Supplementary Fig. 2). Furthermore, the inefficiency of some bacterial inhibitory RNA constructs with one critical TF recruiter of Mi-2/NuRD complex with one embedded Cele42 minirepeat in the RNA interference construct has been noted. However, one RNA interference construct excluding this Cele42 minirepeat gave a very high frequency of the multivulva phenotype, almost phenotypically mimicking lin-1 null mutants (unpublished). In humans, as mentioned previously, HSF-1, MTA-1, and CHD-3 interact mutually in cancerous cells. Taken together, it is tempting to speculate that heat shock stress could be involved in heterochromatin organization, partly through the RNA interference mechanism (Fig. 7). Interestingly, the transcription factor ETS-4, one paralog of LIN-1/ETS-1 in C. elegans and an ortholog of vertebrate SAM-pointed domain-containing Ets transcription factor, is a longevity determinant.84
Extension of Lifespan without Diminishing Health in C. elegans
The lifespan of C. elegans can be extended by several different mechanisms, including calorie restriction, as seen in the let-363/ceTOR and eat-2 mutations, reduced Ins/IGF-1 signaling mutations in daf-2, germline ablation, food-sensing amphid ablation, mitochondrial deficiency isp-1 and clk-1 mutations, and decreased temperature. Reduced Ins/IGF-1 signaling and calorie restriction can increase the lifespan of mice. Rapamycin, a drug that negatively regulates mTOR, could prolong the lifespan in mice. Mutations in the gld-1 gene of C. elegans cause germ cells to proliferate uncontrollably to lethal tumors. In most organisms, tumor susceptibility increases with age. Four different mutations can promote longevity and also suppress tumorigenesis. When gld-1 worms bear mutations in daf-2, eat-2, isp-1, or clk-1 genes, the proliferation rate of germline tumor cells was suppressed. 22 However, remarkably, none of these mutations alone affected the proliferation rate of germ cells in worms. The mice bearing mclk-1+/− are tumor-free.85
It is unlikely that we would achieve both longevity and protection against cancer at the cost of energy or nutrient restriction to cells. Interestingly, hormesis is known to influence aging, because transient heat shock can extend the lifespans of worms.86,87 The lifespan can be also extended through forced expression of HSF-1, and the longevity of stress-induced HSF-1 is obtained by activating downstream lifespan-extending genes, which are small heat shock proteins.88 It has yet to be determined if a mild stress on humans might also extend lifespan. Similar to the aforementioned extreme and healthy longevity, the average lifespan of daf-2 (e1370) mutants grown in an axenic medium was able to achieve a 7.5-fold extension of adult lifespan relative to wild-type controls grown on standard media.89
Many C. elegans sensory and chemosensory receptor mutants are long-lived. This lifespan extension is dependent on daf-16.90 Neurons do not deteriorate during normal aging in C. elegans.91 Children with Hutchinson-Gilford progeria syndrome have normal intelligence and emotional development.92 The nuclear lamina network has not been studied in the brain. Differences in the way lamins are expressed or used in the brain might provide new clues about the mechanisms of aging and laminopathies. Brain neurons differ profoundly from connective tissues that derive from mesenchymal stem cells susceptible to “accelerated aging” in at least one major respect, ie, brain neurons are sheltered from mechanical injuries by the skull. Interestingly, Lamin A is required for mechanosensitive gene expression in cultured mouse fibroblasts.93 Further work on lmn-1 in worm “brains” (ie, equivalent) may shed light on the role of the lmn1/Lamina A/Mi-2/NuRD complex in nuclear architecture in SLAC.
With regard to long-lived or lifespan-shortened mutants, such as eat-2, isp-1, and clk-1, soma-to-germline stem-like transformations in mutants of components of the Mi-2/NuRD complex (eg, LET-418/Mi-2β and MEP-1/KLF4) might influence lifespan. LET-418 depletion could result in dramatic chromatin reprogramming. Previously we performed suppression subtractive hybridization and RNA interference screens to characterize the functions of the targets of LET-418/Mi-2β. Interestingly, we could identify two suppressors of lethality of the let-418 mutant during RNA interference assays against those upregulated LET-418/Mi-2β downstream target genes generated from a suppression subtractive hybridization library (Kaeser and Zhang et al, in preparation). To our limited knowledge, such suppressors have not yet obtained via conventional suppressor screen. Therefore, reduction or depletion of LET-418/Mi-2β activity could result in a shift towards somatic robustness, longevity, and sterility.
In humans, Lamin A binds SREBP1, which is a critical target of DAF-12/Liver X receptor α.94 In C. elegans, as with DAF-21/HSP90, the DAF-12/Liver X receptor α is the critical regulator in the transition from L2 larvae to dauer larvae. Our DAF-12/Liver X receptor α genome-wide chromatin immunoprecipitation (ChIP) analysis of direct targets reveals many important functional modules in different biological mechanisms (Hochbaum, Zhang and Fisher et al in preparation). One is the chromatin remodeling module, including the Mi-2/NuRD complex, the PcG complex and the heterochromatin silencing complex (Hochbaum, Zhang and Fisher et al unpublished). Importantly, neither mutant animals with null alleles nor known loss/gain of function alleles, but only some daf-12 RNAi feeding on DAF-12 (rh274, gain of function) animals have a weak multivulval phenotype in our observations (unpublished), which is similar to the let-418 (loss of function) allele.74 This indicates that chromatin with DAF-12 could be dynamic and tightly controlled, and its histone methylation/demethylation and acetylation/deacetylation could undergo global changes during transition from normal L2 to dauer development in response to extracellular epigenetic cues. These chromatin-remodeling complexes could have instructive and programming roles during the development of C. elegans. Therefore, DAF-12 could be essential for establishing and maintaining pluripotent and multipotent states in some cells, and thus provide insights for 4-2-1/SLAC. In humans, some snip alleles of the Liver X receptor could have a longer lifespan,95 and SIRT1, the longevity gene, deacetylates and positively regulates the Liver X receptor. The deacetylation of LXRs by SIRT1 may ultimately affect atherosclerosis and other age-related diseases involving lipid metabolism.96
Targeting the Mi-2/NuRD Complex in the mTOR Pathway to Extend Longevity
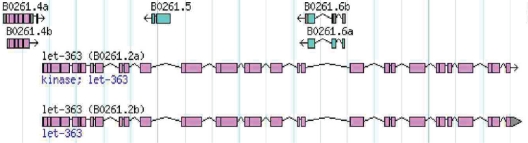
We have previously reported that TOR deficiency in C. elegans more than doubles the natural lifespan of the organism. This new function for TOR signaling in control of ageing may represent a link between nutrition, metabolism, and longevity,97 and this extension of longevity has been recently proven in mice.98 Interestingly, there is one large genomic intron in the open reading frame encoding the nematode let-363/TOR kinase gene that includes three internal opposite-direction encoding genes; B0261.6 shows up in the library of upregulated downstream target genes of let-418 (loss of function) after we performed suppression subtractive hybridization.76 Interestingly, this kind of internal opposite-direction encoding gene even exists in the mTOR kinase gene structures for both mice and humans (data not shown). It is thus tempting to speculate that an opposite transcriptional antisense silencing regulation could be executed in these different species99 (Fig. 8). The human mTOR pathway is known to be involved in tumorigenesis.100 However, in rapidly dividing tumor cells, the demand for nutrients is higher. mTOR pathway mutations may starve tumor cells, inducing a stress response to shut down the cell cycle machinery. However, nematodes bearing a let-363/TOR kinase deficiency are long-lived, but not so healthy. Clearly, further experiments are needed to confirm this hypothesis. Interestingly, starvation protects germline stem cells and extends reproductive longevity in C. elegans.101 It is now tempting to investigate the role of TOR kinase in germline stem cells.
Figure 8.
Putative antisense-silencing gene regulation (adapted from Wormbase). B0261.6, a LET-418/Mi-2β putative target gene, might encode the antisense transcripts to repress the transcription of let-363.
LSD-1 Longevity in C. elegans
In mammalian breast cancer, the histone demethylase LSD-1 is associated with the Mi-2/NuRD complex. The worm LSD-1 orthologs are T08D10.2 and spr-5. The latter is involved in the Notch pathway. In C. elegans, exposure to lithium at clinically relevant concentrations causes a reduction of expression of worm LSD-1 (T08D10.2). Knockdown of T08D10.2 by RNA interference also extends longevity. Lithium may regulate survival by modulating histone methylation and chromatin structure, possibly via the Mi-2/NuRD complex. It would be of interest to know if lithium could extend longevity in humans.102
Mi-2/NuRD Complex and DNA Repair Mechanisms Under Stress
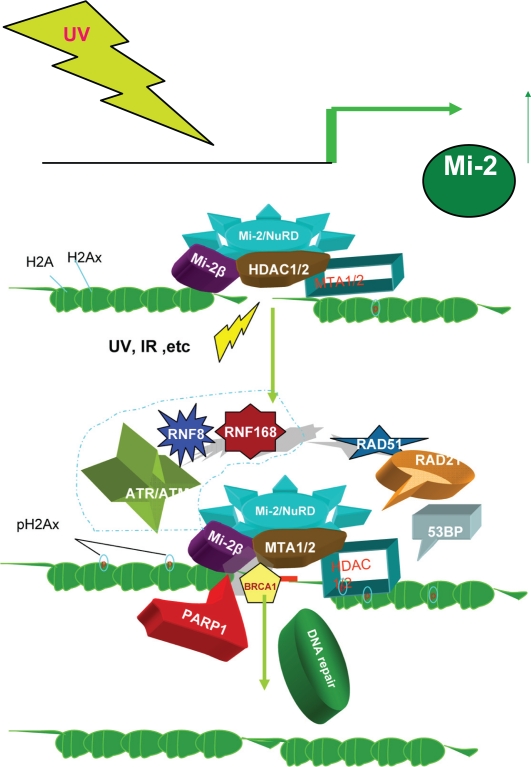
MTA1/2 and Mi-2β/CHD4, components of the core Mi-2β/NuRD complex, have been clearly shown to be involved in the DNA damage response pathway (Fig. 8). The MTA1/Mi-2β/NuRD complex maintains genome stability and orchestrates proper signaling (eg, ATR-Claspin-Chk1 checkpoint signaling, the ATR-H2AX pathway, cell cycle progression, and repair of double strand breaks) and promotes cell survival.103–106
In response to ionizing radiation stress, cells delay cell cycle progression and initiate DNA repair, which is vital for genome integrity. In C. elegans, egr-1 was identified in a genome-wide RNA interference screen as having the ability to protect cells against ionizing radiation. Knockdown of the human homolog of egr-1, MTA2, and Mi-2β/CHD4, led to accumulation of spontaneous DNA damage and increased sensitivity to ionizing radiation. MTA2 and CHD4 accumulate in chromatin tracks containing double strand breaks after laser microirradiation. Furthermore, Mi-2β/CHD4 becomes transiently immobilized on chromatin after ionizing radiation. Knockdown of CHD4 enhanced Cdc25A degradation and p21 (Cip1) accumulation, which resulted in more pronounced cyclin-dependent kinase inhibition and cell cycle delay. Mi-2β/CHD4 mediates poly-(adenosine diphosphate-ribose)-dependent recruitment of the Mi-2/NuRD complex to DNA damage sites. Directly at these double strand breaks, the DNA damage response kinase (ataxia, telangiectasia mutated) phosphorylates Mi-2β/CHD4, and promotes the chromatin response alongside RNF168 ubiquitin ligase to facilitate local ubiquitylation and BRCA1 assembly. In addition, Mi-2β/CHD4 acts as a critical regulator of the G1/S cell-cycle transition by modulating p53 deacetylation. Similarly, MTA1 is implicated in the response to DNA damage induced by ionizing radiation. MTA1 is stabilized and activated in response to ionizing radiation through disruption of COP1-mediated proteolysis.
In response to ultraviolet radiation exposure, the Mi-2 protein level increases in cell culture systems. Ultraviolet light treatment results in better translation through responsive elements in the 5′ untranslated region. Furthermore, radiation to the 5′ untranslated region stabilizes MTA1 in an ATR (ataxia, telangiectasia mutated and Rad3-related)-dependent manner and increases MTA1 binding to ATR, which phosphorylates, activating a number of downstream substrates, such as checkpoint kinase 1 and histone 2A variant X. The depletion of MTA1 results in a defect in the G(2)-M checkpoint and increases cellular sensitivity to ultraviolet-induced DNA damage. Thus, MTA1 is required for the activation of the ATR-Claspin-checkpoint kinase 1 and ATR-histone 2A variant X pathways following ultraviolet light treatment.
Mi-2/NuRD Complex, iPSCs, and Cancer Stem Cells
Cellular plasticity is remarkable. The transfer of nuclei from somatic cells, first in frogs, then in fish, and later in mammals,107–111 as well as cell fusion studies,112 have provided us with considerable information on basic biology. This finally lead to the important technological breakthrough of iPSCs, ie, the forced expression of a cocktail of stemness factors (Oct-4, Sox-2, c-myc, and Klf4) to induce mouse fibroblast cells to show stem cell characteristics, including the expression of pluripotency genes and a normal karyotype contribution to all three germ layers in teratomas and chimeric embryos.113,114 Later, iPSCs were established in human cell lines by using normal or somatic cells from patients. An adult cell can be reprogrammed, altering its gene expression profiles, and hence its fate, to that typical of another cell type. Several distinct nuclear reprogramming approaches including nuclear transfer, cell fusion, transcription-factor transduction, and reprogramming using synthetic molecules like “reversine” can induce nuclei from ‘terminally differentiated’ somatic cells to express genes that are typical of embryonic stem cells.115–117 However, iPSCs carry the risk of causing cells to become cancerous. Different strategies have been developed to create a range of iPSCs, from the human iPSCs with virus-containing transgenes, human iPSCs with factor-free viruses and iPSC made by using recombinant proteins for stemness factors alone, to enhanced human iPSCs with critical gene knockdown/overexpression118 (eg, p53 knockdown, see Fig. 9), and so on. Treatment with an inhibitor of DNA methylation, such as 5-aza-2’-deoxycytidine (5-azadC, AZA), increases the efficiency of iPSCs reprogramming.119 Similar results were obtained with the methyltransferase DNMT1 knockdown containing a short “hairpin”. In addition, the use of other drugs, such as several histone deacetylase inhibitors, eg, VPA, TSA, or SAHA (or BayK8644 or BIX-01294, an inhibitor of the G9a histone methyltransferase), also enhances the efficiency of reprogramming.120 Vitamin C has been shown to enhance reprogramming significantly.
Figure 9.
Model of stress, the Mi-2/NuRD complex and repair of DNA damage. Upper: Ultraviolet light upregulates the level of Mi-2 protein.6 Bottom: Summary of the Mi-2/NuRD mechanism in the response to DNA damage. In response to DNA damage caused by ultraviolet light or ionizing radiation, the Mi-2/NuRD complex is rapidly recruited to the site of damaged DNA and exerts its function in DNA repair by many different mechanisms, including RNF168 level ubiquination, blocking the ongoing transcription, and so on. Adapted with permission from.103
The micro RNA-based strategy for reprogramming somatic cells into pluripotent stem cells has been successful. 121 Micro RNAs are critical in maintaining the pluripotent state, cell lineage specification, and epigenetic modifications of chromatin. Micro RNAs can be used for directed cellular reprogramming and induced cell fate conversion between lineages without reversion to a pluripotent state. In addition, it has been recently shown that selected cardiac transcription factors can directly reprogram fibroblasts to become cardiomyocytes without first becoming a stem/progenitor cell. This has been realized by reprogramming a combination of three developmental transcription factors122 (ie, Gata4, Mef2c, and Tbx5), which rapidly and efficiently reprogramm postnatal cardiac or dermal fibroblasts directly into differentiated cardiomyocyte-like cells. Interestingly, the FOG-2/Mi-2/NuRD complex is the master regulator of Gata4 expression.123 Another novel iPSC method not only avoids genes, but is also more efficient. Chemically modified RNAs transcribed from the four genes, KLF4, c-myc, OCT4, and SOX2 have been introduced into human fibroblast cells. Furthermore, an additional RNA transcript treating these iPSC cells converts these into muscle cells.124
Similarity and Difference Among Stemnessed Human Embryonic Stem Cells, Induced Pluripotent Stem Cells, and Cancer Stem Cells
Many of the earliest stem cell studies were performed on cells isolated from tumors, including research on embryonic carcinoma cells, a type of stem cell derived from teratocarcinoma. Embryonic stem cells isolated from the mouse, and then later from humans, shared not only pluripotency with their induced pluripotent stem cell cousins, but also robust tumorigenicity, because each readily form teratomas. However, iPSCs are predicted to possess tumorigenic potential equal to or greater than that of embryonic stem cells (Fig. 10).
Figure 10.
The model of similarity and difference between stemness pluripotency and tumorigenicity. Biological and molecular links between pluripotency and tumorigenicity are illustrated graphically. In this model, one hypothesis is that the variation/boundary of signatures could be amplified along with differentiation/tumorigenesis from those in compact stemness states.
Abbreviations: EC, embryonic carcinoma cells; hESC, human embryonic stem cells; IPSC, induced pluripotent stem cells; CSC, stem-like tumor cell/cancer stem cells.
The subtle difference between human embryonic stem cells and human iPSCs was revealed by recent cancer vaccine research using stem cells and recent studies of epigenetic memory in stem cell nuclear transplantation, human embryonic stem cells, and iPSCs. The cancer vaccine research showed that an injection of human embryonic stem cells into mice generated a strong antitumor immune response in colon cancer but artificial iPSCs did not (Fig. 10). Moreover, a more recent study124 demonstrated that somatic cell nuclear transfer is more effective at establishing the ground state of pluripotency than factor-based reprogramming that can keep an epigenetic memory of the tissue of origin. This may affect the potential applications of iPSCs in directed differentiation disease modeling or treatment. Basically, human embryonic stem cells and cancer stem cells share some common characteristics (Fig. 10), especially in the way they form and replicate, so an human embryonic stem cell vaccine could fool the immune system into believing that cancer cells are present, and thus initiating an antitumor immune program, ie, immune systems could recognize antigens of tumor cells, triggering an immune response to make antibodies to fight the tumor and consequently reducing tumor growth in the immunized mice. Therefore, immunization with embryonic materials might produce antitumor responses. Moreover, gene profiling experiments have revealed similarities between cancer and embryonic stem cells in mice (Fig. 10). The gene expression signature of embryonic stem cells goes into three functional modules. The Myc nexus, including genes targeted by Myc-interacting proteins, accounts for most of the similarity between embryonic stem cells and cancer cells.153
Although iPSCs have a developmental potential similar to that of embryonic stem cells, several studies have reported distinct gene expression differences between iPSCs and embryonic stem cells of both human and mouse origin.125–127 Human iPSCs developed teratomas more efficiently and faster than human embryonic stem cells,128 and both seem to follow a stochastic model to acquire stemness block (Fig. 10). Another study shows that iPSCs retained epigenetic imprints from human parental retinal pigmented epithelial cells and showed a propensity for spontaneous differentiation back into retinal pigmented epithelial cells after removal of fibroblast growth factor.129 iPSC lines exhibited a marked preference for redifferentiation into retinal pigmented epithelial cells.129 These cells can be reprogrammed to pluripotency, which confirms that they retain a memory of their previous state of differentiation (Fig. 10).
However, Guenther et al130 found that only four genes are consistently differentially expressed between human iPSCs and embryonic stem cells (Fig. 10). There are no significant differences between the genome-wide distributions of the activating H3K4me3 and repressive H3K27me3 histone modifications in human iPSCs and embryonic stem cells, so human iPSCs have accurately reinstalled the transcriptional and epigenetic controls of embryonic stem cells, and there are minimal overt molecular differences between human iPSCs and embryonic stem cells. In a separate study, Newman and Cooper131 observed that human iPSC and embryonic stem cell lines cultured in the same laboratory reproducibly clustered together (Fig. 10). However there are many different signatures in different laboratories, and we cannot rule out that even a slight difference could have a critical biological function, eg, use of a stem cell antitumor vaccine (Fig. 10).
Mi-2/NuRD Complex, Micro RNAs, and SLAC
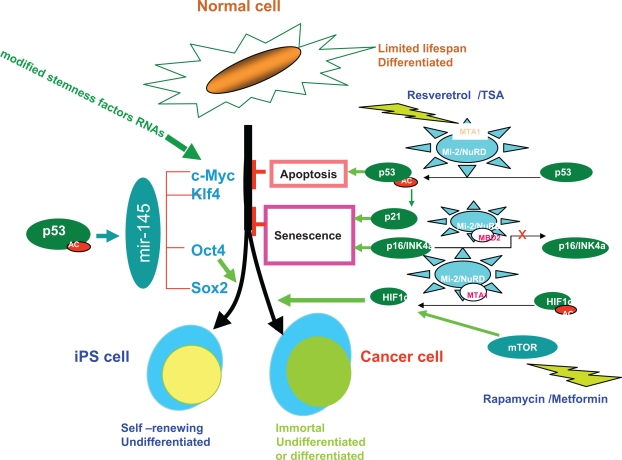
The biological complexity of the human body is enormous, but a simplicity becomes apparent if we turn our attention to epigenetic regulation, specifically Mi-2/NuRD complex and RNAs, such as LINC RNAs and micro RNAs. Micro RNAs were first discovered in genetic screens for regulators of developmental timing in the stem cell-like seam cell lineage in C. elegans. Micro RNAs were found to be expressed in different tissues, and they have been linked to the development of mouse and human stem cells. The Mi-2/NuRD complex can work on micro RNA-145 through deacetylation of p53132 (Fig. 11), a direct target gene of p53 and a repressor of the stemness factors Oct-4, Sox2, and KLF4 in stem cells.
Figure 11.
Schematic representation of the role of the Mi-2/NuRD -p53-micro RNA network in induced pluripotent stem cells and cancer stem cells. Normal fibroblasts, which are mature, differentiated cells, can be reprogrammed into induced pluripotent stem cells or tumor cells by a combination of defined factors. Adapted by permission from Macmillan Publishers Ltd, Nature, 2009;460:1085–6, copyright 2009. Left: The transcription factors c-myc and Klf4 promote reprogramming of fibroblasts into induced pluripotent stem cells in a manner that conceptually parallels their roles in transforming normal cells into tumor cells. Oct4 and Sox2, although overexpressed in cancers, are currently thought to function specifically to promote formation of induced pluripotent stem cells. Right: The p53 tumor suppressor gene is a well known master regulator that helps downregulate the genes required for proliferation and survival. p53 directly or indirectly limits the reprogramming of fibroblasts into induced pluripotent stem cells or into transformed cancer cells by inducing apoptosis, or cellular senescence through its target protein, the cell-cycle inhibitor p21, which promotes arrested growth and cell death in response to various stress signals.
Along similar lines, loss of RB1 expression in fibroblasts results in reprogramming to an embryonic stem cell-like state by upregulation of Sox2, Oct-4, Klf4, and Nanog.133 These reprogrammed cells form spheres and are tumorigenic, exhibiting elevated Zeb1 expression and a high CD44/low CD24 cancer stem cell-like phenotype. Thus, loss of the Rb tumor suppressor may stimulate the emergence of cancer stem cells in concert with changes in micro RNA function.
In C. elegans, let-7 regulates longevity through the RAS pathway,134 which is negatively regulated by the Mi-2/NuRD complex. In humans, cancers may arise from rare self-renewing tumor-initiating cells within a special niche. Because micro RNAs can regulate cell fate, a comparison of micro RNA expression in self-renewing and differentiated cells from breast cancer lines and in breast tumor-initiating cells and nonbreast tumor-initiating cells from first-degree breast cancers shows that let-7 micro RNAs were markedly reduced in breast tumor-initiating cells and increased with differentiation. Increased let-7 paralleled reduced H-RAS and HMGA2, which are known let-7 targets. Let-7 regulates multiple breast tumor-initiating cell stem cell-like properties by silencing more than one target. It has been shown that a lack of HSF-1 made mouse embryonic fibroblasts resistant to H-RASV12D-induced focus formation in Hsf1−/− and wild-type mouse embryonic fibroblasts. HMGA2 links to cellular senescence related to proliferation-associated genes and stem cell ageing.135 HSF-1/Mi-2/NuRD also has important interactions with RNA fields in other ways, especially via large noncoding RNAs (LINCs) such as Xist, HOTAIR, and HSR-1.136 RNA coimmunoprecipitation reveals that a significant proportion of LINC RNAs are physically associated with chromatin-modifying complexes, such as the putative TWIST1/BMI1/PRC1, TWIST1/EZH2/PRC2 complex. The TWIST1/Mi-2/NuRD complex has also been shown to be involved in epithelial-mesenchymal transition. Furthermore, the breast cancer stem cell phenotype is characterized by high expression of CD44, little or no expression of CD24, and importantly, TWIST modulates breast cancer stem cells by transcriptional regulation of CD24 expression. 151 Because HSR-1 and Xist are closely linked to HSF-1/Mi-2/NuRD or its equivalents, it would be interesting to see if and how Mi-2/NuRD complexes function with LINC RNAs in regulating the epigenetic landscape on a genomic scale. It is known that ncRNA SRA binds to p68 RNA helicase137 which is associated with the Mi-2/NuRD complex and plays a role in RNA processing. p68 RNA helicase regulates a transcriptome at the estrogen receptor-regulated pS2 promoter, which is repressed by the HSF-1/CHD-3/Mi-2/NuRD complex in the MCF-7 cancerous cell line. A myosin heavy chain IIB promoter, p68 RNA helicase, together with ncRNA steroid receptor RNA activator,138 stabilizes the BRG-1 protein and the location of the TATA binding protein and RNA polII. Furthermore, HSF1 regulates the multidrug resistance MDR1 expression in vivo (Unpublished) at the heat-induced activation of the MDR1 promoter. The increase of multidrug resistance (MDR)1 could potentially increase the resistance against cytotoxic drugs as one of the major obstacles for chemotherapy of tumors and recurrence of malignancies.
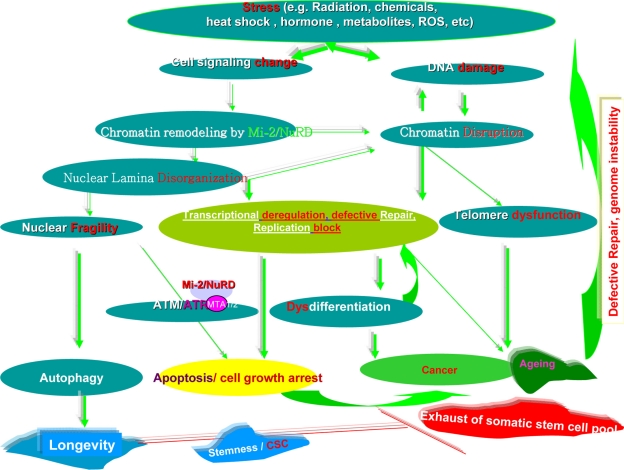
Put simply, a number of stressors can induce changes in cell signaling and/or DNA damage. Chromatin remodelers, such as the Mi-2/NuRD complex, could cause nuclear lamina disorganization and chromatin disruption, which in turn affects DNA damage and makes nuclear fragile. The chromatin disruption could activate the autophagy in a scope and thus promote possibly longevity. Otherwise, disrupting the tertiary structure of lamina, and its subsequent interactions with chromatin domains and higher order chromatin structure, then affects gene transcription, repair of splicing DNA damage, DNA replication, and subsequently cell division. This can also cause disassembly of the nuclear membrane in mitosis and meiosis, and improper assembly at the completion of mitosis and meiosis leading to cell death and/or cell growth arrest. When the critical somatic stem cell pool becomes exhausted, carcinogenesis and ageing could be triggered. In addition, chromatin disruption could cause telomere dysfunction and lead to ageing and cancer. The two disease states would enhance defective repair and genome instability, and thus worsen the DNA damage further. Most of these processes are functionally intertwined (Fig. 12).
Figure 12.
Simplified summary of cross-talk among the Mi-2/NuRD complex, stress, and stemness, longevity/ageing, and cancer. The color green denotes promotion or activation, and the color red indicates inhibition or an abnormality.
Mi-2/NuRD Complex and Cell Metabolism in SLAC
Fuel signals from cell metabolism are crucial for the activities of Mi-2/NuRD complexes in SLAC. For instance, the energy of ATP is essential for remodeling of the Mi-2/NuRD complex, and is generally provided through the glucose-pyruvate axis and the tricarboxylic acid cycle. Cell metabolism also ensures that cells have a reservoir for acetyl-CoA, which is important for the balance of histone acetyltransferase-histone deacetylase.139 The cholesterol-citrate cycle to acetyl-CoA is also one signal of favorable or unfavorable environmental cues for C. elegans to determine its entry into the long-lived but inactive dauer state (ie, a slow life history) or to undergo normal development (ie, a fast life history, Figs. 4 and 5). Glutamine is a key factor that influences the axis of mTOR, hypoxia-inducible factor-1α, and/or p53-involved autophagy. Autophagy generally promotes longevity and dauer formation in C. elegans, and probably also in mammals. In addition, the oncogenic activation of Myc promotes glutamine utilization. C-myc and hypoxia-inducible factor-1 regulate glucose metabolism and stimulate the Warburg effect. Firstly, deregulation of the expression of glutamine transporters and miR-23a/b by Myc targets glutaminase and triggers an addiction to glutamine.152 Isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2)140 mutations could affect methylation patterns via competitive inhibition of histone demethylases by 2-hydroxyglutarate, and this links to the histone demethylases, JMJD2, JHDM1, and/or LSD-1, and their substrate α-ketoglutarate as a cofactor in DNA hypermethylation, eg, methylation of the promoter of the DNA repair gene O6-methylgluguanine-DNA methyltransferase. Of note, IDH3 catalyzes the conversion of isocitrate to α-ketoglutarate in the mitochondria, producing one equivalent of NADH in a NAD+dependent reaction. It is interesting that mutations in IDH1 and IDH2 (but not IDH3) are found in cancer. However, we speculate that the LSD-1/Mi-2/NuRD complex itself could have a chance to function directly on oncometabolites together with IDH3. Furthermore, there could be a cross-talk between the Mi-2/NuRD complex with NAD+related SIRT 1 deacetylation and/or Lat11 (dihydrolipoamide acetyltransferase), a longevity factor in the calorie restriction pathway. Finally, 2-hydroxyglutarate produced by mutated isocitrate dehydrogenase might act through the accumulation of HIF-1α via prolyl hydroxylase inhibition, leading to development of cancer. In C. elegans, fumarylacetoacetate hydrolase, which catalyzes tyrosine metabolism and causes fumarate to cycle into the tricarboxylic acid cycle, is required for a normal life span and has a role in protein aggregation in connection with HSF-1,141 holding the potential for preventing age-related diseases.
Normal cells can undergo aerobic glycolysis and inefficient glutamine breakdown when they are induced to proliferate. When nutrients dwindle, the cells can respond by reactivating oxidative phosphorylation and cease proliferation through the activities of tumor suppressors, such as p53. Cancer cells, which often lack p53 and other tumor suppressors, exhibit “addiction” to glucose and/or glutamine consumption. Moreover, cancer cells make inefficient use of glutamine. In normal cells, glutamine is used for the synthesis of amino acids and nitrogen for de novo nucleotide formation. Cancer cells, in contrast, secrete glutamine-derived nitrogen as waste.152 The cellular uptake of L-glutamine is the rate-limiting step that activates mTOR. Certain tumor cell lines do not need L-glutamine uptake and are primed for mTOR activation. Thus, L-glutamine flux regulates mTOR translation and autophagy to coordinate cell growth and proliferation.143 However, the detailed effects of cell metabolism on Mi-2/NuRD complexes in SLAC remain unclear. For the moment, our proposed model as (Supplementary Fig. 6).
Potential Consequences of Mi2/NuRD Activity in SLAC
Direct reprogramming via a stochastic process could possibly require activity of Mi-2/NuRD complexes. Reprogramming stemness transcription factors (ie, Oct4, Sox2, Klf4, and c-Myc) are associated with the Mi-2/NuRD complex. Following a continuous stochastic process, almost all mouse donor cells eventually give rise to iPSCs with continued growth and transcription factor expression. Overexpression of the stemness factor Lin28 or additional inhibition of the p53/p21 pathway, which could be modulated by the Mi-2/NuRD complex (Figs. 11 and 13), increase the cell division rate and resulted in an accelerated the formation of iPSCs. This is proportional to the increase in cell proliferation.144 The cancer stem cell model could be relatively easily reconciled with the Darwinian selection cancer model because the Mi-2/NuRD complex could provide a novel layer of dynamic influence of reversible factors, such as LSD-1 demethylase and RBP2. In addition to the reversible modifications of DNA methylation and histone acetylation and deacetylation, this arsenal governs the dynamic state of cells and regulates the heterogeneity of cancer cells. Put simply, the different tissues make the cell a specific niche of cellular physical, chemical and physiological components, as well as extracellular and environmental stresses (eg, the basal or luminal breast cell niche). These components could create distinct genomic mutations stochastically and develop a different epigenetic background. Such mutations and epigenetic factors integrate environmental cues, such as nutrients (Figs. 4 and 5), ultraviolet light, ionizing radiation, or mechanical trauma. Collectively, these factors activate “core” functional chromatin remodeling, and the various percentages of cancer stem cell types, eg, those of the MCF-7, SK-BR-3, and MDA-MB-231 breast cancer cell lines. In addition, the Mi-2/NuRD complex has additional specific “isoforms”, including its unique histone acetyltransferase-histone deacetylase supercomplex. RBP2 regulates heterogeneity within cancer cell populations in response to stress, including drug treatment. In drug-sensitive human tumor cell lines, there is a small subpopulation of reversibly “drug-tolerant” cells.146 These have significantly reduced drug sensitivity and maintain viability via engagement of IGF-1 receptor signaling, and RBP2/KDM5A/Jarid1A helps to alter the chromatin state. Treatment with IGF-1 receptor inhibitors or chromatin-modifying agents can selectively ablate the drug-tolerant subpopulation. It remains unknown if such a “drug-tolerant” state is a stem-like one. HSF-1 can directly control the promoter activity of the multidrug resistance gene 1.147 It remains to be determined if the HSF-1/Mi-2/NuRD complex has a role in the regulation of multidrug resistance gene 1. In C. elegans, the RBP2 H3K4 trimethylation complex could regulate lifespan in a germline-dependent manner. Interestingly, the MBD2/Mi-2/NuRD complex represses the promoter activity of p16/INK4a, and inactivation of p16/INK4a can increase longevity in progeria.148 This raises the possibility that epigenetic interference with MBD2/Mi-2/NuRD might switch progeria to longevity.
Figure 13.
Model of soma-to-germline transformation in Caenorhabditis elegans and possible stochastic cancer stem-like formation in mammals. Adapted by permission from Macmillan Publishers Ltd, Nature, 2009;459:1079–84, copyright 2009.
Ongoing Research on the Mi-2/NuRD Complex
With regard to genomic research, the ChIP DSL assay has been applied to the LSD-1/Mi-2/NuRD complex, and an interesting list of target genes has been established. 12 Further ChIP-chip and ChIP-seq on individual components of different Mi-2/NuRD complexes for in depth comparisons are awaited.
Ribonomic research is meanwhile focusing on RNA IP/high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP). The CHD-3 binding proteins, CGI-55 and Ki-1/57, might be involved in nuclear functions along with the remodeling of chromatin. Ki-1/57 may be also involved in modulations of the higher-order structure of chromatin. CGI-55 (also named PAI-RBP1 for the plasminogen activator inhibitor mRNA-binding protein 1) is a mRNA-binding protein.149 The chromodomain of the Mi-2 protein may bind to noncoding RNA,150 and, thus, RCIP and HITS-CLIP could be worthwhile investigating to clarify further the roles of the Mi-2/NuRD complex in RNA fields.
Proteonomics research primarily involves tandem affinity purification walking. An Oct4-based tandem affinity purification walking from Oct4 to Nanog and Esrrb1 has generated a remarkable Oct4-centered expended interactor network, which contains a functional module of remodelers, including the Oct4-associated Mi-2/NuRD complex. This could be applied to the core Mi-2/NuRD complex as well as its derivatives. One question is whether some metabolites and RNAs could hypothetically interact with core or specific Mi-2/NuRD complexes to achieve some biological functions directly.28,29
Specific research questions that are the subject of ongoing research include:
What can we discover about the dramatic changes in the genome-wide distribution of the Mi-2/NuRD complex during cellular senescence, and tissue and organism ageing? How do those changes affect gene expression programs in detail?
Is Mi-2/NuRD involved in senescence-associated heterochromatic foci regulation or DNA replication in SLAC? If so, how?
It is well known that the Mi-2/NuRD complex shares the synMuv B pathway with the HP1/Rb complex and has an antagonistic relationship with mes-2/PcG during embryonic development in C. elegans. What are the relationships among the Mi-2/NuRD, HP1/Rb, and BMI1/EZH2/PcG complexes in mammals? Furthermore, how does the Mi-2/NuRD complex coordinate with other chromatin remodelers in 4-2-1/SLAC?
Does any close relationship exist between the nuclear stress body and Mi-2/NuRD bodies, given that they seem to be physically tightly connected?154 Are there any evolutionary common characteristics between the p-granule in C. elegans and the nuclear stress body in mammals?
There are clearly different “isoforms” of Mi-2/NuRD complexes. How do the levels, assembly and activities of these different Mi-2/NuRD or Mi-2/NuRD-like complexes (eg, NODE) regulate 4-2-1/SLAC?
Do the Mi-2/NuRD complexes function differently in subpopulations of cancer stem cells and noncancer stem cells, and how do they differ?
What is the nature of the cross-talk between the Mi-2/NuRD complex and environmental toxin-induced oxidative DNA damage during cancer or age-related disease progression?
What are the endogenous sources of chemically reactive species? Is the origin of stem cells and cancer cells the same or not? Although they have plenty of similarities, a quick guess is that they do not. Stem cells could behave like “bacteria” and the cancer cells could be rather like phages, viruses, or prions of parasites and hijack the machinery of life from the host. Where and how do the stemlike traits originate in the defective Mi-2/NuRD complex? Some answers have been proposed, but how well do they correlate? The first hypothesis is that TWIST1/Mi-2/NuRD complex ⇒ BMI1 ⇒ stem-like traits. The epithelial-mesenchymal transition is associated with the acquisition of stem cell-like characteristics. The epithelial-mesenchymal transition inducer directly and transcriptionally targets the stemness PRC1 (Polycomb complex) protein Bmi1 that promotes self-renewal of certain stem cell populations.152 The second hypothesis is that Mi-2/NuRD complex ⇒ DNA repair defects ⇒ forced regeneration ⇒ stem-like traits.
How about the bona fide CpG island DNA demethylase? Could the LSD-1 in Mi-2/NuRD complex perform any job for this?155,156
Conclusion
To use a simile, the Mi-2/NuRD is like democracy on the planet. Its “core” always seems to be same. The different tissue- or cell-specific Mi-2/NuRD complexes contribute significantly to 4-2-1/SLAC by dynamically repressing or activating transcriptional activity and instructing the high-order chromatin structure within the same paradigm or “core”; then extensions of expended scope. (Supplementary Fig. 7) The activity of the Mi-2/NuRD complexes is the hub of human physiology and pathology in SLAC. If our comprehensive understanding of SLAC can be likened to a necklace, the chromatin remodeling activities are the pearls. A further view is here put forward of the “core” Mi-2/NuRD complex by employing indepth current high-sensitivity “omics” platforms, eg, ChIP-seq and ChIP-chip for genomics, “tandem affinity purification walking” for proteomics,157 and HITS–CLIP, RIP -seq for ribonomics,158–160 to generate a large amount of data and then apply computation tools, such as rough-set soft computing to extract the hub information, the functional module and the cause-effect regulatory network by using packages such as DAVID, IPA, and Metacore161 to glean novel information about the modulation on the “core” in certain contexts. Clearly, the arrival of the Human Proteome Project era, a preferred shift from “differential expression” to “differential networking” and emerging bioinformatic tools will provide us with significant breakthroughs. To take a specific example, because of the longevity of neurons and their special protection inside the skull, priority could be given to SLAC-associated neuron research. For the moment, one possibility in the near future is to target the Mi-2/NuRD complex and/or its closely related determinants in SLAC, such as the critical steps of mechanisms like histone deacetylation, histone demethylation, and abnormal DNA methylation. For example, pharmacological agents can successfully modulate the differentiation state of a tumor. Moreover, cancer stem cells can be eliminated or functionally antagonized by inducing their differentiation. Thus, “differentiation-inducing” agents, such as salinomycin or histone deacetylase inhibitors, may have therapeutic value. We could also target the key pathways, such as the TGF-β and Wnt pathways, to eliminate cancer stem cells by strong activation of antiapoptotic signaling, such as those mediated by PI3K and nuclear factor-kB. Because the role of micro RNAs in cancer stem cell maintenance is now becoming fully appreciated, therapeutic delivery of micro RNAs may represent an additional potential strategy to disrupt cancer. Ideally, our efforts might reverse an entire program of events to blunt tumor growth and/or reverse and delay ageing (Supplementary Fig. 7). We look forward to the next decade and the likelihood of many more exciting discoveries that will rely heavily on the involvement and ingenuity of biologists and clinicians in both basic and clinic research fields using at least C. elegans and mammals. In everyday life, we could choose some of the anticancer, antiageing foods, such as red wine or grapes (containing resveratrol), ginseng, strawberries and similar fruits, and supplements/drugs, such as rapamycin, metformin, ginkgo bilboa. For the time being, the most prudent Mi-2/NuRD “epidrug”-related advice for healthy longevity is to avoid extremes, do not be “greedy”, keep a balance between Yin and Yang (the Chinese medicine principle), ie, work, drink, eat, and exercise reasonably. Simple, smart choices of food intake and activity level could positively influence the chance of living a long healthy life.
With the unlocking of more and more secrets from “friends” like C. elegans and even bacteria, and solving the mystery of the powerful reversal enzymes in epigenetic regulation of dynamic activities of Mi-2/NuRD complexes, ageing could be “stopped”.162 Although we have not discovered the “fountain of youth”, if we can extrapolate from the conclusion of “nearly 10-fold extension of both median and maximum adult lifespan in age-1(mg44) animals relative to its control wild type N2 DRM C. elegans”, we may be able to live remarkably long lives, and even 1000 years may become feasible.163
Supplementary Information
Acknowledgments
The author is indebted to all members of the laboratories of Drs. Mueller, Fisher, Calderwood, and Brunschwig for sharing their unpublished, personal communications and insightful discussions. Thanks are also extended to the Switzerland Federal Government and CSC fellowship support.
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Rosch PJ. Handbook of Stress, Medicine, and Health. Boca Raton, FL: CRC Press Inc; 1996. Stress and cancer: Disorders of communication, control, and civilization. [Google Scholar]
- 2.Cao L, Liu X, Lin EJ, Wang C, et al. Environmental and genetic activation of a brain-adipocyte BDNF/leptin axis causes cancer remission and inhibition. Cell. 2010;142:52–64. doi: 10.1016/j.cell.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blagosklonny MV. Why human lifespan is rapidly increasing: Solving “longevity riddle” with “revealed-slow-aging” hypothesis. Aging (Albany NY) 2010;2:177–82. doi: 10.18632/aging.100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David AR, Zimmerman MR. Cancer: An old disease, a new disease or something in between? Nature Reviews Cancer. 2010;10:728–33. doi: 10.1038/nrc2914. [DOI] [PubMed] [Google Scholar]
- 5.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Li Y. The expanding Mi-2/NuRD complexes: A schematic glance. Proteomics Insights. 2010;3:79–109. [Google Scholar]
- 7.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 8.Ramírez J, Hagman J. The Mi-2/NuRD complex: A critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–6. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- 9.Peric-Hupkes D, Meuleman W, Pagie L, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng HH, Bird A. Histone deacetylases: Silencers for hire. Trends Biochem Sci. 2000;25:121–6. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 11.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Zhang H, Chen Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 13.Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–84. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–9. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 16.Lendahl U, Orrenius S, Brenner S, Horvitz R, Sulston J. Genetic regulation of organ development and programmed cell death. Lakartidningen. 2002;99:4026–32. Swedish. [PubMed] [Google Scholar]
- 17.Bernards R. The Nobel Prize in Physiology or Medicine for 2006 for the discovery of RNA interference. Ned Tijdschr Geneeskd. 2006;150:2849–53. Dutch. [PubMed] [Google Scholar]
- 18.Chalfie M. The 2009 Lindau Nobel Laureate meeting: Martin Chalfie, Chemistry 2008. J Vis Exp. 2010;36:1570. doi: 10.3791/1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruvkun G. The perfect storm of tiny RNAs. Nat Med. 2008;14:1041–5. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 20.Johnson TE. For the special issue: The nematode Caenorhabditis elegans in aging research. Exp Gerontol. 2006;41:887–9. doi: 10.1016/j.exger.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Kenyon C. My adventures with genes from the fountain of youth. Harvey Lect. 2004–2005:100, 29–70. [PubMed] [Google Scholar]
- 22.Pinkston-Gosse J, Kenyon C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet. 2007;39:1403–9. doi: 10.1038/ng.2007.1. [DOI] [PubMed] [Google Scholar]
- 23.Kirienko NV, Mani K, Fay DS. Cancer models in Caenorhabditis elegans. Dev Dyn. 2010;239:1413–48. doi: 10.1002/dvdy.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nimmo RA, Slack FJ. An elegant miRror: MicroRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–18. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–9. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 26.Koch HB, Zhang R, Verdoodt B, et al. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle. 2007;6:205–17. doi: 10.4161/cc.6.2.3742. [DOI] [PubMed] [Google Scholar]
- 27.Feng B, Jiang J, Kraus P, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 28.Van den Berg DL, Snoek T, Mullin NP, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–81. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardo M, Lang B, Yu L, et al. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–95. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tai MH, Chang CC, Kiupel M, et al. Oct4 expression in adult human stem cells: Evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 31.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–82K. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 33.Lauberth SM, Rauchman M. A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. J Biol Chem. 2006;281:23922–31. doi: 10.1074/jbc.M513461200. [DOI] [PubMed] [Google Scholar]
- 34.Kanga J, Shakyaa A, Tantina D. Stem cells, stress, metabolism and cancer: A drama in two Octs. Trends Biochem Sci. 2009;34:491–9. doi: 10.1016/j.tibs.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Bultman S, Gebuhr T, Yee D, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 36.Yan Z, Wang Z, Sharova L, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–65. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–91. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–55. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 39.Ng SY, Yoshida T, Georgopoulos K. Ikaros and chromatin regulation in early hematopoiesis. Curr Opin Immunol. 2007;19:116–22. doi: 10.1016/j.coi.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–66. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida T, Hazan I, Zhang J, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–89. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in. C elegans Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Rao S, Chu J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–8. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 44.Cui M, Kim EB, Han M. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2006;2:e74. doi: 10.1371/journal.pgen.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takasaki T, Liu Z, Habara Y, et al. MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development. 2007;134:757–67. doi: 10.1242/dev.02771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Kennedy S, Conte D, Jr, et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–7. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- 47.Sun G, Alzayady K, Stewart R, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323–7. doi: 10.1038/nature09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–6. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 50.Sternberg PW. Developmental biology. A pattern of precision. Science. 2004;303:637–8. doi: 10.1126/science.1094409. [DOI] [PubMed] [Google Scholar]
- 51.Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, Shi Y. Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol Cell Biol. 2002;22:3024–34. doi: 10.1128/MCB.22.9.3024-3034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 2005;24:2613–23. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component MBD3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 54.Kaji K, Nichols J, Hendrich B. MBD3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–32. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 55.Zhu D, Fang J, Li Y, Zhang J. MBD3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS One. 2009;4:e7684. doi: 10.1371/journal.pone.0007684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prokocimer M, Davidovich M, Nissim-Rafinia M, et al. Nuclear lamins: Key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–85. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumaran RI, Muralikrishna B, Parnaik VK. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J Cell Biol. 2002;159:783–93. doi: 10.1083/jcb.200204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spann TP, Goldman AE, Wang C, Huang S, Goldman RD. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J Cell Biol. 2002;156:603–8. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mancini MA, Shan B, Nickerson JA, Penman S, Lee WH. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci U S A. 1994;91:418–22. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jagatheesan G, Thanumalayan S, Muralikrishna B, Rangaraj N, Karande AA, Parnaik VK. Colocalization of intranuclear lamin foci with RNA splicing factors. J Cell Sci. 1999;112:4651–61. doi: 10.1242/jcs.112.24.4651. [DOI] [PubMed] [Google Scholar]
- 61.Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–85. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 62.Maherali N, Sridharan R, Xie W, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 64.Kim JB, Zaehres H, Wu G, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–50. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 65.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 66.Bru T, Clarke C, McGrew MJ, Sang HM, Wilmut I, Blow JJ. Rapid induction of pluripotency genes after exposure of human somatic cells to mouse ES cell extracts. Exp Cell Res. 2008;314:2634–42. doi: 10.1016/j.yexcr.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paradisi M, McClintock D, Boguslavsky RL, Pedicelli C, Worman HJ, Djabali K. Dermal fibroblasts in Hutchinson-Gilford progeria syndrome with the lamin A G608G mutation have dysmorphic nuclei and are hypersensitive to heat stress. BMC Cell Biol. 2005;6:27. doi: 10.1186/1471-2121-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrés V, González JM. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol. 2009;187:945–57. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halaschek-Wiener J, Brooks-Wilson A. Progeria of stem cells: Stem cell exhaustion in Hutchinson-Gilford progeria syndrome. J Gerontol A Biol Sci Med Sci. 2007;62:3–8. doi: 10.1093/gerona/62.1.3. [DOI] [PubMed] [Google Scholar]
- 70.Csoka AB, English SB, Simkevich CP, et al. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004;3:235–43. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 71.Whetstine JR, Ceron J, Ladd B, Dufourcq P, Reinke V, Shi Y. Regulation of tissue-specific and extracellular matrix-related genes by a class I histone deacetylase. Mol Cell. 2005;18:483–90. doi: 10.1016/j.molcel.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–5. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 73.Warner HR, Sierra F. Models of accelerated ageing can be informative about the molecular mechanisms of ageing and/or age-related pathology. Mech Ageing Dev. 2003 May;124:581–7. doi: 10.1016/s0047-6374(03)00008-3. [DOI] [PubMed] [Google Scholar]
- 74.Von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Müller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–84. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- 75.Guerry F, Marti CO, Zhang Y, Moroni PS, Jaquiéry E, Müller F. The Mi-2 nucleosome-remodeling protein LET-418 is targeted via LIN-1/ETS to the promoter of lin-39/Hox during vulval development in C. elegans. Dev Biol. 2007;306:469–79. doi: 10.1016/j.ydbio.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y. Identification of differentially expressed target genes of human nucleosome remodelling Mi-2 orthologue LET-418 in C. elegans. 2006. University of Fribourg, PhD Thesis.
- 77.Khaleque MA, Bharti A, Gong J, et al. Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene. 2008;27:1886–93. doi: 10.1038/sj.onc.1210834. [DOI] [PubMed] [Google Scholar]
- 78.Valgardsdottir R, Chiodi I, Giordano M, et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423–34. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shumaker DK, Dechat T, Kohlmaier A, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Enukashvily NI, Donev R, Waisertreiger IS, Podgornaya OI. Human chromosome 1 satellite 3 DNA is decondensed, demethylated and transcribed in senescent cells and in A431 epithelial carcinoma cells. Cytogenet Genome Res. 2007;118:42–54. doi: 10.1159/000106440. [DOI] [PubMed] [Google Scholar]
- 81.Eymery A, Calllanan M, Vourch C. The secret message of heterochromatin new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol. 2009;53:259–268. doi: 10.1387/ijdb.082673ae. [DOI] [PubMed] [Google Scholar]
- 82.Faulkner J, Keirstead HS. Human embryonic stem cell-derived oligodendrocyte progenitors for the treatment of spinal cord injury. Transpl Immunol. 2005;15:131–42. doi: 10.1016/j.trim.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 83.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 84.Thyagarajan B, Blaszczak AG, Chandler KJ, Watts JL, Johnson WE, Graves BJ. ETS-4 is a transcriptional regulator of life span in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001125. doi: 10.1371/journal.pgen.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2010;67:1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–9. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–4. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–5. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 89.Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Extending lifespan in C. elegans. Science. 2004;305:1238–9. doi: 10.1126/science.305.5688.1238c. [DOI] [PubMed] [Google Scholar]
- 90.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 91.Haithcock E, Dayani Y, Neufeld E, et al. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:16690–5. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerindeficient cells. J Cell Biol. 2005;170:781–91. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McKenna NJ, O’Malley BW. Nuclear receptors I. Cell. 2010;142:822. doi: 10.1016/j.cell.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 94.Mooijaart SP, Kuningas M, Westendorp RG, et al. Liver X receptor alpha associates with human life span. J Gerontol A Biol Sci Med Sci. 2007;62:343–9. doi: 10.1093/gerona/62.4.343. [DOI] [PubMed] [Google Scholar]
- 95.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 96.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Müller F. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 97.Kapahi P, Chen D, Rogers, et al. With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–65. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Burtner CR, Kennedy BK. Progeria syndromes and ageing: What is the connection? Nat Rev Mol Cell Biol. 2010;11:567–78. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- 99.Carver BS, Tran J, Chen Z, et al. ETS rearrangements and prostate cancer initiation. Nature. 2009;457:E1. doi: 10.1038/nature07738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nardella C, Carracedo A, Alimonti A, et al. Differential requirement of mTOR in postmitotic tissues and tumorigenesis. Sci Signal. 2009;27:2ra2. doi: 10.1126/scisignal.2000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Angelo G, Van Gilst MR. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science. 2009;326:954–8. doi: 10.1126/science.1178343. [DOI] [PubMed] [Google Scholar]
- 102.McColl G, Killilea DW, Hubbard AE, Vantipalli MC, Melov S, Lithgow GJ. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–7. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li DQ, Kumar R. Mi-2/NuRD complex making inroads into DNA-damage response pathway. Cell Cycle. 2010;9(11) doi: 10.4161/cc.9.11.11735. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Larsen DH, Poinsignon C, Gudjonsson T, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–40. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–9. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–9. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gurdon JB. Adult frogs derived from the nuclei of single somatic cells. Dev Biol. 1962;4:256–73. doi: 10.1016/0012-1606(62)90043-x. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Y, Zhu X. Progress of fish gene technology research in China. Aquaculture Asia. 2002;7:15–6. [Google Scholar]
- 109.Yu L, Zuo W, et al. Cell-engineering grass carp produced by the combination of electric fusion and nuclear transplantation. J Fishery China. 1996;20:314–8. [Google Scholar]
- 110.Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–8. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- 111.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–6. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 112.Weimann JM, Johansson CB, Trejo A, Blau HM. Stable reprogrammed heterokaryons form spontaneously in Purkinje neurons after bone marrow transplant. Nat Cell Biol. 2003;5:959–66. doi: 10.1038/ncb1053. [DOI] [PubMed] [Google Scholar]
- 113.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1(1):39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 114.Schambach A, Cantz T, Baum C, Cathomen T. Generation and genetic modification of induced pluripotent stem cells. Expert Opin Biol Ther. 2010;10:1089–103. doi: 10.1517/14712598.2010.496775. [DOI] [PubMed] [Google Scholar]
- 115.Ding S, Schultz PG. A role for chemistry in stem cell biology. Nat Biotechnol. 2004;22:833–40. doi: 10.1038/nbt987. [DOI] [PubMed] [Google Scholar]
- 116.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–5. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao R, Daley GQ. From fibroblasts to iPS cells: Induced pluripotency by defined factors. J Cell Biochem. 2008;105:949–55. doi: 10.1002/jcb.21871. [DOI] [PubMed] [Google Scholar]
- 118.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–12. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 121.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–61. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roche AE, Bassett BJ, Samant SA, Hong W, Blobel GA, Svensson EC. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol. 2008;44:352–60. doi: 10.1016/j.yjmcc.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–23. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Marchetto MCN, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dolgin E. Gene flaw found in induced stem cells. Nature. 2010;464:663. doi: 10.1038/464663a. [DOI] [PubMed] [Google Scholar]
- 128.Gutierrez-Aranda I, Ramos-Mejia V, Bueno C, et al. Human induced pluripotent stem cells develop teratoma more efficiently and faster than human embryonic stem cells regardless the site of injection. Stem Cells. 2010;28:1568–70. doi: 10.1002/stem.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hu Q, Friedrich AM, Johnson LV, Clegg DO. Memory in induced pluripotent stem cells: Reprogrammed human retinal pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem Cells. 2010;28:1981–91. doi: 10.1002/stem.531. [DOI] [PubMed] [Google Scholar]
- 130.Guenther MG, Frampton GM, Soldner F, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–57. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–62. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 132.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 133.Liu Y, Clem B, Zuba-Surma EK, et al. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell. 2009;4:336–47. doi: 10.1016/j.stem.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 135.Hammond SM, Sharpless NE. HMGA2, microRNAs, and stem cell aging. Cell. 2008;135:1013–6. doi: 10.1016/j.cell.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–60. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 137.Caretti G, Lei EP, Sartorelli V. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: Eclectic regulators of disparate biological functions. Cell Cycle. 2007;6:1172–6. doi: 10.4161/cc.6.10.4228. [DOI] [PubMed] [Google Scholar]
- 138.Carter CL, Lin C, Liu CY, Yang L, Liu ZR. Phosphorylated p68 RNA helicase activates Snail1 transcription by promoting HDAC1 dissociation from the Snail1 promoter. Oncogene. 2010;29:5427–36. doi: 10.1038/onc.2010.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 140.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–74. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 141.Fisher AL, Page KE, Lithgow GJ, Nash L. The Caenorhabditis elegans K10C2.4 gene encodes a member of the fumarylacetoacetate hydrolase family: A Caenorhabditis elegans model of type I tyrosinemia. J Biol Chem. 2008;283:9127–35. doi: 10.1074/jbc.M708341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–6. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hanna J, Saha K, Pando B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yamagoe S, Kanno T, Kanno Y, et al. Interaction of histone acetylases and deacetylases in vivo. Mol Cell Biol. 2003;23:1025–33. doi: 10.1128/MCB.23.3.1025-1033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vilaboa NE, Galán A, Troyano A, de Blas E, Aller P. MDR1 regulation of multidrug resistance 1 (MDR1)/P-glycoprotein gene expression and activity by heat-shock transcription factor 1 (HSF1) J Biol Chem. 2000;275:24970–6. doi: 10.1074/jbc.M909136199. [DOI] [PubMed] [Google Scholar]
- 148.Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci U S A. 2001;98:4990–5. doi: 10.1073/pnas.101617298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lemos Taíla A, Passos Dario O, Nery Flávia C, Kobarg Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 2003;533:14–20. doi: 10.1016/s0014-5793(02)03737-7. [DOI] [PubMed] [Google Scholar]
- 150.Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407(6802):405–9. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 151.Vesuna F, Lisok A, Kimble B, Raman V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia. 2009;11:1318–28. doi: 10.1593/neo.91084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dang CV. MYC, microRNAs and glutamine addiction in cancers. Cell Cycle. 2009;8:3243–5. doi: 10.4161/cc.8.20.9522. [DOI] [PubMed] [Google Scholar]
- 153.Rothenberg ME, Clarke MF, Diehn M. The Myc connection: ES cells and cancer. Cell. 2010;143:184–6. doi: 10.1016/j.cell.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 154.Helbling Chadwick L, Chadwick BP, Jaye DL, Wade PA. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009;118:445–57. doi: 10.1007/s00412-009-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010 Sep;11(9):607–20. doi: 10.1038/nrm2950. Epub 2010 Aug 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Karberg S. Switching on Epigenetic Therapy. 2009;139(6):1029–31. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 157.Lanz RB, O’Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 2010;107:2431–6. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Deshpande AM, Dai YS, Kim Y, et al. Cdk2ap1 is required for epigenetic silencing of Oct4 during murine embryonic stem cell differentiation. J Biol Chem. 2009;284:6043–7. doi: 10.1074/jbc.C800158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang Y. ETS-Fusions networking, triggering and beyond. Genetics and Epigenetics. 2010:31–4. [Google Scholar]
- 160.Zhao J, Ohsumi TK, Kung JT, et al. Genome-wide Identification of Polycomb-Associated RNAs by RIP-seq. Molecular Cell. 2010;40(960):939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang Y. Rough set soft computing cancer classification and network: One stone, two birds. Cancer Inform. 2010;9:139–45. doi: 10.4137/cin.s4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shahrestani P, Mueller LD, Rose MR. Does aging stop? Curr Aging Sci. 2009;2:3–11. doi: 10.2174/1874609810902010003. [DOI] [PubMed] [Google Scholar]
- 163.Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.