Abstract
Background
Human papillomavirus (HPV) positive cases of squamous cell carcinoma of the head and neck (SCCHN) have a much better disease outcome compared to SCCHN cases lacking HPVs. Differences in microRNA (miRNA) expression may affect their clinical outcomes.
Methods
miRNA expression was studied using microarrays and quantitative RT-PCR in HPV-16 positive and HPV-negative SCCHN cell lines. The role of HPV-16 E6 and E7 oncogenes in altering miRNA expression was investigated using human foreskin keratinocytes (HFKs).
Results
MiRNAs miR-363, miR-33 and miR-497 were upregulated while miR-155, miR-181a, miR-181b, miR-29a, miR-218, miR-222, miR-221 and miR-142-5p were downregulated in HPV-positive cells compared to both HPV-negative SCCHN and normal oral keratinocytes. HPV-16 E6 oncogene altered miRNA expression in HFKs and in an HPV-16 positive cell line with E6 knockdown using siRNA.
Conclusions
MiRNAs differentially expressed in the presence of HPV-16 may provide biomarkers for SCCHN and identify cellular pathways targeted by this virus.
Keywords: Human papillomaviruses, SCCHN, microRNA
Squamous cell carcinoma of the head and neck (SCCHN) ranks sixth among cancers worldwide.1 Many of these cases are associated with heavy consumption of alcohol and/or tobacco use, which over time induce mutations in essential genetic pathways that regulate the cell cycle. However, human papillomavirus (HPV) type 16 DNA has been found in up to 30 percent of these cancers, most often in the oropharynx region, and such cases of SCCHN are often found in individuals without the risk factors of alcohol and tobacco use.1, 2 The HPV-positive SCCHN subset has increased in the past 10 years.2 Because of this demographic shift and distinct clinical behavior, the association and relevance of HPV in SCCHN is under intense investigation.
Characteristics of HPV-associated SCCHN are very different from HPV-negative SCCHN, causing disputes whether these cancers should be classified as distinct tumors.3 HPV-positive oral tumors often exhibit loss of cell cycle control proteins, including pRb and cyclin D1, whereas these two proteins are commonly overexpressed in HPV-negative oral tumors.1, 3 One of the most common tumor suppressor proteins, p53, is mutated in up to half of oral cancers, but is very rarely mutated in HPV-positive SCCHN, and tumors with a high viral load have a better prognosis compared to tumors with a low viral load or tumors that are HPV-negative.1, 3 Patients with HPV-positive oral tumors have a better response to chemotherapy, radiation, and surgery,3 and have evidence of immune activation against viral antigens,4 despite having frequent metastasis to regional lymph nodes.3 The biological basis for the differential behavior of HPV-positive SCCHN is not understood.
Micro (mi) RNAs are small, ∼22 nt long, chromosome-encoded single-stranded RNAs that are commonly associated with negative regulation of gene expression.5 MiRNAs are transcribed and exported to the cytoplasm where further processing takes place, and the mature miRNA strand is incorporated into the RNA-induced silencing complex (RISC).5 The miRNA guides the RISC to the 3′ untranslated region of its target mRNA where, depending upon the degree of complementarity, the miRNA either translationally represses the mRNA or targets it for degradation.5 MiRNA dysregulation has been implicated in many different types of human cancers.6, 7
Previous reports have shown altered miRNA profiles in head and neck cancers compared to the normal oral tissue.8-11 MiRNAs with high expression in the tumors compared to the normal oral tissue included miR-21, while miR-125b was downregulated.8, 10, 11 Basal miRNA expression in nine head and neck cancer cell lines found that 33 miRNAs were expressed at a high level and 22 miRNAs were expressed at a low level.12 Interestingly, one of these cell lines, UM-SCC47, is HPV-16-positive.13 In all nine cell lines, let-7a, miR-16, miR-21, and miR-205 were highly expressed, and miR-342, miR-346, and miR-373* were expressed at low levels.12 Although these studies show alterations in miRNA levels in head and neck cancer, they do not address the role of HPVs. Since the number of cases of HPV-16-positive SCCHN have been increasing in the past 10 years,2 and the characteristics of HPV-positive and HPV-negative SCCHN support distinction between these cancers,3 we sought to analyze the miRNA profiles in HPV-positive and HPV-negative SCCHN cell lines.
In this study, we demonstrate that miRNA expression profiles in HPV-16-positive SCCHN cells are distinctly different from those in HPV-negative SCCHN cells and in normal oral keratinocytes (NOKs) that have been immortalized by activation of h-TERT. Using human foreskin keratinocytes expressing either the HPV-16 E6 or E7 oncogene, we also demonstrate that expression of the E6 oncogene results in upregulation of miR-363 and downregulation of miR-181a, mR-218 and miR-29a. Furthermore, siRNA knockdown of HPV-16 E6 in the HPV-positive SCCHN cell line SCC2 reduced expression of miR-363.
Materials and Methods
Cell lines
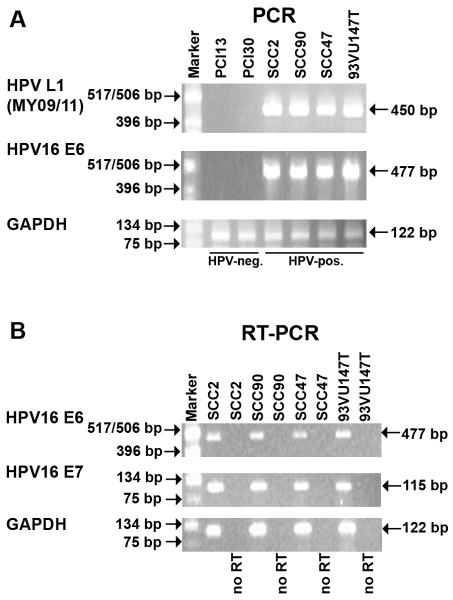
The cell lines used in this study are described in Table 1. Two HPV-16 positive SCCHN cell lines, UD-SCC-2 (gift from Dr. Henning Bier, University of Dusseldorf)14 and UPCI:SCC90,15 and two HPV-negative oropharyngeal SCCHN cell lines, PCI-13 and PCI-30 (gifts from Dr. Theresa Whiteside, UPCI)16 were used for miRNA expression profile analysis. The HPV-16-positive SCCHN cell lines UM-SCC4713, 17 (gift from Dr. Thomas Carey, University of Michigan) and 93-VU-147T18 (gift from Dr. Hans Joenje, VU Medical Center Van der Boechorststraat 7, The Netherlands) were used with the above cell lines for validation of the miRNA microarrays. The UPCI:SCC90, UM-SCC-47 and 93-VU-147T cell lines contain integrated HPV-16 DNA while the HPV-16 status (integrated vs. episomal) of the UD-SSC-2 cell line is not known.15, 18, 19 However, all the HPV-16 positive cell lines were shown to express the viral E6 and E7 genes (see Fig. 1B). The UD-SCC-2, UPCI:SCC90, UM-SCC47, PCI-13, and PCI-30 cell lines were grown in Dulbecco's Modified Eagle's Medium (Lonza, Walkersville, MD, USA) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2% L-glutamine at 37°C in the presence of 5% CO2. The 93-VU-147T cell line was grown in DMEM/F12 medium (MediaTech, Manassas, VA, USA) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2% L-glutamine at 37°C in the presence of 5% CO2. Normal oral keratinocytes (NOKs) that were immortalized by activation of h-TERT20 were grown in defined keratinocyte serum-free medium (Gibco, Grand Island, NY, USA) supplemented with bovine pituitary extract and 1% penicillin/streptomycin at 37°C in the presence of 5% CO2. Primary human foreskin keratinocytes (HFKs) that were transduced with a LXSN-based retroviral vector expressing high-risk HPV-16 E6 or E7 were grown in EpiLife medium (Invitrogen, Carlsbad, CA, USA,) supplemented with human keratinocyte growth supplements (Invitrogen) and 1% penicillin/streptomycin at 37°C in the presence of 5% CO2. HPV-status of samples. The SCCHN cell lines were confirmed to be either HPV-positive or HPV-negative by PCR analysis using the MY09/MY11 primer set, which amplifies a conserved region of the HPV L1 gene.15 Although the HPV-positive SCCHN cell lines have previously been characterized 13-15, 17, 18, we confirmed the HPV status of these cells. UD-SCC-2, UPCI:SCC90, UM-SCC47, and 93-VU-147T were further confirmed to contain HPV-16 DNA by PCR using primers that amplify a 477-bp region of the HPV-16 E6 gene using 5′-ATGCACCAAAAGAGAACTGC-3′ as the forward primer and 5′-TTACAGCTGGGTTTCTCTAC-3′ as the reverse primer. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a loading control using 5′-AGGGGAGATTCAGTGTGGTG-3′ as the forward primer and 5′-GGCCTCCAAGGAGTAAGACC-3′ as the reverse primer, amplifying a 122-bp region. All PCR reactions were performed as described previously.15 The PCR amplified DNA was analyzed by agarose gel electrophoresis.
TABLE 1.
SCCHN Cell Line Characteristics
| SCCHN Sample | TNM Stage | Specimen Site | Gender | HPV Status | p53 gene |
|---|---|---|---|---|---|
| PCI-13 | T4N1M0 | Oral cavity | Male | HPV-negative | E286K |
| PCI-30 | T3N1M0 | Oral cavity | Male | HPV-negative | wt |
| UD-SCC-2 | T1N2M0 | Hypopharynx | Male | HPV-16 | wt |
| UPCI:SCC90 | T2N1M0 | Base of Tongue | Male | HPV-16 | wt |
| UM-SCC47 | T3N1M0 | Lateral Tongue | Male | HPV-16 | wt |
| 93-VU-147T | T4N2 | Floor of mouth | Male | HPV-16 | wt |
TNM stage, tumor, node, metastasis; wt, wild-type
Fig. 1.
PCR and RT-PCR analysis of SCCHN cell lines. (A) HPV-typing of cell lines by PCR. (B) RT-PCR analysis of HPV-16 E6 and E7 expression in the HPV-positive SCCHN cell lines. No RT, no reverse transcriptase added.
RNA isolation and RT-PCR analysis
Total RNA was isolated from all the cell lines grown to 90% confluency using the Ultraspec™ RNA Isolation System (Biotecx, Houston, TX, USA). DNase-I-treated total RNA (1 μg) of UD-SCC-2, UPCI:SCC90, UM-SCC47, and 93-VU-147T was subjected to RT-PCR analysis for expression of the HPV-16 E6 and E7 oncogenes using the Advantage® Clontech RT-for-PCR Kit (Clontech, Mountain View, CA, USA) according to the manufacturer's instructions. The HPV-16 E6 gene was amplified using the primer set described above, and expression of the HPV-16 E7 gene was done using the forward primer 5′-CAGCTCAGAGGAGGAGGATG-3′ and the reverse primer 5′-GCACAACCGAAGCGTAGAGT-3′, amplifying a 115-bp region. Expression of the GAPDH gene was used as a control, using the primer set described above. The PCR products were analyzed by agarose gel electrophoresis. The HFK-16E6 and HFK-16E7 cell lines were also confirmed to be expressing the intended oncogene via RT-PCR as described above.
miRNA microarray analysis
Small RNAs (<200nt) were enriched from total RNA from 2 HPV-positive SCCHN cell lines (UD-SCC-2 and UPCI:SCC90) and 2 HPV-negative SCCHN cell lines (PCI-13 and PCI-30) using the RNeasy Mini Kit and the RNeasy MinElute Clean Up Kit (Qiagen, Valencia, CA, USA). The small RNA fractions were validated on a 15% acrylamide gel. Small RNA fractions obtained from 5 μg of total RNA were labeled with AlexaFluor 647 (Invitrogen) using the mirVana™ Labeling Kit and hybridized (in duplicate) to the mirVana™ miRNA Bioarrays V2 (Ambion, Austin, TX, USA). These Bioarrays contained 662 antisense oligonucleotides, spotted in quadruplicate, which included 328 known human miRNAs, 152 predicted miRNAs (ambi-miRs), 266 mouse miRNAs (114 unique miRNAs), and 238 rat miRNAs (46 unique miRNAs). The arrays were hybridized with labeled miRNAs at 42°C overnight. Each array was subsequently washed once in low stringency wash solution followed by twice in high stringency wash solution, and dried by centrifugation. The arrays were immediately scanned using GenePix 4000B scanner and the median fluorescent intensities minus the background fluorescence were obtained using the GenePix Pro 6.0 software. The median fluorescent intensities of each spot on the bioarrays were log2 transformed and normalized using the mean intensities within the array and the global mean adjustment between the arrays by the GEDA program (http://bioinformatics.upmc.edu/GE2/GEDA.html). Significance Analysis of Microarray (SAM) program version 1.21 (http://www-stat.stanford.edu/∼tibs/SAM/) was used to perform a t-test to obtain the differential miRNA expression patterns of each sample. MiRNAs with at least a two-fold change in expression with a q-value (false discovery rate) of zero were considered to have significant changes in their expression between the samples.
siRNA knockdown of HPV-16 E6 and transfection assays
The role of HPV-16 E6 in altered miRNA expression in HPV-positive SCCHN cell lines was analyzed using double-stranded siRNA against HPV-16 E6 (siRNA 209 complementary to E6 positions 277 to 298, sense sequence 5′-UCCAUAUGCUGUAUGUGAUTT-3′, Dharmacon, Lafayette, CO, USA).21, 22 The HPV-positive SCCHN cell line SCC2 was seeded (1.5 × 105) into six-well plates, and after 24 hours transfected with 125 nM siRNA using Lipofectamine™ 2000 Reagent (Invitrogen) and OPTI-MEM® I (Gibco). BLOCK-iT fluorescent oligo (Invitrogen) was used as a negative control siRNA (it has no human homologous sequences) as well as a transfection efficiency control. Cells were harvested after 72 hours, and RNA extractions were done as previously described.
Real-Time Quantitative RT-PCR
Array data were confirmed by quantitative real-time RT-PCR (qRT-PCR) using the TaqMan® MicroRNA Reverse Transcription Kit and the TaqMan® MicroRNA Assays (Applied Biosystems, Foster City, CA, USA) and the Real-Time thermocycler iQ5 (Bio-Rad, Hercules, CA, USA). These assays utilize stem-loop primers designed to amplify only the mature miRNA. DNase I-treated total RNA (5 ng) was used for each reaction, and all reactions were done in triplicate. Assays were performed according to the manufacturer's instructions and the miRNA levels were normalized to the small nucleolar (sno) RNU43 levels. Relative expression levels of the miRNAs were calculated using the 2-ΔΔCT values.23 Statistical analysis was done via a two-tailed t-test.
HPV-16 E6 expression in siRNA knockdown experiments was confirmed via qRT-PCR using the QuantiTect SYBR Green PCR kit (Qiagen) according to the manufacturer's instructions. The E6 gene was amplified using the forward primer 5′-AGCGACCCAGAAAGTTACCA - 3′ and the reverse primer 5′-GCATAAATCCCGAAAAGCAA-3′, amplifying a 134bp region. The E6 mRNA levels were normalized to the GAPDH gene, using the primer set described above. DNase I-treated total RNA (1 μg) was used for each reaction, and all the reactions were done in triplicate. E6 mRNA levels were normalized to the GAPDH levels, and relative expression levels were calculated using the 2-ΔΔCT values.23 Statistical analysis was done via a two-tailed t-test.
Results
MicroRNA expression in SCCHN cell lines
The UD-SCC-2, UPCI:SCC90, UM-SCC47 and 93-VU-147T cell lines were confirmed to be HPV-positive by PCR analysis using the MY09/MY11 primers (Fig. 1A). These cell lines were further confirmed to contain HPV-16 DNA by PCR using E6 gene primers (Fig. 1A). Finally, the expression of the HPV-16 E6 and E7 genes in these cell lines was confirmed by RT-PCR analysis (Fig. 1B). We then analyzed miRNA expression in two HPV-16 positive (UD-SCC-2 and UPCI:SCC90) and two HPV-negative (PCI-13 and PCI-30) SCCHN cell lines utilizing miRVana™ miRNA Bioarrays V2. MiRNA analysis showed that 129 human miRNAs were expressed in both of the HPV-negative cell lines (PCI-13 and PCI-30), with miR-21, miR-16 and miR-29a being the most highly expressed (Supplementary Table 1). The HPV-16 positive cell lines (UD-SCC-2 and UPCI:SCC90) both expressed 216 human miRNAs, indicating a general upregulation of miRNA expression in the presence of HPV-16 DNA. The miRNAs with high basal expression included miR-205, miR-16, and miR-21 (Supplementary Table 2).
MicroRNA expression is altered in HPV-16-positive SCCHN cell lines
We compared miRNA expression profiles in two HPV-16 positive cell lines, SCC2 and SCC90, with that of two HPV-negative cell lines, PCI13 and PCI30. Two human miRNAs, miR-363 and miR-33, as well as the rat miRNA miR-497 (which differs from human miRNA-497 by just one nucleotide), were upregulated in HPV-positive cell lines compared to the HPV-negative cell lines (Table 2). Eight human miRNAs and one predicted human miRNA were downregulated in HPV-positive cell lines (Table 2). A comparison of miRNA expression between individual cell lines showed that 2 miRNAs were downregulated in SCC2 compared to PCI13 (Supplementary Table 3), whereas 6 miRNAs were overexpressed in SCC2 compared to PCI30 (Supplementary Table 4). When SCC90 was compared to PCI13, 4 miRNA were overexpressed and 3 miRNA were underexpressed (Supplementary Table 5). On the other hand, 10 miRNA were overexpressed in SCC90 compared to PCI30 (Supplementary Table 6). MiR-363 was upregulated in SCC90 compared to both the PCI13 and PCI30 cell lines, and in SCC2 compared to PCI30 (Supplementary Tables 4-6). We also found that miR-181a was downregulated in both SCC2 and SCC90 compared to PCI13 (Supplementary Tables 3 and 5). Although there were differences in miRNA expression in individual SCCHN cell lines, several miRNAs including miR-363 and miR-181a were similarly altered in both the HPV-positive cell lines in individual as well as pair-wise comparisons with the two HPV-negative cell lines.
TABLE 2.
MiRNAs Differentially Expressed in HPV-16-Positive SCCHN Cell Lines Compared to HPV-Negative SCCHN Cell Lines
| MiRNA | Fold Changea |
|---|---|
| Overexpressed | |
| hsa_miR_363 | 5.16 |
| rno_miR_497 | 3.20 |
| hsa_miR_33 | 1.99 |
| Underexpressed | |
| hsa_miR_155 | -7.60 |
| hsa_miR_181a | -7.34 |
| hsa_miR_181b | -6.76 |
| hsa_miR_29a | -4.68 |
| hsa_miR_218 | -4.22 |
| hsa_miR_222 | -3.69 |
| hsa_miR_221 | -3.38 |
| hsa_miR_142_5p | -3.20 |
| ambi_miR_13232 | -3.10 |
miR, microRNA; hsa, human; rno, rat; ambi, Ambion predicted;
Mean fold changes in HPV-16 positive SCCHN cell lines UD-SCC-2 and UPCI:SCC90 compared to HPV-negative SCCHN cell lines PCI-13 and PCI-30. The q-values of all miRNAs were 0.
HPV-16-positive SCCHN cell lines have altered microRNA expression as compared to both HPV-negative SCCHN cell lines and immortalized normal oral keratinocytes
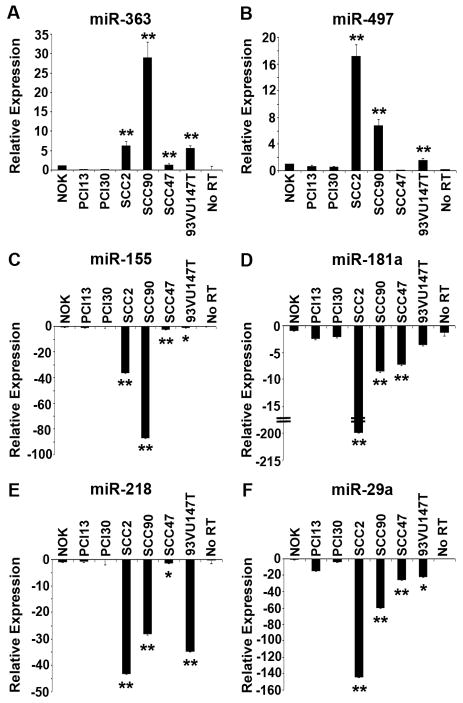
To validate the microarray data, we carried out qRT-PCR analysis for selected miRNAs that were found to be differentially expressed in SCC2 and SCC90 (HPV-positive) cell lines compared to PCI13 and PCI30 (HPV-negative). For this, we used four HPV-16 positive SCCHN cell lines (two that were included in the array analysis and two that were not) and 2 HPV-negative SCCHN cell lines. In order to exclude miRNA profiles only associated with squamous differentiation or immortalization, we also utilized a normal oral keratinocyte (NOK) cell line that has been immortalized by activation of h-TERT.20 The most overexpressed miRNA in the HPV-positive cells based on the array analysis was miR-363. The qRT-PCR results showed higher expression of miR-363 in HPV-positive cell lines SCC2 (6.3-fold), SCC90 (28.9-fold), SCC47 (1.3-fold), and 93-VU-147T (5.5-fold) compared to NOK cells (Fig. 2A). The expression of miR-363 in the HPV-negative cell lines was reduced 12-fold in PCI13 cells and 7 fold in PCI30 cells compared to the NOK cells (Fig. 2A). MiR-363 was upregulated in the above four HPV-positive SCCHN cell lines by 61.7-, 283.1-, 12.8-, and 54.4-fold, respectively, compared to the average expression in PCI13 and PCI30, with p<0.01 (Fig. 2A). Since miR-363 is part of a cluster of miRNAs, we carried out qRT-PCR analysis for two other miRNAs in the cluster, miR-106a and miR-92a. There was no difference in expression of these miRNAs in the HPV-positive SCCHN cell lines compared to the HPV-negative SCCHN cell lines (data not shown). The Ambion mirVana™ miRNA microarray includes a probe for both rat and human miR-497. The rat miR-497 differs from the human miR-497 by the addition of one adenosine at its 3′ end.24 Since the rat miR-497, but not human miR-497, was significantly altered in the HPV-positive samples as compared to the HPV-negative samples in our array analysis, we also carried out qRT-PCR analysis for this miRNA. These results showed that human miR-497 was upregulated in three HPV-positive cell lines, SCC2 (17.2-fold), SCC90 (6.8-fold), and 93-VU-147T (1.61) compared to the NOK cells (Fig. 2B). This miRNA was slightly downregulated in HPV-negative PCI13 (1.8-fold) and PCI30 (2.0-fold) cells relative to the NOK cells (Fig. 2B). MiR-497 was upregulated in the above three HPV-positive SCCHN cell lines by 32.6-, 13.0-, and 3.1-fold, respectively, compared to the average expression in PCI13 and PCI30, with p<0.01 (Fig. 2B).
Fig. 2.
Quantitative real-time RT-PCR validation of miRNA expression data in four HPV-positive and two HPV-negative SCCHN cell lines, and transformed normal oral keratinocyte (NOK) cells. (A) miR-363. (B) miR-497. (C) miR-155. (D) miR-181a. (E) miR-218. (F) miR-29a. No RT, no reverse transcriptase added. Intensity values are relative to the NOK cells, which were arbitrarily assigned a value of 1 or -1. The p values for the HPV-positive cell lines compared to the two HPV-negative SCCHN cell lines are indicated by ** (p<0.01) and * (p<0.05).
The array results also showed that 8 human miRNAs, and one predicted human miRNA were downregulated in two HPV-positive cell lines, SCC2 and SCC90, compared to the HPV-negative cell lines (Table 2). The downregulation of a few selected representative miRNAs in four HPV-positive cell lines as compared to both the HPV-negative cell lines and NOK cells was confirmed by qRT-PCR. MiR-155 was downregulated in SCC2 (36.7-fold), SCC90 (87.4-fold), SCC47 (2.9-fold), and 93-VU-147T (1.4-fold) compared to the NOK cells (Fig. 2C). This miRNA was downregulated in the four HPV-positive cell lines by 42.7-, 101.7-, 3.4-, and 1.6-fold, respectively, compared to the average expression in the PCI13 and PCI30 cell lines, with p<0.01 for SCC2, SCC90, and SCC47 and p<0.05 for 93VU147T compared to PCI13 and PCI30 (Fig. 2C). MiR-181a was downregulated in SCC2 (212.3-fold), SCC90 (8.7-fold), SCC47 (7.3-fold), and 93-VU-147T (3.36-fold) compared to the NOK cells (Fig. 2D). This miRNA was downregulated in the above four HPV-positive cell lines by 91.4-, 3.7-, 3.1-, and 1.6-fold, respectively, compared to the average expression in the PCI13 and PCI30 cell lines, with p<0.01 for SCC2, SCC90, and SCC47 compared to PCI13 and PCI30 (Fig. 2D). Similarly, miR-218 was downregulated in SCC2 (43.3-fold), SCC90 (28.5-fold), SCC47 (1.4-fold), and 93-VU-147T (35.1-fold) compared to the NOK cells (Fig. 2E). This miRNA was downregulated in the above four HPV-positive cell lines by 141.9-, 93.4-, 4.6-, and 114.7-fold, respectively, compared to the average expression in the PCI13 and PCI30 cell lines, with p<0.01 for SCC2, SCC90, and 93VU147T and p<0.05 for SCC47 compared to PCI13 and PCI30 (Fig. 2E). Finally, miR-29a was downregulated in SCC2 (144.8-fold), SCC90 (59.9-fold), SCC47 (26.5-fold), and 93-VU-147T (22.3-fold) compared to the NOK cells (Fig. 2F), while this miRNA was downregulated 18.3-, 7.6-, 3.4-, and 2.8-fold, respectively, compared to the average values of PCI13 and PCI30 cell lines, with p<0.01 for SCC2, SCC90, and SCC47 and p<0.05 for 93VU147T compared to PCI13 and PCI30 (Fig 2F).
The HPV-16E6 oncogene alters MicroRNA expression
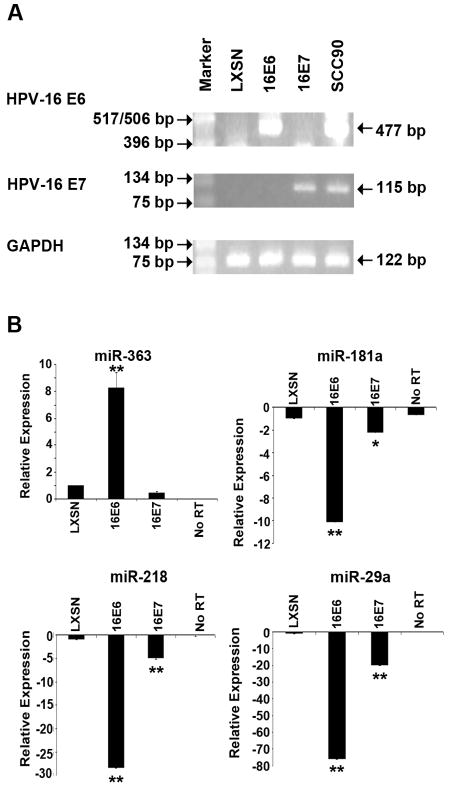
In order to test whether the altered miRNA expression in the HPV-positive SCCHN cell lines was due to the expression of the HPV-16 oncogenes, we utilized primary human foreskin keratinocytes (HFKs) transduced with either the empty vector (LXSN) or vectors expressing the high-risk HPV-16 E6 or E7. Analysis of RNA samples from HFKs containing the empty LXSN vector by qRT-PCR showed that it did not express the HPV oncogenes while the HFK-16E6 and HFK-16E7 cell lines expressed the appropriate oncogene (Fig. 3A). We then tested the relative expression levels of four miRNAs, miR-363, miR-181a, miR-218, and miR-29a that were significantly affected in HPV-positive cell lines (see Fig. 2). QRT-PCR analysis showed that miR-363 (upregulated in HPV-positive SCCHN cell lines compared to both HPV-negative SCCHN cell lines and NOK cells) was upregulated in the HFK-16E6 cell line, with p<0.01, but not in the HFK-16E7 cell line (Fig. 3B). Similarly, miR-181a, miR-218, and miR-29a (downregulated in HPV-positive SCCHN cell lines compared to both HPV-negative SCCHN cell lines and NOK cells) were downregulated in the HFK-16E6 cell line, with p<0.01. These miRNAs were either not affected or affected to a smaller extent in the HFK-16E7 cell line (Fig. 3B). These results showed that expression of the E6 oncogene of HPV-16 is associated with upregulation of miR-363 and downregulation of miR-181a, miR-218 and miR-29a.
Fig. 3.
RT-PCR and qRT-PCR analysis of HFK cell lines. (A). RT-PCR analysis of the HPV-16 E6 and E7 oncogene expression in the respective HFK cell lines. The UPCI:SCC90 cell line, known to express the HPV-16 E6 and E7 genes, was used as a positive control. (B). QRT-PCR analysis of miR-363, miR-181a, miR-218, and miR-29a in the HFK cell lines. No RT, no reverse transcriptase added. Intensity values are relative to the HFK-LXSN cells, which were arbitrarily assigned a value of 1 or -1. The p values for the HPV-16 E6 and E7 expressing HFK cell lines compared the HFK-LXSN cell line are indicated by ** (p<0.01) and * (p<0.05).
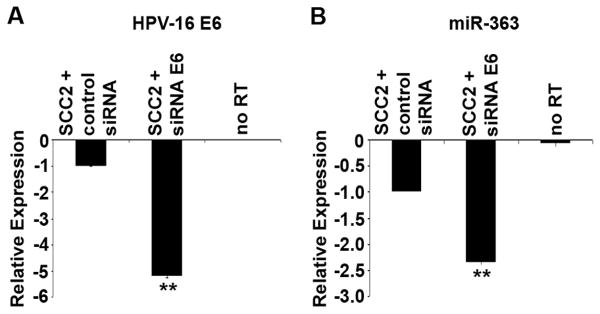
Since the expression of the HPV-16 E6 oncogene altered miRNA expression in the HFK cells, siRNA knockdown of HPV-16 E6 was done in the HPV-positive SCCHN cell line SCC2. The qRT-PCR results showed that reduction in the levels of E6 were accompanied by a reduction in miR-363 levels (Fig. 4). These results suggest that E6 is involved in the upregulation of miR-363 in HPV-positive cell lines.
Fig. 4.
qRT-PCR analysis of HPV-positive SCCHN cell line SCC2 transfected with siRNA against HPV-16 E6. (A). qRT-PCR analysis of HPV-16 E6 oncogene expression in the HPV-positive cell line SCC2 upon transfection with a negative control siRNA or with siRNA against E6. (B). qRT-PCR analysis of miR-363 expression in cells transfected with a negative control siRNA or with siRNA against E6. No RT, no reverse transcriptase added. Intensity values are relative to the SCC2 cells transfected with a negative control siRNA, which were arbitrarily assigned a value of -1. The p-values for the SCC2 + siRNA E6 cells compared to the SCC2 with control siRNA are indicated by ** (p<0.01).
Discussion
HPV-positive and HPV-negative SCCHN have distinctly different clinical outcomes, and the expression and mutation status of important cell cycle control proteins are very different.1, 3 Our data show that several cellular miRNAs are also differentially expressed in HPV-positive SCCHN cell lines as compared to HPV-negative SCCHN cell lines. Many of these miRNAs were also found to be differentially expressed between the HPV-positive SCCHN cell lines and transformed oral keratinocytes lacking HPV DNA. Similarly, expression of the high-risk HPV-16 E6 oncogene in HFKs was strongly associated with changes in miRNA expression similar to that seen in the HPV-positive SCCHN cell lines, while siRNA knockdown of HPV-16 E6 reversed this effect for miR-363. These results suggest that HPV-16, and in particular the E6 oncogene, may be involved in altering the levels of several cellular miRNAs.
Altered regulation of cellular miRNAs has been observed in several types of human cancers6, 7, 25 as well as upon oncogenic viral infections, including hepatitis B and C,26 Epstein-Barr virus,27 human T-cell lymphotrophic virus 1,28 and HPV-16.25, 29, 30 Recent studies have analyzed miRNA expression in SCCHN,8, 9, 12, 31-33 and found that miRNA profiles in SCCHN are different compared to normal oral tissue. However, currently there is no information available on differential miRNA expression between HPV-positive and HPV-negative SCCHN. Since HPV infection has been shown to play a significant role in the etiology and prognosis of SCCHN,1, 3 we wished to study the effect of HPV-16 infection in cellular miRNA dysregulation in SCCHN.
Very few HPV-positive SCCHN cell lines have been described in the literature. Of four such cell lines that are available, we utilized two (UD-SCC-2 and UPCI:SCC90) to compare their miRNA expression profiles to that of two HPV-negative SCCHN cell lines (PCI13 and PCI30) by microarray analysis. We further utilized the two additional HPV-16 positive SCCHN cell lines (UM-SCC47 and 93-VU-147T) to validate the miRNA data obtained in the above comparison. MiRNA microarray analysis showed that three miRNAs (human miR-363 and miR-33, and rat miR-497) were upregulated and eight known and one predicted miRNAs were downregulated in the HPV-positive SCCHN cell lines compared to the HPV-negative SCCHN cell lines (Table 2 and Fig. 2). The miRNA microarray analysis was used as a screening tool, and the data was subsequently validated via qRT-PCR. Similar to our current results, we have previously found that qRT-PCR analysis is much more sensitive than the miRNA microarrays and the fold-difference in qRT-PCR assays is usually much greater.34 MiR-363 was specifically upregulated in HPV-16 positive SCCHN cell lines compared to the HPV-negative SCCHN cell lines as well as NOK cells (Table 2, Fig. 2A), suggesting a possible role of HPV-16 in altering the levels of this miRNA. Furthermore, experiments with HFKs showed that expression of the HPV-16 E6 oncogene increased the levels of miR-363 (Fig. 3B) and siRNA knockdown of E6 reversed this effect (Fig. 4). Interestingly, miR-363 is part of the oncogenic miR-17∼92 family of clusters, which is composed of three clusters of miRNAs, miR-17∼92, miR-106a∼363, and miR-106b∼25 and thought to have evolved through a series of deletions and duplications.35 Other members of this family of miRNA clusters have been implicated in cancers, including small cell lung cancer,36 B cell lymphoma,37 and T cell leukemia.38 MiRNAs in this family have similar or identical seed sequences,35 and since the seed sequence of a mature miRNA contributes significantly to its specificity for its target mRNA5, it has been hypothesized that miRNAs in the miR-17∼92 family may have similar functions.35 MiR-363 has identical seed sequences to miR-92-1, miR-92-2, and miR-25.24 MiR-92-2 and miR-25 are also overexpressed in pancreatic, prostate, and stomach cancers.39 Recently, miR-25 has been shown to be upregulated in gastric cancers where it targets p57, an essential tumor suppressor.40 Since miR-363 and miR-25 have the same seed sequence, and miR-25 is involved in cell cycle disruption,40 it is possible that miR-363 may also be involved in the dysregulation of cell cycle in HPV-associated SCCHN. The qRT-PCR analysis for miR-106a and miR-92a did not show any differences in expression between the HPV-positive and HPV-negative SCCHN cell lines (data not shown). This is not surprising since many miRNAs in a cluster have independent promoters. Eric Rassart's group has shown that the miR-106-363 cluster of miRNAs in mice is located downstream of the Kis2 gene.38 This gene has three different transcription start sites and it appears to encode the primary miRNAs of the 106-363 cluster. Also, the radiation leukemia virus (RadLV) is commonly integrated close to the Kis2 locus. In mice, RadLV-induced tumors had varied expression of miRNAs in the miR-106-363 cluster, indicating that they may not be transcribed from the same promoter.38 Also, in gastric cancer, miR-363 was shown to be downregulated compared to the normal tissue, whereas all of the other miRNAs in the miR-106a∼363 cluster were upregulated.40 Thus, while miR-363 is overexpressed in HPV-positive SCCHN cells compared to HPV-negative SCCHN cells, it is not surprising that we did not see a difference in expression of miR-106a and miR-92a between these cell lines.
Our results also show downregulation of several miRNAs in HPV-associated SCCHN cell lines as compared to both HPV-negative SCCHN and NOK cell lines, including miR-155, miR-181a, miR-218, and miR-29a (Table 2, and Figs. 2 and 3B). We have recently demonstrated that the HPV-16 E6 oncogene downregulates miR-218 expression in HPV-16 positive cervical carcinomas.34 Furthermore, we showed that miR-218 targets LAMB3, and downregulation of miR-218 by the E6 oncogene results in overexpression of LAMB3 in cervical carcinoma cells.34 We found that expression of HPV-16 E6 in HFK cells also reduced the levels of miR-218 (Fig. 3B). The downregulation of miR-218 in both HPV-positive cervical and oropharyngeal cancer cell lines suggests that HPV-16 may target cellular pathways common to these two types of cancers. Although it is documented that p53 expression activates miR-34a41 and miR-34a levels are reduced in HPV-16 positive cervical cancer,30 we did not find a statistically significant difference between miR-34a levels between HPV-positive and HPV-negative SCCHN cell lines. While all the HPV-positive cell lines used in our study are p53 wt, the HPV-negative cell line PCI-13 has a p53 mutation while PCI-30 has wt p53 (Table 1). There are several possible reasons for our observations on miR-34a. For example, since the p53 pathway is complex, it is possible that a single miRNA may be subject to multiple regulatory mechanisms.
Viral infections have been implicated in altered cellular miRNA expression. In human B lymphocytes infected with the Epstein Barr Virus (EBV), elevated levels of miR-155 help in viral persistence by reducing NF-κB signaling.42 It is intriguing that miR-155 was found to be downregulated in the presence of HPV-16 in our studies (Table 2, and Figs. 2C and 3B). There have been other studies on miRNA expression in head and neck cancers that have found miR-155 and miR-181a to be upregulated in oral cancer compared to normal oral tissue.9, 31, 32 However, when we compared HPV-positive and HPV-negative SCCHN cell lines, these miRNAs were downregulated in the presence of HPV-16 DNA. Future studies should define the relationship between reduced levels of these miRNAs in HPV-positive SCCHN.
Our studies showed that miR-181a and miR-29a were downregulated in HPV-positive SCCHN cells compared to HPV-negative SCCHN and NOK cells (Table 2, Figs. 2D, 2F and 3B). The levels of these two miRNAs also decrease upon expression of the HPV-16 E6 oncogene in HFKs (Fig. 3B), suggesting a role for E6 in downregulation of these miRNAs. The miR-181 family is known to be highly expressed in the brain43 and is involved in thymocyte development,44 but its role in other tissues is less well-understood. Our data is the first to show a downregulation of miR-181a and miR-181b in HPV-positive SCCHN cell lines compared to HPV-negative SCCHN cell lines (Table 2, Figs. 2D and 3B). Overexpression of miR-181a and miR-181b has been shown to induce apoptosis and inhibit growth and invasion in glioma cells.45 Further studies on the roles of the miR-181 family may elucidate roles of these miRNAs in the different characteristics seen in HPV-positive and HPV-negative SCCHN. MiR-29a has been shown to interact with viral proteins. For example, miR-29a targets the HIV-1 Nef protein and interferes with viral replication.46 MiR-29a also targets p85 and CDC42, which are negative regulators of p53.47 Since the HPV-16 E6 protein promotes the degradation of the p53 protein,48 it is possible that downregulation of miR-29a by E6 may further reduce p53 levels upon persistent HPV infection. The precise role of HPV infection in cellular miRNA dysregulation, and the role of HPVs in SCCHN development which also contributes to a better prognosis for these cancers as compared to their HPV-negative counterpart will be the subject of future studies.
Supplementary Material
Acknowledgments
We thank Dr. Henning Bier, University of Dusseldorf, for the UD-SCC-2 cell line, Dr. Thomas Carey, University of Michigan, for the SCC47 cell line, Dr. Hans Joenje, VU Medical Center Van der Boechorststraat 7, The Netherlands, for the 93-VU-147T cell line, and Dr. Theresa Whiteside, University of Pittsburgh Cancer Institute, for the PCI-13 and PCI-30 cell lines.
Contract grant sponsor: NIDCR RO1 DE016406 (Dr. Khan). NIH training grants 2T32 AI049820 (Molecular Microbial Persistence and Pathogenesis) and 5T32 GM065100 (Biotechnology Training Grant).
Abbreviations
- HPV
human papillomavirus
- miRNA
microRNA
- SCCHN
squamous cell carcinoma of the head and neck
References
- 1.Tran N, Rose BR, O'Brien CJ. Role of human papillomavirus in the etiology of head and neck cancer. Head Neck. 2007;29:64–70. doi: 10.1002/hed.20460. [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 3.Vidal L, Gillison ML. Human papillomavirus in HNSCC: recognition of a distinct disease type. Hematol Oncol Clin North Am. 2008;22:1125–1142. vii. doi: 10.1016/j.hoc.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Albers A, Abe K, Hunt J, et al. Antitumor activity of human papillomavirus type 16 E7-specific T cells against virally infected squamous cell carcinoma of the head and neck. Cancer Res. 2005;65:11146–11155. doi: 10.1158/0008-5472.CAN-05-0772. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 8.Ramdas L, Giri U, Ashorn CL, et al. miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head Neck. 2009;31:642–654. doi: 10.1002/hed.21017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong TS, Liu XB, Chung-Wai Ho A, Po-Wing Yuen A, Wai-Man Ng R, Ignace Wei W. Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. Int J Cancer. 2008;123:251–257. doi: 10.1002/ijc.23583. [DOI] [PubMed] [Google Scholar]
- 10.Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hui AB, Lenarduzzi M, Krushel T, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 12.Tran N, McLean T, Zhang X, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 13.Bradford CR, Zhu S, Ogawa H, et al. P53 mutation correlates with cisplatin sensitivity in head and neck squamous cell carcinoma lines. Head Neck. 2003;25:654–661. doi: 10.1002/hed.10274. [DOI] [PubMed] [Google Scholar]
- 14.Gwosdz C, Balz V, Scheckenbach K, Bier H. p53, p63 and p73 expression in squamous cell carcinomas of the head and neck and their response to cisplatin exposure. Adv Otorhinolaryngol. 2005;62:58–71. doi: 10.1159/000082473. [DOI] [PubMed] [Google Scholar]
- 15.Ferris RL, Martinez I, Sirianni N, et al. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur J Cancer. 2005;41:807–815. doi: 10.1016/j.ejca.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 16.Chikamatsu K, Nakano K, Storkus WJ, et al. Generation of anti-p53 cytotoxic T lymphocytes from human peripheral blood using autologous dendritic cells. Clin Cancer Res. 1999;5:1281–1288. [PubMed] [Google Scholar]
- 17.Brenner JC, Graham MP, Kumar B, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2009 doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steenbergen RD, Hermsen MA, Walboomers JM, et al. Integrated human papillomavirus type 16 and loss of heterozygosity at 11q22 and 18q21 in an oral carcinoma and its derivative cell line. Cancer Res. 1995;55:5465–5471. [PubMed] [Google Scholar]
- 19.Gupta AK, Lee JH, Wilke WW, et al. Radiation response in two HPV-infected head-and-neck cancer cell lines in comparison to a non-HPV-infected cell line and relationship to signaling through AKT. Int J Radiat Oncol Biol Phys. 2009;74:928–933. doi: 10.1016/j.ijrobp.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piboonniyom SO, Duensing S, Swilling NW, Hasskarl J, Hinds PW, Munger K. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 2003;63:476–483. [PubMed] [Google Scholar]
- 21.Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- 22.Tang S, Tao M, McCoy JP, Jr, Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godshalk SE, Bhaduri-McIntosh S, Slack FJ. Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell Cycle. 2008;7:3595–3600. doi: 10.4161/cc.7.22.7120. [DOI] [PubMed] [Google Scholar]
- 28.Yeung ML, Yasunaga J, Bennasser Y, et al. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;68:8976–8985. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Wang HK, McCoy JP, et al. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang SS, Jiang WW, Smith I, et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 33.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 34.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 37.Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 38.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 39.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YK, Yu J, Han TS, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu F, Weidmer A, Liu CG, Volinia S, Croce CM, Lieberman PM. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. 2008;82:10436–10443. doi: 10.1128/JVI.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miska EA, Alvarez-Saavedra E, Townsend M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 45.Shi L, Cheng Z, Zhang J, et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 46.Ahluwalia JK, Khan SZ, Soni K, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 48.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.