Abstract
The eukaryotic cell nucleus displays a high degree of spatial organization, with discrete functional subcompartments that provide microenvironments where specialized processes take place. Concordantly, the genome also adopts defined conformations that, in part, enable specific genomic regions to interface with these functional centers. Yet the roles of many subcompartments and the genomic regions that contact them have not been explored fully. More fundamentally, it is not entirely clear how genome organization impacts function, and vice versa. The past decade has witnessed the development of a new breed of methods that are capable of assessing the spatial organization of the genome. These stand to further our understanding of the relationship between genome structure and function, and potentially assign function to various nuclear subcompartments. Here, we review the principal techniques used for analyzing genomic interactions, the functional insights they have afforded and discuss the outlook for future advances in nuclear structure and function dynamics.
Keywords: genome organization, nuclear structure and function, chromosome conformation capture, next generation sequencing
Transcription of the genome is fundamentally regulated at three distinct yet inter-dependent levels. Firstly, trans-acting regulatory factors are attracted to specific DNA sequences to ensure genes are transcribed in the right cells, at the right time [1]. The second level of control is achieved through modification of both the DNA and the nucleosomes that package it into chromatin [2]. These epigenetic marks can act to recruit activating or repressive protein complexes, and modulate activity by effecting chromatin compaction. Finally, non-random organization of the genome within the nucleus appears to influence transcription [3]. Indeed, the nucleus is a highly structured and compartmentalized organelle. Many subcompartments have been identified by microscopy and biochemical analysis; some of these are relatively well characterized, while the functions of others remain elusive. It is likely that specific regions of the genome contact different subcompartments to carry out their individual roles.
Arrangement of the genome such that certain regions contact the various nuclear subcompartments is perhaps predictable, yet interestingly, it is a responsive, adaptable and cell-type specific process. At interphase, individual chromosome territories occupy discrete positions in the nucleus [4–6], although at least a certain degree of intermingling between neighboring territories is evident [7]. Each chromosome often adopts a preferred position within the nucleus, and displays specific preferences for neighboring chromosomes. Remarkably, this organization is cell-type specific [8], implying that chromosomal positioning may reflect or direct differential gene expression. Indeed, changes in the preferred neighboring of chromosomes occur as progenitor cells differentiate into mature cells [9]. The speed with which the whole chromosome repositioning takes place is astonishing; Bridger and colleagues have observed large-scale chromosome movements in fibroblast nuclei within 15 min of quiescence-inducing serum starvation [10]. Significantly, these movements require energy and actin/myosin polymerization, suggesting that genome reorganization is a directed process. The tight relationship between genome conformation and gene activity is further supported by a study that artificially and reversibly tethered a genomic region to the repressive nuclear periphery, resulting in transcriptional suppression [11, 12], although this is not always the case [13]. Therefore, it appears likely that genomic rearrangements can play a driving role in gene expression changes, rather than merely reflecting a passive consequence.
Over the past 30 years, the limelight has been fixed upon the contributions to genetic control by specific DNA elements, and subsequently chromatin structure modifications, yet the involvement of nuclear organization has remained enigmatic and in the shadows. The major advances have come mostly through microscopy-based analysis, which have been instrumental in the identification and characterization of nuclear subcompartments, and the non-random organization of the genome. However, these efforts have been hampered by both resolution limitations of light microscopy and difficulties in identifying the DNA elements with which the subcompartments interface.
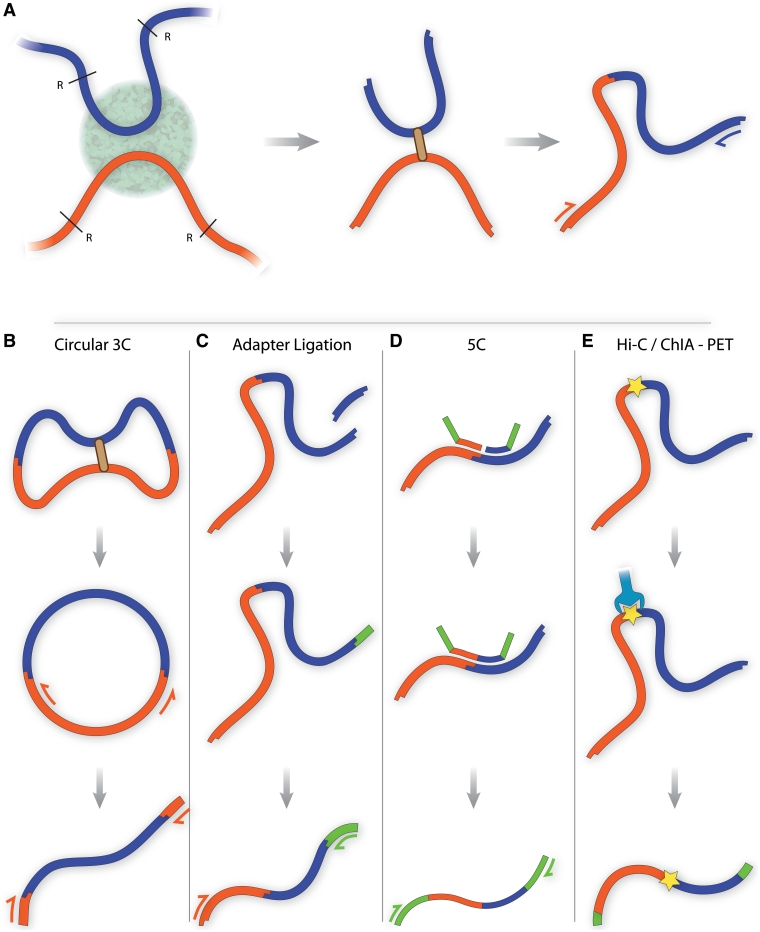
An important technological breakthrough occurred in 2002, when new methodologies were developed to study genomic spatial arrangements of the genome. Firstly, Dekker and colleagues seized upon the ‘Nuclear Ligation Assay’ from the Seyfred laboratory [14, 15] to develop the chromosome conformation capture method (3C) (Figure 1A), a DNA ligation-based proximity assay that was used to examine the structural organization of yeast chromosomes in the nucleus [16]. This provided the first description of a genomic conformation in situ.
Figure 1:
(A) Traditional chromosome conformation capture (3C). DNA sequences co-localising within a region of the nucleus are fixed using formaldehyde and cut using a restriction endonuclease such as Bgl II or Hind III. Cut DNA ends are joined in dilute conditions, favoring ligation events within a fixed complex. Cross-links are reversed and DNA is purified. PCR and quantitative PCR can be used with specific primers to detect expected products in a quantitative manner. (B) Circularized 3C, as carried out by Zhao et al. and Lomvardas et al. [24, 25]. Würtele et al. and Simonis et al. use a second round of restriction digestion and ligation to reduce the DNA circle size [26, 27] (data not shown). DNA sequences are subjected to a prolonged ligation step to form circles. Nested PCR using primers within the known ‘bait’ sequence is performed, yielding a library with known end fragments containing unknown interacting sequences. (C) Adapter ligation 3C. A second restriction enzyme digestion step creates a sticky end near the ligation junction. An adapter sequence is ligated to this sticky end, enabling PCR amplification of products using primers within the ‘bait’ sequence and adapter. (D) 5C. Multiple ligation-mediated amplification primers containing adapter sequence are annealed directly adjacent to restriction sites of interest. Those paired at ligation junctions are ligated, enabling PCR amplification of the ligation products using the adapter sequences present within the primers. (E) Hi-C/ChIA-PET. All ligation junctions are labeled using a biotin tag. Complexes are purified using streptavidin, and adapters are ligated to the library to enable massive parallel sequencing.
3C was rapidly adapted to measure long-range interactions in mammalian cells. Along with a separate, newly developed method called RNA FISH TRAP, which identifies the genomic spatial arrangements at the site of a specific transcript [17], 3C was used to capture the close, physical association that occurs between the β-globin gene promoter and its locus control region (LCR) located 50 kb upstream [18]. This result was significant because it provided strong evidence that distal regulatory elements act upon their target promoters through direct contact, while looping out intervening sequences. This highlighted that gene regulation occurs in three dimensions. Whereas RNA FISH TRAP is technically rather challenging, the relative simplicity of 3C has led to its widespread adoption in studies of long-range interactions at numerous gene loci. The linear distance separating specific interacting regions can be staggering; some cis-interacting partners are located over a megabase apart [19]. Even more remarkable, 3C experiments have suggested specific interactions can occur between loci on separate chromosomes [20, 21]. However, not all interactions detected by 3C necessarily indicate a specific and functional contact between loci. Detected interactions may also occur through co-associations at shared nuclear subcompartments, such as actively transcribed genes at transcription factories [22, 23].
A frustrating limitation of 3C is that it relies upon detection of interacting loci using specific polymerase chain reaction (PCR) primers, yet within a 3C library, a full gamut of genome-wide interactions is present. This precludes the detection of unexpected, yet important interactions. Several groups have addressed this limitation in an effort to identify all the loci that interact with a specific sequence of interest. In essence, two general approaches have emerged, each with varying degrees of success in detecting unpredicted interactions.
The first approach can be categorized generally as circularized 3C (Figure 1B). Here, DNA circles that contain the ‘bait’ sequence and an interacting partner sequences are studied. The DNA circles are either formed naturally through the 3C procedure [24, 25], or generated during subsequent steps by a second restriction enzyme digestion and ligation [26, 27]. Finally, the interacting partner sequences are amplified by inverse PCR and identified by either microarray or sequencing. Würtele and Chartrand used their ‘open-ended 3C’ assay to investigate the spatial environment of the HoxB1 gene during its induction [27]. Zhao et al. [25] used a technique termed 4C (circular 3C or 3C-on-chip) to uncover extensive networks of epigenetically regulated chromosomal interactions, both in cis and in trans. Work by Simonis et al. [26] using 4C also gave insight into nuclear organization by showing that active and inactive chromatin domains form distinct interaction clusters, which is in keeping with the long-held view of active and silent chromatin being spatially segregated into euchromatin and heterochromatin, respectively. Interestingly, circularized 4C approaches detect relatively few long-range cis and trans interactions compared with other 4C methodologies, although it is uncertain why this is the case.
The second approach used to capture interactions of a specific locus can be classified as ‘adapter 3C’ methodologies (Figure 1C). Like some iterations of circular 3C, these techniques involve a second restriction enzyme digestion step to cut the DNA within the interaction partner sequence; however, they diverge when an adapter sequence is ligated to the sticky end. The library is amplified using primers that hybridize within the adapter and bait sequences. The method was first employed by Ling et al. [28] in their ‘associated chromosome trap’ (ACT) assay, which identified three potential interaction partners of an imprinting control region within the Igf2-H19 locus on mouse chromosome 7. Of the three interactions, one was intrachromosomal and known previously, while the remaining two were interchromosomal and novel. However, the assay failed to detect other regions known to interact with the Igf2-H19 locus [29, 30], suggesting that the list of targets identified by the screen was not comprehensive; again, the underlying reasons for this are unclear [31].
A related technique was developed by Schoenfelder et al. [32], called enhanced ChIP 4C (e4C), which incorporates two major modifications to the method used by Ling et al., namely immunoprecipitation and biotin enrichment. Immunoprecipitation has been employed before in 3C-derived methods; both the ChIP loop and combined 3C-ChIP cloning (or 6C) assays contain an immunoprecipitation step before or after ligation, respectively [33, 34]. The enriched ligation products are either detected by specific PCR primers, or cloned and sequenced. In the e4C studies, an antibody that recognizes the transcription-initiating form of RNA polymerase II was used to enrich for the transcribed regions of the genome, to focus on interactions that occur within transcription factories. Secondly, further enrichment is achieved by annealing a biotinylated, bait-specific primer and primer extending into the 3C-ligated interaction partner sequences. By isolating the biotinylated bait ligation fragments on streptavidin-coated magnetic beads, library complexity is greatly reduced, which greatly increases the sensitivity for detection of specific interactions involving a region of interest. Indeed, Schoenfelder et al. [32] found a higher incidence and richer assortment of long-range cis and trans interactions between genes than had previously been detected by other ‘4C’-like incarnations. Significantly, they found evidence that genes preferentially co-associate with distinct subsets of other genes, which appears to be in part a reflection of the shared trans-acting factors that regulate them, and highlights that transcription may act as a major influencing force on genome organization.
Efforts of late have moved toward capturing multiple interacting genomic regions concurrently to provide a more comprehensive view of genome organization. Dostie et al. [35] made the first step, developing 5C (chromosome conformation capture carbon copy) to map the interactions that occur within a given gene locus. This technique uses ligation-mediated amplification, where oligonucleotide primers that anneal immediately adjacent to 3C restriction sites are ligated together to generate an interaction library of fusion oligonucleotides that can be assayed by sequencing or microarray (Figure 1D). Dostie and colleagues applied 5C to the human β-globin gene locus, where they confirmed previously studied interactions and also identified new ones. While there will be practical and financial limitations to the degree of 5C multiplexing that is achievable, this method remains suitable to obtain a detailed, yet comprehensive structure within a given gene locus.
More recently, methodologies have been devised to capture all interactions throughout the genome simultaneously. The Genome Conformation Capture (GCC) technique used by Rodley et al. [36] to capture yeast chromosome interactions simply involved next-generation sequencing of the entire 3C library, without selection or enrichment of ligation products. This is feasible due to the small genome size of yeast; however, GCC is unlikely to be applied in organisms with larger genomes. Three groups have designed methods to enrich for the 3C ligation junctions using biotinylated nucleotide or oligonucleotide tag sequences in between interacting ligated fragments, which is used to purify ligation junctions (Figure 1E). The Hi-C technique developed by Lieberman-Aiden et al. [37] incorporated biotin-based ligation selection to provide a population-average, whole-genome conformation snapshot of human nuclei with one megabase resolution, and gives insight into the organization of active and inactive chromatin domains, chromosome folding and preferred spatial arrangements of individual chromosome territories. The method applied by Duan et al. [38] was similar, where a modified circular 3C technique was combined with biotin enrichment to generate a three-dimensional model of the less complex haploid yeast genome with kilobase resolution, thus revealing the folding pattern of chromosomes and complexity of interchromosomal interactions. The third technique, ChIA-PET (chromatin interaction analysis with paired-end tag sequencing) differs most significantly from Hi-C and modified circular 3C by the method of generating fragments (sonication as opposed to restriction enzyme digestion), and its inclusion of an immunoprecipitation step to selectively identify the genomic loci interactions that occur within the context of a specific protein [39], much like ChIP Loop, 6C and e4C. Fullwood and colleagues used ChIA-PET to map the genomic interactions that co-associate with estrogen receptor alpha binding. This is an exciting development that may be capable of identifying interactions that take place at various nuclear subcompartments, which can provide clarification of the significance of many poorly understood nuclear bodies [40].
Of course, the utility of 3C derivatives to study nuclear organization would be limited had it not been for the genome sequencing projects to unambiguously map the positions of interacting loci, and powerful multiplexing technologies to capture a multitude of genomic interactions concurrently. The initial derivatives of 3C employed custom-made microarrays to assess the frequency of interactions [26, 35]. Gradually, this is being superseded by high-throughput next-generation sequencing, which can provide a greater versatility between experiments, and an ever-increasing output capacity. Certainly, such capacity will be needed to attain an adequate depth of coverage, given the extreme complexity of interactions that are likely to be present in Hi-C and ChIA-PET interaction libraries.
The development of these latest truly genome-wide interaction capture assays can offer a snapshot of whole-genome behavior in its entirety, and will be crucial to our understanding of structure–function relationships of the genome. However, these methods are unlikely to supersede e4C or 5C, which will provide a much more specific and detailed analysis of interactions from the perspective of a limited number of representative loci. Indeed, 3C will continue to be applied to study specific interactions, as well as to confirm those detected via the derivative methods. Akin to the use of different objective lenses on a microscope, the combination of methods can potentially provide a holistic view of genome organization, ranging from a detailed, localized view to an all-encompassing, global representation.
RNA FISH TRAP, which was used in parallel with 3C to capture β-globin gene promoter locus control region interactions, has not had such an immense impact on the genome organization studies as 3C. However, there is a considerable potential for its application to study unique aspects of nuclear organization. RNA FISH TRAP was used to investigate how the Air non-coding transcript represses other genes that reside within the locus [41]. The Air transcript was shown to localize over the promoter regions of these genes, and nucleate the recruitment of transcriptional repressors. This methodology can certainly be applied to other regulatory non-coding RNA species, such as the X chromosome-coating Xist transcript, to elucidate their mode of recruitment and function. Incorporation of next-generation sequencing of RNA FISH TRAP libraries may also ascertain the localization pattern of such non-coding RNA, to determine if interactions are restricted to genes in cis, or if they can occur in trans, as has been observed with the HOTAIR transcript [42], perhaps through nucleated non-coding RNA subcompartments.
This new breed of techniques will open the door to understanding how and why the genome is organized, and provide insight into the roles of many nuclear subcompartments. However, it is important to be mindful of the limitations that accompany it. Libraries for 3C and its derivatives are typically prepared from tens of millions of cells, from which a population-averaged genome conformation will be obtained. While this is useful to explore many generalities of genomic interactions, the specific contexts of interactions such as alternate configurations and mutual exclusivity of interactions will be lost. Single-cell analysis may not yet be feasible for 3C applications, but analysis by microscopy may fill the niche. Developments in the field of light microscopy continue to push against the barriers of resolution limits, and may be capable of distinguishing alternate genomic conformations [43]. In addition, higher resolution electron spectroscopic imaging can be used to show how the genome interfaces with various nuclear subcompartments [44, 45]. Yet to truly understand the functional significance of genome interactions at both a local and global scale, it will be important to integrate these ‘interactomic’ datasets with the ever-accumulating transcriptomic (coding and non-coding RNA) and epigenomic (DNA and histone modifications) datasets, along with genome-wide binding profiles of key regulator proteins. An improved understanding of how they interface and influence each other should make clearer the causal relationships which will undoubtedly be key during cell fate decisions, development and disease.
Key Points.
Spatial organization of the genome has a considerable impact on nuclear function.
Development of novel techniques for detecting chromatin interactions is rapidly advancing an understanding of functional nuclear organization.
Coupling these techniques with the power of next-generation sequencing allows for the collection and collation of large datasets with which complex nuclear arrangements can be inferred.
Acknowledgements
We wish to thank S. Schoenfelder for useful comments in the manuscript. Cameron Osborne is a Leukemia and Lymphoma Research Bennett Fellow. Philip Ewels is supported by a BBSRC studentship, and Alice Young is supported by a MRC studentship.
Biographies
Cameron Osborne studied Biology at Simon Fraser University, Vancouver, Canada. After a PhD with Peter Cockerill, Adelaide, Australia, he worked in the laboratories of James Ellis, Toronto, Canada, and Peter Fraser, Cambridge, UK. He now investigates the relationship between genome organization and cancer.
Philip Ewels studied Biochemistry at the University of Bristol, graduating in 2008. He is now studying for a PhD with the University of Cambridge at the Babraham Institute under the supervision of Cameron Osborne.
Alice Young studied Natural Sciences at the University of Cambridge, graduating in 2006. She worked with François Spitz, Heidelberg, Germany, before starting a PhD at the Babraham Institute under the supervision of Cameron Osborne.
References
- 1.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 2.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2010;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 3.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 4.Bolzer A, Kreth G, Solovei I, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manuelidis L. Individual interphase chromosome domains revealed by in situ hybridization. Hum Genet. 1985;71:288–93. doi: 10.1007/BF00388453. [DOI] [PubMed] [Google Scholar]
- 6.Schardin M, Cremer T, Hager HD, et al. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum Genet. 1985;71:281–7. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- 7.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuroda M, Tanabe H, Yoshida K, et al. Alteration of chromosome positioning during adipocyte differentiation. J Cell Sci. 2004;117:5897–903. doi: 10.1242/jcs.01508. [DOI] [PubMed] [Google Scholar]
- 9.Szczerbal I, Foster HA, Bridger JM. The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma. 2009;118:647–63. doi: 10.1007/s00412-009-0225-5. [DOI] [PubMed] [Google Scholar]
- 10.Mehta IS, Amira M, Harvey AJ, et al. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11:R5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlan LE, Sproul D, Thomson I, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy KL, Zullo JM, Bertolino E, et al. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–7. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 13.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–6. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 15.Gothard LQ, Hibbard JC, Seyfred MA. Estrogen-mediated induction of rat prolactin gene transcription requires the formation of a chromatin loop between the distal enhancer and proximal promoter regions. Mol Endocrinol. 1996;10:185–95. doi: 10.1210/mend.10.2.8825558. [DOI] [PubMed] [Google Scholar]
- 16.Dekker J, Rippe K, Dekker M, et al. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 17.Carter D, Chakalova L, Osborne CS, et al. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–6. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 18.Tolhuis B, Palstra RJ, Splinter E, et al. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 19.Lettice LA, Heaney SJ, Purdie LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–35. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 20.Spilianakis CG, Lalioti MD, Town T, et al. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 21.Xu N, Donohoe ME, Silva SS, et al. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–6. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 22.Osborne CS, Chakalova L, Brown KE, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 23.Osborne CS, Chakalova L, Mitchell JA, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomvardas S, Barnea G, Pisapia DJ, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Z, Tavoosidana G, Sjolinder M, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 26.Simonis M, Klous P, Splinter E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 27.Wurtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 2006;14:477–95. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 28.Ling JQ, Li T, Hu JF, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–72. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 29.Kurukuti S, Tiwari VK, Tavoosidana G, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 31.Krueger C, Osborne CS. Raising the curtains on interchromosomal interactions. Trends Genet. 2006;22:637–9. doi: 10.1016/j.tig.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Schoenfelder S, Sexton T, Chakalova L, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiwari VK, Cope L, McGarvey KM, et al. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–9. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horike S, Cai S, Miyano M, et al. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 35.Dostie J, Richmond TA, Arnaout RA, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodley CD, Bertels F, Jones B, et al. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet Biol. 2009;46:879–86. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan Z, Andronescu M, Schutz K, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fullwood MJ, Liu MH, Pan YF, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborne CS, Eskiw CH. Where shall we meet? A role for genome organisation and nuclear sub-compartments in mediating interchromosomal interactions. J Cell Biochem. 2008;104:1553–61. doi: 10.1002/jcb.21750. [DOI] [PubMed] [Google Scholar]
- 41.Nagano T, Mitchell JA, Sanz LA, et al. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 42.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heintzmann R, Ficz G. Breaking the resolution limit in light microscopy. Brief Funct Genomic Proteomic. 2006;5:289–301. doi: 10.1093/bfgp/ell036. [DOI] [PubMed] [Google Scholar]
- 44.Eskiw CH, Rapp A, Carter DR, et al. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J Cell Sci. 2008;121:1999–2007. doi: 10.1242/jcs.027250. [DOI] [PubMed] [Google Scholar]
- 45.Ahmed K, Dehghani H, Rugg-Gunn P, et al. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]