Abstract
Pluripotent embryonic stem (ES) cells are specialized cells with a dynamic chromatin structure, which is intimately connected with their pluripotency and physiology. In recent years somatic cells have been reprogrammed to a pluripotent state through over-expression of a defined set of transcription factors. These cells, known as induced pluripotent stem (iPS) cells, recapitulate ES cell properties and can be differentiated to apparently all cell lineages, making iPS cells a suitable replacement for ES cells in future regenerative medicine. Chromatin modifiers play a key function in establishing and maintaining pluripotency, therefore, elucidating the mechanisms controlling chromatin structure in both ES and iPS cells is of utmost importance to understanding their properties and harnessing their therapeutic potential. In this review, we discuss recent studies that provide a genome-wide view of the chromatin structure signature in ES cells and iPS cells and that highlight the central role of histone modifiers and chromatin remodelers in pluripotency maintenance and induction.
Keywords: embryonic stem cells, induced pluripotent stem cells, reprogramming, epigenetics, chromatin structure, differentiation
INTRODUCTION
Embryonic stem (ES) cells are pluripotent cells that are derived from the inner cell mass of the pre-implantation embryo at the blastocyst stage. Their most distinguishable features are their capacity to differentiate in cell types derived from the three germ layers: endoderm, mesoderm and ectoderm and to indefinitely self-renew in vitro [1, 2]. The pluripotency capacity of ES cells agrees with the requirement of dynamic genomic organization to support their functional plasticity.
CHROMATIN STRUCTURE OF PLURIPOTENT STEM CELLS
ES cells and nuclear dynamics
From the chromosome territory occupation and genome distribution inside the nucleus, it is clear that the epigenome is dynamic and, that among other processes, it contributes to gene expression and cell differentiation [3–5]. ES cells present a different nuclear architecture and dynamics than differentiated cells [6], indicating that ES cells experience drastic and progressive changes during the differentiation process.
ES cell nuclei are larger than those of differentiated cells, globally, ES cells have a more relaxed chromatin configuration and particular epigenetic features. When differentiation programs are turned on, a gradual and organized redistribution of the genome occurs inside the nucleus, resulting in a rapid reorganization of large areas of the genome that acquire heterochromatin conformation [7]. Indeed, it has been proposed that, the regulated formation of heterochromatin is one of the most critical signals for differentiation [8].
Then, ES cells’ chromatin is globally more de-condensed as compared with differentiated cells and has particular epigenetic features (see below).
Chromatin modifications: histone modifications and histone variants
The recent development of genome-scale chromatin analyses, in particular for a large set of histone covalent modifications, has changed our vision about the chromatin structure forming the skeleton of genes and surrounding intergenic regions, including regulatory elements [9]. Such modifications contribute to the establishment of the ES cell global chromatin configuration and impact on gene expression regulation; ES cell self-renewal and differentiation [10, 11].
Indeed, the capacity of ES cells to respond to differentiation stimuli and acquire a particular cell fate might be determined by a very specific epigenetic trait known as bivalent chromatin. Bivalent chromatin domains are enriched in histone H3 tri-methylated and di/tri-methylated at lysines 4 and 27 (H3K27me3 and H3K4me2/me3), respectively [12–15]. H3K27me3 and H3K4me are marks associated with transcriptionally inactive and active chromatin, respectively (Figure 1). These opposing marks are thought to provide bivalent genes, which are expressed at basal levels in ES cells, with the plasticity to reach full expression potential or be repressed upon activation of specific differentiation programs. Indeed, many of the genes in bivalent domains encode for transcription factors directing tissue-specific differentiation programs. This chromatin organization suggests that histone modifiers inducing H3K27me3 and H3K4me3 have a key function in maintaining pluripotency [16, 17]. Importantly, bivalent chromatin is not the only epigenomic trait associated with ES cells (Figure 1).
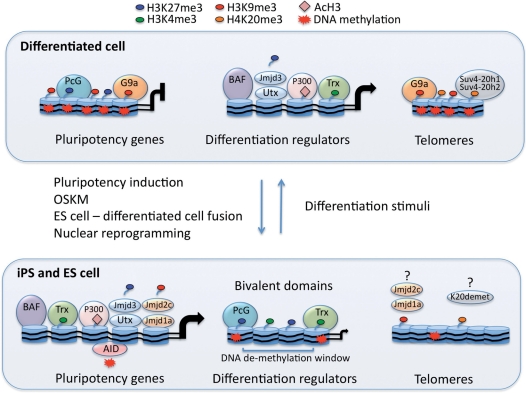
Figure 1:
Model of chromatin reorganization in pluripotency induction. Histone and DNA modifiers participate in the establishment of a generally relaxed and plastic chromatin structure needed for pluripotency induction and maintenance. On the other hand, these epigenetic regulators also control gene expression programs determining cell fate and regulating differentiation. Upon cell reprogramming by OSKM induction, ES cell-differentiated cell fusion or nuclear reprogramming, the pluripotency-associated genes transition from an inactive to an active stage. In differentiated cells, pluripotency regulators are kept repressed by the action of the PcG of proteins via the histone methyltransferase Ezh2, which catalyzes H3K27me3 and G9a, which catalyzes H3K9me3. Additionally, gene repression is ensured by heavy DNA methylation. Upon pluripotency induction, H3K27me3 and H3K9me3 are removed likely by the histone methyltransferases Jmjd3 and Utx and by Jmjd2c and Jmjd1a, respectively, while DNA methylations are removed by AID. Simultaneously, members of the Trx group of proteins introduce H3K4me3, while P300/CBP acetylates histones and ATP-dependent chromatin remodelers of the BAF complex shift nucleosomes, promoting establishment of transcriptionally permissive chromatin and gene expression activation (curved arrow). Differentiation regulators, actively expressed in differentiated cells, are poorly methylated and have a permissive chromatin structure favored by enzymes mediating H3K4me3, histone acetylation and ATP-dependent chromatin remodelers. Upon pluripotency induction, the differentiation regulators shift to a chromatin configuration characterized by the presence of the bivalent marks H3K27me3 and H3K4me3. This chromatin configuration allows for basal gene expression (small curved arrow), while poising genes for repression or activation in future cell fate decisions. Additionally, establishment of DNA demethylation windows poises differentiation regulators for gene activation during cell differentiation. Telomeres are heavily methylated and enriched in repressive histone marks H3K9me3 and H4K20me3, which are catalyzed by Suv4-20h1 and 2. Upon pluripotency induction, telomere length increases along with decreased histone and DNA methylation levels. Whether H3K9 and H4K20 demethylases participate in telomere remodeling is not known.
Epigenetic silencing associated with histone lysine 9 methylation also contributes to the ES cell maintenance. It is known that globally, H3K9me2 and H3K9me3 histone marks, associated with repressive chromatin, are maintained at low levels in ES and they become enriched in differentiated cells (Figure 1) [6]. Ng and collaborators showed that the H3K9me demethylases Jmjd1a and Jmjd2c are important for ES cell self-renewal [18]. Notoriously, Oct4 positively regulates the expression of these histone demethylases, which maintain the Tcl1 and Nanog genes (two key transcription factors for self-renewal in ES cells) in an open chromatin configuration by H3K9me2 and H3K9me3 demethylation, respectively [18]. Furthermore, the down regulation of Oct4 during differentiation favors decreased Jmjd1a and Jmjd2c transcription, facilitating the incorporation of H3K9me2 and H3K9me3 and the epigenetic silencing of pluripotency-associated genes. Thus, histone demethylases play a key function in ES cell pluripotency maintenance and differentiation.
Another relevant aspect of ES cell epigenetics is the incorporation of histone variants. Allis and collaborators recently demonstrated that the histone variant H3.3 interacts with active and repressed genes in ES cells, in a HIRA-dependent manner [19]. HIRA is a histone chaperone specific for histone H3.3 that mediates replication-independent nucleosomes assembly [20] and appears to limit ES cell differentiation [6], suggesting that indeed H3.3 might influence the ES cell status. Other complexes have been found to deposit H3.3 in ES cells. The death domain-associated protein (Daxx) and the α-thalassemia X-linked mental retardation protein (ATRX) deposit H3.3 at constitutive heterochromatin in murine ES cells [21]. However, if ATRX-Daxx and its associated deposition of H3.3 have a function in pluripotency remains to be addressed.
The human ES cell DNA methylome
DNA methylation is important for establishing the dynamic chromatin configuration of the genome in pluripotent ES cells, and for coordinating genomic reorganization during cell differentiation. DNA methylation and Polycomb-repressive proteins (PcG) are both required for pluripotency; they impede premature expression of differentiation regulators [22]. However, although DNA methylation is critical in early embryonic differentiation, cellular memory and development [23], its function in stem cell pluripotency and differentiation remains a topic of intense discussion.
ES cells apparently tolerate loss of both de novo and maintenance DNA methyltransferases [24, 25]. With just 0.6% methylation of CpG dinucleotides, Dnmt3a−/− and Dnmt3b−/− ES cells cannot initiate differentiation efficiently, but remain viable and pluripotent, as indicated by the presence of alkaline phosphatase and Oct4 expression [24]. Similarly, a triple knockout ES cell line for Dnmt1, Dnmt3a, and Dnmt3b grew robustly and maintained its undifferentiated characteristics [25]. These observations suggest that DNA methylation is not essential for ES cell pluripotency, but rather for ES cell differentiation. In addition, active promoters in murine ES cells are heavily methylated and 36% of genes with methylated promoters can still be expressed. Promoters bound by Nanog or Oct4 are examples of this trait [26]. Thus, DNA methylation by itself does not suffice for gene repression in ES cells and pluripotency maintenance.
More recently, genome-wide DNA methylation analyses uncovered distinct and dynamic epigenetic profiles in stem cells as compared with differentiated cells [27, 28]. For instance, DNA methylation is associated with the majority (87%) of repressed genes that do not overlap with bivalent chromatin domains in ES cells [29]. Thus, DNA methylation constitutes a relevant repressive mechanism for genes not influenced by bivalent chromatin in ES cells.
DNA methylation is also necessary for the epigenetic silencing of key pluripotency transcription factors needed for ES cell differentiation (Figure 1). Indeed, pluripotency-associated genes like Nanog1 and Zfp42/ are unmethylated and expressed in ES cells, while they are silenced and methylated in mouse fibroblasts [29]. Furthermore, the DNA methyltransferases Dnmt3a and Dnmt3b target the Oct3/4 and Nanog promoters in differentiated ES cells [30].
More recently a single-base resolution map of DNA methylation in human ES cells was generated [31]. An unexpected result was the significant methylation of non-CpG-enriched DNA, with varied distributions of methyl marks on mCHG or mCHH (where H represents C, T or A). Moreover, non-CpG DNA methylation represents ∼25% of the ES cell DNA methylation and is underrepresented in binding sites for Nanog, Sox2 and Oct4 transcription factors, enriched in exons, introns and 3′-untranslated regions [31]. Importantly, the DNA methylation distribution in ES cells is different from that in differentiated cells, in which non-CpG methylation is lost, suggesting that non-CpG methylation may participate in cell differentiation and that it might be a signature for pluripotency [31].
DNA methylation is linked to PcG-complexes-mediated repression. However, evidence suggests that this is not always the case. For example a genomic scale comparison of genes targeted by PcG complexes and those enriched on DNA methylation showed that both sets of genes were not strongly associated in ES cells [29]. Thus, DNA methylation and PcG-mediated repression can act as independent silencing mechanisms. However, this is still in debate. In cancer cells, DNA methylation is linked to PcG components [32]. For instance, EZH2 acts in concert with DNA methyltransferases. In contrast, other reports suggest that DNA methylation and PcG complexes act independently [33, 34]. These results and others suggest that EZH2 is not the main means for DNA methylation recruitment in cancer cells [35]. Indeed, the majority of the H3K27me3 occupied genes lack DNA methylation. Moreover, recent studies determined that targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a, but not de novo DNA methylation [35]. In conclusion, at this point the mechanisms targeting DNA methylation in undifferentiated cells are poorly understood. Identifying targets in which repression is associated with PcG-dependent or PcG-independent DNA methylation in ES cells would further our understanding of the function of different repressive chromatin configurations in establishing the pluripotency transcriptional network, as well as in determining cell lineages.
ATP-dependent chromatin remodeling complexes in embryonic stem cells
The ATP-dependent chromatin remodeling complexes are multiprotein complexes of variable compositions. Using energy from ATP hydrolysis, they relocate nucleosomes through sliding mechanisms and nucleosome eviction [36], induce changes in nucleosomes conformation and favor the interchange of canonical histones by histone variants [20, 37]. By these activities, chromatin-remodeling complexes contribute to gene expression activation or repression and label defined sectors of the genome through the incorporation of histone variants. ATP-dependent chromatin remodeling complexes are mainly grouped in the SWI/SNF, ISWI, CHD and INO80 families [38].
In addition to DNA methylation and PcG-mediated regulation, the ATP-dependent chromatin remodeling complexes participate in regulating the ES cell chromatin structure (Figure 1), self-renewal capacity and differentiation. In ES cells these complexes cooperate with pluripotency factors in gene expression regulation [1]. A large-scale RNA interference screen against regulatory factors and chromatin components relevant for ES cell maintenance identified Brg1, which is the ATPase of the SWI/SNF complex. Indeed Brg1 knockdown results in loss of the capacity of ES cells to self-renew [39, 40]. Furthermore, Brg1 interacts and co-localizes with the pluripotency transcription factors Nanog, Oct4 and Sox2 at their target genes [41, 42]. Interestingly, Brg1 binds to a significant number of lineage-associated genes that have bivalent histone marks in ES cells, suggesting that the repressive activity of Brg1 is relevant for cell fate determination [42]. In support of this notion, Brg1 depletion impairs ectodermal and mesodermal determination [43]. In addition, Baf250a or Baf250b, which are subunits of the SWI/SNF complex known as Baf (Brg1 associated factor), are also important for ES cell maintenance and differentiation [44]. Several other remodeling complexes are necessary for stem cell pluripotency. For instance, Chd1 (chromodomain-helicase-DNA-binding protein 1), a component of the mammalian ISWI complex, maintains an open chromatin conformation and is required for pluripotency maintenance and induction [45, 46].
The NURD (Mi-2/nucleosome remodeling and deacetylase) complex is associated with ATP-dependent nucleosome remodeling and histone deacetylation activities that mediate gene repression [47]. The NURD component MBD3 (methyl-CpG-binding domain protein) is indispensable for silencing of pluripotency-associated factors, ES cell commitment into developmental lineages [48] and embryo development [49].
In summary, ATP-dependent chromatin remodeling enzymes are required for ES cell self-renewal, pluripotency and cell differentiation into particular lineages. Whether ATP-dependent chromatin remodelers perform hierarchical functions in the remodeling of ES cell chromatin would insight into the epigenetic control of pluripotency. Determining the genome-wide occupation and target genes of different ATP-dependent chromatin remodelers in ES cells and during induction and progression of differentiation should help resolve this issue.
Polycomb group of proteins in human embryonic stem cells
Transcriptional repression via the Polycomb repressor complexes (PRC) is important for maintaining the pluripotent state. PRCs are mostly conserved from Drosophila to human [50]. The PRC2 is recruited to genomic sites via interaction with DNA-binding factors (like YY1) [51] and mainly catalyzes H3K27me3. This histone mark provides the recognition signal for PRC1 incorporation, resulting in induction of a repressive chromatin configuration that can be segregated through cell generations [52]. In ES cells, the PRC2 complex occupies bivalent chromatin domains (Figure 1). Thus, an important function of PRC2 is to keep cell differentiation regulators repressed to maintain pluripotency [53]. At the same time genes repressed by PRC2 are marked by H3K4me3 and remain poised for activation upon differentiation induction (Figure 1).
Accumulated evidence supports a dual function for PcG in ES cells. PcG proteins are required to maintain pluripotency and progenitor stem cells populations, in part, by epigenetically regulating key genes linked to the cell-cycle control and cell proliferation, such as p16INK4a and p19ARF [54]. The resolution of bivalent histone marks upon cell differentiation induction implies that histones have to be demethylated either at K4 or K27 in a regulated manner. In this regard, JMJD3 and UTX have been identified as H3K27me2/3 demethylases that might counteract Polycomb-mediated epigenetic silencing and favor transcriptional activation of lineage-specific groups of genes [55].
Similar to what happens upon loss or reduction of DNA methylation, lack of the polycomb members Ring1B or Eed in ES cells results in lineage-specific gene derepression [56–58]. These transcriptional changes destabilize ES cells, but surprisingly, they do not affect their self-renewal properties. In addition, ES cells lacking members of either PRC1 or PRC2 can differentiate in vitro. Similarly, Eed-deficient ES cells retain pluripotency, as they form teratomas in mice [59]. Thus, members of the PcG of proteins appear to be dispensable for maintaining the ES cell state and for ES cell differentiation. Yet, these complexes contribute to establishing the global chromatin environment in ES cells, raising the possibility that PcG proteins act in concert with other epigenetic mechanisms in pluripotency regulation.
The PRC1 complex mono-ubiquitylates H2A at lysine 119 and induces gene repression [60]. Despite the fact that PRC2-mediated H3K27me3 serves as docking site for PRC1, whether ubiquitylated H2A has a function in maintenance of bivalent domains, pluripotency maintenance or cell fate acquisition remains to be explored.
Another function of the PcG, which is poorly explored in the context of ES cells, is the formation of high-order structures through multiple long-range chromatin interactions or looping that occlude access of regulatory factors to their target sequences [61].
In addition to PcG, the Trithorax (TrxG) group of proteins participates in the epigenetic regulation of ES cells. To some extent, this group of proteins antagonizes the activity of PcG proteins. TrxG forms a complex in which the histone methyltransferase MLL1 induces H3K4me3 methylation, which is an open chromatin mark. The activity of MLL1 complexes in transcriptional activation is complemented by the SWI/SNF or the NURF ATP-dependent chromatin remodeling complexes [62].
A critical aspect of the action of the Polycomb and Trithorax (TrxG) groups of proteins is how such regulatory complexes are recruited to their genomic target regions in ES cells. Although there are no clear proposals, three possibilities have been discussed. The first one and the less documented, is the existence of highly specific DNA binding elements analogous to the Drosophila Polycomb response elements (PREs), which might be recognized by PcG members. To our knowledge, only two mammalian PRE sequences have been identified [63, 64], but whether PcG proteins occupy such sites in ES cells or not is an open question. The second proposal is that PcG is recruited via interaction with transcription factors and associated co-factors. One of the most studied PcG protein is YY1, the vertebrate homolog of Drosophila PHO, which is a transcription factor that can recruit PcG complexes. However, this function of YY1 remains controversial, as its capabilities as recruiter cannot be generalized [55]. Two other factors that interact with DNA have been associated with PRC2 recruitment in ES cells. JARID2, a histone demethylase, binds DNA through its C-terminal domain and co-occupies genomic regions with PRC2 complexes. Moreover, depletion of JARID2 negatively affects the interaction of PRC2 with its target genes. In one proposal, JARID2, which is catalytically inactive [65], acts as enhancer or attenuator of the activity of the PcG complexes [55]. The other factor, PCL2/MTF2 (Polycomb-like 2/metal response element-binding transcription factor 2), is the homolog of Drosophila Polycomb-like (dPcl) and associates with the PRC2 complex in ES cells [66, 67]. Like JARID2, PCL2 co-localizes with PRC2 in a subset of PcG target genes in ES cells and promotes H3K27me3, suggesting that PCL2 might function in regulating the pluripotency transcriptional network. Interestingly, the pluripotent transcription factors Oct4 and Nanog interact with the Pcl2 gene promoter in ES cells and the Pcl2 relative abundance decreases upon differentiation [67]. Finally, the third component recruiting PcG and TrxG proteins to their target sites along the genome are the non-coding RNAs [68]. HOTAIR (Hox antisense Intergenic RNA), the most striking example, corresponds to a 2.2-kb non-coding RNA, which is transcribed from the HOXC locus in the human chromosome 12 in fibroblasts and recruits PRC2 to the HOXD locus on chromosome 2 via interaction with the PRC2 member SUZ12 [69]. A recent report demonstrated that HOTAIR over-expression promotes cancer metastasis [70]. Thus, HOTAIR or related non-coding RNAs might contribute to gene repression in ES cells by recruiting PcG proteins.
Two novel non-coding RNAs that can recruit the PcG complexes to specific locations have been recently described. ANRIL, a 30–40 kb long non-coding RNA, expands over the INK4a/ARF/INK4b locus [71]. The association of ANRIL with PcG of proteins is mediated by CBX7 (chromobox 7), a component of the PRC1 complex that binds ANRIL. CBX7 and ANRIL are expressed at elevated levels in prostate cancer tissues. The other non-coding RNAs are small RNAs of 50–200 nucleotides, which are transcribed from the 5′-non-transcribed region of Polycomb target genes in primary T cells and ES cells [72]. A stem–loop structure formed by these short-RNAs interacts with the PRC2 complex through SUZ12, mediating repression of Polycomb target genes. Importantly, such short RNAs are depleted from polycomb target genes upon initiation of cell differentiation and transcriptional activation [72].
MicroRNAs are essential for controling pluripotency. Indeed, ES cells lacking proteins that mediate microRNA biogenesis exhibit defects in proliferation and differentiation [73]. On the other hand epigenetic regulators target microRNAs in ES cells. Indeed, the H3K27 histone methyltransferase and PcG member EZH2 represses the expression of miR-214 in skeletal muscle and ES cells [74]. Interestingly, once miR-214 is expressed a negative feedback loop is created; in which miR-214 targets the EZH2 3′-UTR, reducing EZH2 levels and promoting ES cell differentiation [74].
In summary, the PcG complexes perform diverse functions over a varied number of target genes in ES cells. This underscores the requirement for better understanding how PcG complexes are recruited in a regulated manner to specific locations in the genome in order to unveil the epigenetic mechanisms of pluripotency, cell fate acquisition and cell differentiation.
CHROMATIN STRUCTURE OF INDUCED PLURIPOTENT STEM CELLS
Induction of an ES cell-like stage
The therapeutic potential held by ES cells prompted for the understanding of the regulatory pathways behind pluripotency. A groundbreaking discovery defined a set of four transcription factors, whose forced expression is sufficient to reprogram mouse embryonic fibroblasts (MEFs) into pluripotent cells known as induced pluripotent stem cells or iPS cells [75, 76]. These factors are Oct4 (Oct3/4, Pou5f1), Sox2, Klf4 and c-myc, often referred to as OSKM. Subsequent research demonstrated the capacity of these factors to reprogram a variety of differentiated cell types into iPS cells [75–80].
iPS cells are remarkably similar to ES cells, in terms of cell morphology and pluripotent capabilities. Both cell types form teratomas containing tissues from endodermal, mesodermal and endodermal origin when introduced into immunocompromised mice. In addition, they are both capable for somatic and germline contributions in chimeric mice when injected into blastocysts and share the most stringent criteria of pluripotency, they can produce viable mice by tetraploid complementation [81–83]. Furthermore, ES cells and iPS cells employ the same molecular mechanisms to maintain expression of the pluripotency regulator Nanog and pluripotency properties via activin/nodal signaling [84]. Moreover, the two cell types share very similar global gene expression profiles [85–89]. The global epigenetic landscape, as indicated by the distribution of histone modifications and DNA methylation, are also very similar between ES and iPS cells [11, 90–92]. These similarities, besides absence of the ethical issues raised by ES cells, highlight the potential of iPS cells as suitable substitutes for ES cells in regenerative medicine [93]. However, the process of iPS generation is still slow and inefficient, stressing the need to understand the molecular mechanisms driving de-differentiation. In this regard, re-programming into a pluripotent state, either by nuclear transfer, cell fusion or transcription factors induction [94], is characterized by genome-wide chromatin reorganization into a more permissive environment for transcription [89–92], pointing to epigenetic control of chromatin structure as central for pluripotency induction. Supporting this notion, histone modifiers and their recruiters, including large intergenic non-coding RNAs (lincRNAs) [95], as well as a DNA de-methylation enzyme, participate in the activation of pluripotency regulators and are critical for reprogramming induction [96]. Our current knowledge on the chromatin structure of iPS cells derived mainly from studies, here discussed, addressing distribution of histone modifications as well as genome-wide and gene-focused DNA methylome analyses.
Histone modifications in iPS cells
Genome-wide occupancy maps of two histone modifications: H3K4me3 and H3K27me3, associated with transcriptionally active and repressive chromatin, respectively, have been generated to understand the global chromatin environment of ES and iPS cells (Figure 1). Analysis of these marks has been particularly informative, as their distributions correlate well with global gene expression profiles, suggesting that these marks might have a relevant function in establishing the pluripotent gene expression program [91]. The distributions of these marks near promoters and in intergenic regions are remarkably similar in ES and iPS cells, but differ significantly from that of the iPS parental MEFs [92]. This indicates that MEFs suffer global chromatin rearrangements during reprogramming to recapitulate the ES cell’s chromatin conformation. Indeed, over 97% of promoters with high CpG contents lacking H3K4me3 in MEFs regain this mark in iPS cells and bivalent domains are reestablished by 80% in high CpG promoters and by 95% in loci encoding developmental transcription factors [97]. Moreover, the variation of these histone marks between ES and iPS cells is not greater than that observed within ES or iPS cell lines [11]. The similarities in the distributions of H3K4me3 and H3K27me3 between ES and iPS cells suggest that both are important for pluripotency induction. However, the distribution pattern of H3K27me3 seems to be more dissimilar among MEFs, iPS and ES cells than that of H3K4me3, which is more conserved, suggesting that reprogramming is mainly associated with H3K27me3 and highlighting the relevance of the polycomb complexes in this process [91]. Accordingly, ES cells deficient in the members of the PRC2 Eed, Suz12 and the H3K27 methyltransferase Ezh2, failed to efficiently reprogram human lymphocytes to a pluripotent state in cell fusion experiments [98].
In contrast to the notion that reprogramming is mainly associated with H3K27me3, integrative genomic analyses incorporating studies on histone methyl marks and DNA methylation distribution suggest that chromatin modifiers catalyzing H3K4me3 and H3K27me3 are both relevant for direct reprogramming to a pluripotent state. This is characterized by opening of global chromatin structure, re-activation of pluripotency regulators and simultaneous polycomb-mediated repression of developmental regulators [97]. Thus, chromatin modifiers that mediate global gene activation and repression are key for pluripotency induction (Figure 1). This opens the possibility for the involvement of histone methyltransferases catalyzing other histone modifications in reprogramming. Indeed, enzymes inducing histone acetylation and H3K9me3, which promote gene expression activation and repression, respectively, are required for induction of pluripotency. The activity of the histone acetyltransferase complex P300/CBP promotes expression of pluripotency regulators and is critical for OSKM-mediated iPS cell derivation [99]. On the other hand, H3K9me, induced by the histone methyltransferase G9a, is associated with Oct4 inactivation [100], suggesting that this mark could also act as a barrier for reprogramming. Accordingly, inhibition of G9a by a small molecule BIX-01294, which induces a decrease of H3K9me2 levels [101], can replace for Oct4 in transcription factor-induced pluripotency [75, 102, 103], furthermore, knockdown of G9a or over-expression of the H3K9 demethylase Jhmd2a induce activation of an Oct4-GFP reporter in cell fusion experiments [103], however, whether G9a directly antagonizes pluripotency induction remains to be tested [104].
Telomeres are required for chromosome stability during cell division, shorten during cell aging and are enriched in methylated DNA and H3K9me3 and H4K20me3 (Figure 1). These histone marks might limit telomere length by impeding access to telomerase [105], which is required for iPS generation [106]. H3K9me and H4K20me, but not DNA methylation, decreases at telomeres and pericentromeric repeats, while telomere length increases to levels comparable to ES cells in iPS cells [106]. This evidence suggests that histone methyltransferases inducing H3K9me and H4K20me might be key for telomere length regulation and pluripotency induction. This implies the possible involvement of H3K9 and H4K20 demethylases in these processes; however, these possibilities remain to be explored.
As for other chromatin modifiers, how histone-modifying complexes get recruited to their targets genes is poorly understood. Recent evidence demonstrating the requirement of a lincRNA for pluripotency induction provides insight into this issue [95]. LincRNAs regulate gene expression via recruitment of Polycomb complexes to target genes [107, 108] and their expression pattern distinguishes ES from iPS cells [95]. A group of 10 lincRNAs are enriched in iPS cells, as compared with ES cells, suggesting that lincRNAs are closely associated with induction of the transcriptional program regulating pluripotency. Indeed, expression of the lincRNA-RoR (lincRNA-regulator of reprogramming) is controlled by OCT4. Furthermore, knockdown of lincRNA-RoR in fibroblasts inhibited, while its over-expression increased reprogramming efficiency [95]. Thus, lincRNA-RoR is important for pluripotency induction, but whether this function is related to recruitment of Polycomb proteins remains to be explored. Addressing this possibility could shed light on the general mechanisms for regulated target selection by histone modifiers and further our understanding on the epigenetic mechanisms controlling reprogramming towards a pluripotent state.
Chromatin remodelers in iPS cell generation
Induction of the pluripotent state requires accessible chromatin [109]. Not surprisingly, ATP-dependent chromatin-remodeling complexes are important for acquisition of the pluripotent state [110]. Indeed, the BAF (Brg1-associated factors) complex maintains Oct4 expression and is required for ES cell renewal and pluripotency [43, 45, 46]. In addition, the members of the BAF complex, Brg1 and Baf155, improve the efficiency of OSKM-induced pluripotency and can substitute for c-Myc in the process. Accordingly, Brg1 and Baf155, in combination with OSK, favored a significant enrichment of H3K4me3 on the Tcf3, Oct4A, Oct4B and Lefty2 promoters; and of H3K9Ac on Tcf3 and Lefty2 promoters, as compared with OSK alone. Simultaneously, H3K27me3 decreased on the promoter region of the pluripotency gene Sall4 [111].
How chromatin-remodeling complexes get recruited to their targets to remodel chromatin structure and aid in pluripotency induction is poorly understood. However, a clue might come from a recent study showing that Klf4 physically interacts with the BAF complex members BRG1 and BRM, which knockdown reduces OSKM-induced pluripotency [109]. Thus, chromatin-remodeling complexes might be recruited to their targets via interaction with pluripotency-associated transcription factors; however, this hypothesis remains to be formally tested.
DNA demethylation in iPS cells
The promoter regions of Oct4 and Nanog, as well as other pluripotency regulators, are methylated in MEFs and become demethylated during reprogramming to a pluripotent state. Moreover, pluripotency induction is more efficient after transfer of somatic nuclei into an enucleated oocyte [97], in which DNA demethylation occurs immediately [112], as compared with pluripotency induction by transcription factors induction, in which DNA de-methylation occurs after weeks of culture [92]. In addition, partially reprogrammed cells with incomplete repression of lineage-specific transcription factors and remodeling of histone modifications also have persistent DNA hypermethylation [97] and treatment of these cells with 5-aza-cytidine, a DNA methyl transferase inhibitor or knockdown of the DNA methyltransferase Dnmt1 results in fully reprogrammed cells [97]. Thus, DNA methylation poses a major barrier for reprogramming. In support of this notion, reprogramming of adult murine cells results in iPS cells with residual DNA methylation patterns matching that or their parental cells. In addition iPS cells tend to differentiate towards related lineages, suggesting that DNA methylation provides a means for epigenetic memory for cell origin [113].
In addition to its requirement for pluripotent regulators reactivation, DNA demethylation might have an important function in the response of pluripotent cells to differentiation stimuli. In agreement with this idea, demethylation of DNA windows on enhancers of developmental regulators is required for gene activation [114]. A recent study provided more evidence for this effect of DNA demethylation. By examining differentially methylated regions (DMRs) in ES, iPS cells and their parental fibroblasts, on a genome-wide scale [115], it identified differentially hypermethylated and hypomethylated cytosine-phospate-guanine (CpG) island shores in human iPS cell lines as compared with their parental fibroblasts. Interestingly, roughly equal levels of hypermethylated and hypomethylated CpG island shores were found in both cell types. However, differential enrichment of hyper- and hypo-methylated CpG island shores distinguished iPS cells from their parental fibroblasts [115]. In iPS cells, more DMRs were hypomethylated and not hypermethylated than in fibroblasts and were associated with bivalent chromatin marks, which identify developmental regulators. In addition, hypomethylated CpG island shores overlapped with binding sites for POU5F1, NANOG and SOX2 [115]. These results support the notion that global DNA methylation remodeling is required for the acquisition of pluripotency.
In apparent contradiction, comparison of the methylation patterns on approximately 66 000 CpG sites in human fibroblasts, ES and iPS cells revealed that globally iPS and hES were more methylated than fibroblasts and that iPS cells were more methylated than hES cell lines. However, a small fraction of CpG sites located at pluripotency-associated genes was hypomethylated in pluripotent cells [116]. These results suggest that the balance between DNA methylation and demethylation are highly regulated during reprogramming and support the requirement of demethylation of pluripotency-associated genes for this process. Thus, DNA must be demethylated for epigenetic memory resetting, reactivation of the pluripotent transcriptional program and might be required for proper response of pluripotent cells to differentiation stimuli, highlighting the necessity to uncover the mechanisms driving DNA demethylation. In this regard, a recent study showed strong evidence suggesting that the cytocine deaminase AID (activation induced cytidine demethylation), which induces DNA demethylation [117], is required for sustained expression of human NANOG and OCT4 and the onset of reprogramming towards pluripotency (Figure 1) [118]. How AID is targeted to pluripotency-associated genes and therefore how selective DNA de-methylation takes place are unknown.
CONCLUSIONS AND FUTURE DIRECTIONS
Histone methyltransferases, chromatin-remodeling complexes and DNA demethylation-mediating enzymes are important for global chromatin resetting to the plastic state needed for induction and maintenance of pluripotency. These global changes imply reprogramming the expression of multiple genes and raise questions on the mechanisms for selective recruitment of chromatin modifiers to target genes. Evidence suggests that recruitment of chromatin modifiers is modulated by interaction with transcription factors as well as non-coding RNAs. Given the relevance of chromatin and DNA modifiers in pluripotency induction, uncovering the global protein–protein and protein–RNA interactions of these modifiers, is of utmost importance to understand the epigenetic mechanisms controlling reprogramming. High throughput approaches, like the use of protein arrays [119], will be instrumental aids in undertaking this challenging task.
Key Points.
Globally relaxed chromatin underlies ES and iPS cell plasticity, which allows for pluripotency maintenance and simultaneous priming of cell-specific genes for activation or repression upon differentiation.
Histone, DNA modifiers and ATP-dependent chromatin remodelers act genome-wide to establish the ES and iPS chromatin environments and thus are essential for pluripotency maintenance, induction and response to differentiation stimuli.
Identification of chromatin remodelers’ recruiters and their genome-wide targets in ES and iPS cells is of utmost importance for understanding pluripotency and reprogramming.
FUNDING
This work was supported by the Dirección General de Asuntos del Personal Académico-Universidad Nacional Autónoma de México (IN209403 and IN214407) and Consejo Nacional de Ciencia y Tecnología (CONACyT: 42653-Q and 58767). PDO was supported by the California Institute Regenerative Medicine.
Acknowledgements
The authors thank Gary Howard for editorial assistance.
Biographies
Paul Delgado-Olguín is a postdoctoral fellow in the Gladstone Institute of Cardiovascular Disease in San Francisco. His research focuses on uncovering the epigenetic mechanisms underlying cardiac cell differentiation, development and maintenance.
Félix Recillas-Targa is Head of the Department of Molecular Genetics at the Instituto de Fisiología Celular in the Universidad Nacional Autónoma de México. His laboratory investigates the effects of chromatin structure in development and gene regulation at distinct levels, ranging from regulatory elements to chromatin domain formation and maintenance.
References
- 1.Keenen B, de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol. 2009;219:1–7. doi: 10.1002/jcp.21654. [DOI] [PubMed] [Google Scholar]
- 2.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–71. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 3.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 4.Chakalova L, Debrand E, Mitchell JA, et al. Replication and transcription: shaping the landscape of the genome. Nat Rev Genet. 2005;6:669–77. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- 5.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2007;7:540–6. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 6.Meshorer E, Yellojoshula D, George E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasser S. Visualizing chromatin dynamics in interphase nuclei. Science. 2001;296:1412–6. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 8.Grigoryev SA, Bulynko YA, Popova EY. The end adjusts the means: heterochromatin remodeling during terminal cell differentiation. Chromosome Res. 2006;14:53–69. doi: 10.1007/s10577-005-1021-6. [DOI] [PubMed] [Google Scholar]
- 9.Barski A, Cuddapah S, Cui KR, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Frampton GM, Sodner F, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–57. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan G, Tian S, Nie J, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Bilodeau S, Kagey MH, Frampton GM, et al. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–9. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landeira D, Sauer S, Poot R, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–24. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alder O, Lavial F, Helness A, et al. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137:2483–92. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 17.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh YH, Zhang W, Chen X, et al. Jmjd1a and Jmjd2c histone Lys 9 demathylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–57. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg AD, Banaszynski LA, Noh K-M, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tagami H, Ray-Gallet D, Almouzni G, et al. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 21.Lewis PW, Elsaesser SJ, Noh KM, et al. Daxx is an H3.3-specific histone chaperone and cooperate with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–80. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med. 2010;207:2287–95. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol. 2009;10:526–37. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 24.Jackson M, Krassowska A, Gilbert N, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–71. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsumura A, Hayakawa T, Kumaki Y, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cell. 2006;11:805–14. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 26.Fouse SD, Shen Y, Pellegrini M, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex and histone H3K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–9. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fouse SD, Shen Y, Pellegrini M, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–9. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farthing CR, Ficz G, Ng RK, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JY, Pu MT, Hirasawa R, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–59. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–21. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vire E, Brenner C, Deplus D, et al. The polycomb group protein EZH2 directly controld DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 33.McGarvey KM, Greene E, Fahrner JA, et al. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 34.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 35.Rush M, Appanah R, Lee S, et al. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics. 2009;4:404–14. doi: 10.4161/epi.4.6.9392. [DOI] [PubMed] [Google Scholar]
- 36.Dechassa ML, Sabri A, Pondugula S, et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts CW, Orkin SH. The SWI/SNF complex-chromatin and cancer. Nat Rev Cancer. 2004;4:133–42. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 38.Saladi SV, de la Serna IL. ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev. 2010;6:62–73. doi: 10.1007/s12015-010-9120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–74. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self renewal and pluripotency. Proc Natl Acad Sci USA. 2009;106:5181–6. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–9. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 42.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–28. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 43.Ho L, Jothi R, Ronan JL, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009;106:5187–91. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan Z, Wang Z, Sharova L, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–65. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaspar-Maia A, Alajem A, Polesso F, et al. Chd1 regulated open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–8. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landry J, Sharov AA, Piao Y, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue Y, Wong J, Moreno GT, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 48.Kaji K, Caballero IM, MacLeod R, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 49.Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–32. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 50.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–42. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 51.Caretti G, Di Padova M, Micales B, et al. The polycomb EZH2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–38. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen KH, Bracken AP, Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 53.Lee TI, Jenner RG, Boyer LA, et al. Control of develomental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–98. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178:219–29. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Endoh M, Endo TA, Endoh T, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–24. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 58.van de Stoop P, Boutsma EA, Hulsman D, et al. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouase embryonic ítem cells. PLoS One. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kallin EM, Cao R, Jothi R, Xia C, et al. Genome-wide uH2Alocalization analysis highlughts Bmi-1-dependent deposition of the mark at repressed genes. PLoS Genet. 2009;5:e1000506. doi: 10.1371/journal.pgen.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mateos-Langerak J, Cavalli G. Polycomb group proteins and long-range gene regulation. Adv Genet. 2008;61:45–66. doi: 10.1016/S0065-2660(07)00002-8. [DOI] [PubMed] [Google Scholar]
- 62.Schuettengruber B, Chourrout D, Vervoort M, et al. Genome regulation by Polycomb and Trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Sing A, Pannell D, Kararskakis A, et al. A vertebrate Polycomb response element governs segmentation of the posterior hind brain. Cell. 2009;138:885–97. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 64.Woo CJ, Kharchenko PV, Daheron L, et al. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Closs PAC, Christensen J, Agger K, et al. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–40. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li G, Margueron R, Ku M, et al. Jarid2 and PRC2 partners in regulating gene expression. Genes Dev. 2010;24:368–80. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker E, Chang WY, Hunkapiller J, et al. Polycomb-like 2 associates with PRC2 and regulates-transcriptional networks during mouse embryonic stem cell self-renewal and commitment. Cell Stem Cell. 2010;1:71–86. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther. 2010;12:214–22. [PMC free article] [PubMed] [Google Scholar]
- 69.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yap KL, Li S, Muñoz-Cabello AM, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kandere A, Viiri K, Aráujo CC, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex 2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mallanna SK, Rizzino A. Emerging role of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Develop Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Juan AH, Kumar RM, Marx JG, et al. Mir-214-dependent regulation of the polycomb protein EZH2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 77.Liu H, Zhu F, Yong J, et al. 2008) Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–90. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 78.Li W, Wei W, Zhu S, et al. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–9. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 79.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 80.Liao J, Cui C, Chen S, et al. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–15. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 81.Okita K, Ichisaka T, Yamanaka S. Generation of germlinecompetent induced pluripotent stem cells. Nature. 2007;448:313–17. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 82.Zhao XY, Li W, Lv Z, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 83.Zhao XY, Li W, Lv Z, et al. Viable fertile mice generated from fully pluripotent iPS cells derived from adult somatic cells. Stem Cell Rev. 2010;6:390–7. doi: 10.1007/s12015-010-9160-3. [DOI] [PubMed] [Google Scholar]
- 84.Vallier L, Mendjan S, Brown S, et al. Activin/Nodal signaling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–49. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 86.Lowry WE, Richter L, Yachechko R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–8. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 88.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 89.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–23. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 91.Maherali N, Sridharan R, Xie W, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 92.Hanna J, Cheng AW, Saha K, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA. 2010;107:9222–7. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 94.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–12. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhutani N, Brady JJ, Damian M, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–7. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pereira CF, Piccolo FM, Tsubouchi T, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–56. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 99.Mali P, Chou BK, Yen J, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cell. 2010;28:713–20. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–94. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 101.Kubicek S, O’Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–81. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 102.Shi Y, Do JT, Desponts C, et al. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–8. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 103.Shi Y, Desponts C, Do JT, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–74. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 104.Ma DK, Chiang CH, Ponnusamy K, et al. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–41. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marión RM, Blasco MA. Telomere rejuvenation during nuclear reporgramming. Curr Opin Genet Dev. 2010;20:190–6. doi: 10.1016/j.gde.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 106.Marión RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–53. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao J, Sun BK, Erwin JA, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mak AB, Ni Z, Hewel JA, et al. A lentiviral functional proteomics approach identifies chromatin remodeling complexes important for the induction of pluripotency. Mol Cell Proteomics. 2010;9:811–23. doi: 10.1074/mcp.M000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fazzio TG, Panning B. Control of embryonic stem cell identity by nucleosome remodeling enzymes. Curr Opin Genet Dev. 2010;20:500–4. doi: 10.1016/j.gde.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singhal N, Graumann J, Wu G, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–55. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 112.Santos F, Hendrich B, Reik W, et al. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 113.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu J, Watts JA, Pope SD, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–38. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deng J, Shoemaker R, Xie B, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;4:353–60. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rai K, Huggins IJ, James SR, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 2008;135:1201–12. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bhutani N, Brady JJ, Damian M, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–7. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaushansky A, Allen JE, Gordus A, et al. Quantifying protein-protein interactions in high throughput using protein domain microarrays. Nat Protoc. 2010;5:773–90. doi: 10.1038/nprot.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]