Abstract
Chromatin modifications at both histones and DNA are critical for regulating gene expression. Mis-regulation of such epigenetic marks can lead to pathological states; indeed, cancer affecting the hematopoietic system is frequently linked to epigenetic abnormalities. Here, we discuss the different types of modifications and their general impact on transcription, as well as the polycomb group of proteins, which effect transcriptional repression and are often mis-regulated. Further, we discuss how chromosomal translocations leading to fusion proteins can aberrantly regulate gene transcription through chromatin modifications within the hematopoietic system. PML–RARa, AML1–ETO and MLL-fusions are examples of fusion proteins that mis-regulate epigenetic modifications (either directly or indirectly), which can lead to acute myeloblastic leukemia (AML). An in-depth understanding of the mechanisms behind the mis-regulation of epigenetic modifications that lead to the development and progression of AMLs could be critical for designing effective treatments.
Keywords: chromatin, epigenetics, transcription, leukemia, polycom
CHROMATIN AND EPIGENETICS
Packaging DNA into chromatin in the eukaryotic nucleus is essential for the storage of large genomes and also provides a basis for regulating gene expression, allowing the accessibility of the transcription machinery to DNA to be modulated [1, 2]. The core histone proteins (H2A, H2B, H3 and H4) form the H2A/H2B and H3/H4 heterodimers that are assembled into octamers. Approximately 146 bp of DNA are wrapped twice around this structure to form the fundamental structure of chromatin, the nucleosome. Progressive coiling of nucleosomes leads to the formation of higher order chromatin structures [3]. Nucleosomes are connected by linker DNA, which provides a binding site for multiple proteins, including the fifth histone protein, the linker histone H1. H1 sits at the base of each nucleosome near the DNA entry-and-exit site, and its interaction with the linker DNA promotes chromatin compaction [4].
The N-terminal tails of all core histones, as well as the C-terminus of histone H2A, protrude from the tightly bound nucleosome, allowing them to be targeted by chromatin-modifying enzymes, such as acetyltransferases, methytransferases and kinases. The posttranslational modifications catalyzed by these enzymes modulate the chromatin structure and thereby influence the transcriptional competence of the underlying DNA as well as perpetuate the epigenetic memory, which is the propagation of the transcriptional activity of a gene during cell replication. Histone posttranslational modifications can usually be grouped as leading to either gene activation or gene repression: acetylation and phosphorylation are associated with gene activation, whereas sumoylation, deimination and proline isomerization have been implicated in repression [5]. However, histone methylation or ubiquitination can affect promoter activity in either ways, depending on the targeted amino acid residue. In addition, DNA can also be modified by the addition of methyl groups at the CpG dinucleotides, which invariably leads to gene repression. Acetylation and deacetylation of histones, as well as methylation of histones and DNA, are often mis-regulated during tumorigenesis. The mechanisms of these modifications are discussed in the following sections.
Histone acetylation and deacetylation
Acetylation of histones is a reversible process, and in general, acetylation leads to transcriptional activation while deacetylation correlates with gene repression. Histone acetyltransferases (HATs) usually modify more than one lysine within one protein, but some enzymes have a limited specificity for a specific lysine [5]. HATs are divided into three main families: GNAT, MYST and CBP/p300 [6]. Histone deacetylases (HDACs) can be grouped into three distinct classes: the class I HDACs (HDAC1, 2, 3 and 8) and class II HDACs (HDAC4, 5, 6, 7, 9 and 10), and the more recently discovered class III NAD-dependent enzymes of the Sir family. Very few of these enzymes show specificity for a particular lysine and usually affect several acetyl groups. HDACs are involved in many signaling pathways and are part of multiple repressive chromatin complexes, such as N-CoR, SMRT and Sin3 [7–9].
Histone methylation
Histone methyltransferases (HMTs) are much more specific than the acetyltransferases. They usually transfer a methyl group to a single lysine or arginine, thus leading to transcriptional activation or repression depending on the modified amino acid residue. Methylation in lysine 4 (K4), K36 and K79 of histone H3, and asymmetric dimethyl arginine 3 (R3) of histone H4, has been associated with transcriptional activation, whereas methylation at K9 and K27 of histone H3, as well as K20 and symmetric dimethylation at R3 of histone H4, has been commonly implicated in transcriptional repression [5, 10]. Moreover, some specific modifications can trigger activation or repression depending on the localization inside the gene [11].
DNA methylation
DNA methylation is a dynamic epigenetic mark usually associated with repressed chromatin that plays a key role in differentiation, self-renewal and homeostasis [12, 13]. DNA methylation is a covalent modification of DNA catalyzed by DNA methyltransferase enzymes (DNMTs) that occurs on the cytosine within CpG dinucleotides, the so-called CpG regions [14]. Aberrant DNA cytosine methylation can alter gene expression by disrupting the ability of transcription factors to bind to their target DNA sequences [15] or by creating docking sites for methylation-specific transcriptional repressors as methyl-CpG binding domain proteins [16, 17]. These repressors can, in turn, recruit histone-modifying complexes such as HMTs, Sin3, N-CoR or HDACs, thereby leading to gene silencing [16–18].
POLYCOMB GROUP OF PROTEINS
Polycomb group (PcG) of proteins are a class of epigenetic repressors that play an essential role in the control of normal development and cell fate. Originally identified in Drosophila melanogaster as repressors of the homeobox (Hox) genes, they are highly conserved in vertebrates [19]. PcG proteins are assembled into multimeric complexes termed polycomb repressive complexes (PRCs). In mammals, two families of PRCs have been identified to date, termed PRC1 and PRC2. Although both these complexes bind to and covalently modify histone tails, they have different biological functions.
PRC2 is involved in chromatin compaction and gene silencing, functioning mainly by catalyzing the trimethylation of K27 of histone H3. In addition to the HMTs enhancer of zeste (EZH1 and EZH2), the core components of PRC2 complexes are the suppressor of zeste-12 (SUZ12) and the embryonic ectoderm development protein [20–22]. Other proteins, such as PCL, RBBP4/7 and JARID, have also been found to be associated to PRC2 [23–29], although they are not essential for complex formation and stability. Rather, they are involved in the modulation of PRC2’s enzymatic activity. Due to the main role of PcG proteins in the control of cell fate and self-renewal, it is not surprising that this complex has been widely associated with carcinogenesis [30]. Thus, a tight regulation of the epigenetic marks is fundamental to assure correct gene transcription and to prevent a pathologic state.
THE HEMATOPOIETIC SYSTEM: PHYSIOLOGY AND PATHOLOGY
Hematopoiesis is the formation of cellular blood components. The production of terminally differentiated blood cells follows a tightly regulated hierarchical scheme, with the hematopoietic stem cells (HSCs) at the top of the hierarchy [31]. The HSCs are responsible for the life-long production of blood, balancing differentiative divisions that generate the different mature blood cell types with self-renewal divisions that result in additional HSCs [32]. In adult humans, the turnover of cells within the hematopoietic system is estimated to be close to 1 trillion cells per day, which is enabled by the hierarchical multiplying hematopoietic scheme that allows amplification of this enormous quantity of terminally differentiated cells to be precisely regulated [33].
Hematological malignancies are the types of cancer that affect blood, bone marrow and lymph nodes. As the three are intimately connected through the immune system, a disease affecting one of the three will often affect the others as well. Chromosomal translocations are uncommon in solid tumors but are a common cause of hematological neoplasms [34], which leads to a different approach in diagnosis and treatment. Hematological neoplasms are traditionally classified as those located mainly in the blood (leukemia) or in lymph nodes (lymphomas). The most common adult leukemia is acute myeloid leukemia (AML), which is characterized by an aberrant proliferation and accumulation of immature myeloid progenitor cells that can affect the bone marrow, peripheral blood and other tissues as spleen or liver [35]. The FAB classification system of the subtypes of acute leukemia, which is one of the most widely used systems, is based on the morphology, type, maturation and cytochemical and immunophenotypic behavior of the leukemic blasts [36]. The AML classification by the FAB system is detailed in Table 1.
Table 1:
FAB classification of AML subtypes
| Type | Name | Impaired differentiation |
|---|---|---|
| M0 | Acute myeloblastic leukemia, with minimal differentiation | Granulocytic |
| M1 | Acute myeloblastic leukemia, without maturation | Granulocytic |
| M2 | Acute myeloblastic leukemia, with maturation | Granulocytic |
| M3 | Acute promyelocytic leukemia | Granulocytic |
| M4 | Acute myelomonocytic leukemia | Granulocytic and monocytic |
| M4eos | Myelomonocytic together with bone marrow eosinophilia | Granulocytic and monocytic |
| M5 | Acute monoblastic leukemia (M5a) or acute monocytic leukemia (M5b) | Monocytic |
| M6 | Acute erythroid leukemias | Erythroid and granulocytic |
| M7 | Acute megakaryoblastic leukemia | Megakaryocytic |
Despite the fact that AML is the most frequent type of leukemia in adults, it is still the one with the lowest survival rate [37]. The incidence of AML increases with age, and older patients have worse treatment outcomes than younger patients. As an acute leukemia, AML progresses rapidly and is typically fatal within weeks or months if left untreated. With a few exceptions, response to treatment is unsatisfactory, and prognosis is generally poor with current therapies.
The majority of AML cases are associated with non-random chromosomal translocations that often result in gene rearrangements and oncofusion protein formation [34]. In fact, AML is the most extensively cytogenetically characterized human neoplastic disorder, representing 33% of all hematological malignancies and 27% of all malignant disorders with chromosomal abnormalities reported [38]. Many of the gene rearrangements involve a locus encoding a transcriptional activator of genes necessary for myeloid differentiation with a transcriptional protein that is capable of interacting with a corepressor complex [39]. The resulting fusion protein retains the DNA-binding motifs of the wild-type transcriptional activator and also gains the capacity to interact with corepressors through its fusion moiety (Figure 1). This leads to an aberrant silencing of target genes necessary for myeloid development, which acts in concert with other types of genetic anomalies to trigger leukemic transformation. Therefore, leukemia is an end development of a multistage process that is driven by an accumulation of genetic and epigenetic changes. The most common recurrent balanced aberrations, and their corresponding oncofusion proteins, are described in Table 2 [38, 40].
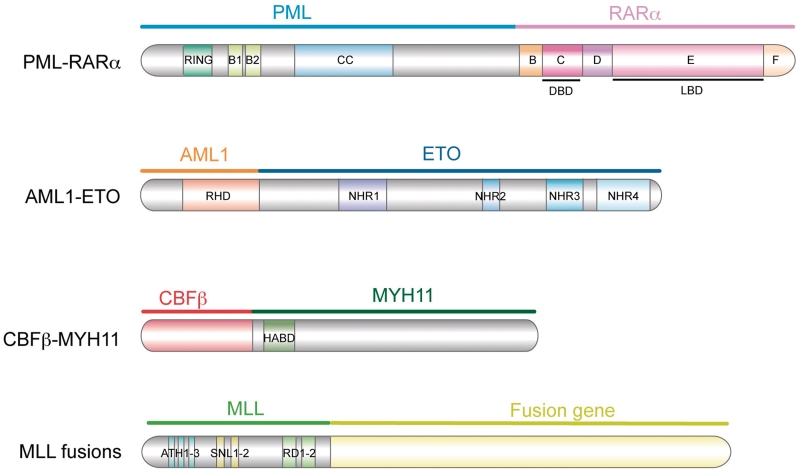
Figure 1:
Structure of the most frequent fusion proteins in AML. PML–RARα, AML1–ETO, CBFβ-MYH11 and the different chromosomal translocations involving the N-terminus domain of MLL are the main AML-associated oncofusion proteins. Ring, really interesting gene; B1 and B2, B-boxes 1 and 2; CC, coiled-coil; DBD, DNA-binding domain; LBD, ligand-binding domain; RHD, Runt homology domain; NHR, nervy homology regions; HABD, high-affinity binding domain; ATH, AT-hook motifs; SNL, speckled nuclear localization sites; RD, repression domain. Note that the MLL fusion is not depicted in scale.
Table 2:
AML-associated oncofusion proteins
| Chromosomal translocation | Oncofusion protein | Occurrence (% of AML) | Prognosis | FAB |
|---|---|---|---|---|
| t(8;21)(q22;q22) | AML1–ETO | 5–12 | Favorable | M2 |
| t(15;17)(q22;q21) | PML–RARα | 6–15 | Favorable | M3 |
| inv(16)(p13q22) | CBFβ–MYH11 | 3–10 | Favorable | M4 |
| der(11q23) | MLL-fusions | 5–8 | Variable | M4/M5 |
| t(9;22)(q34;q11) | BCR–ABL1 | 1–2 | Adverse | M1/M2 |
| t(6;9)(p22;q34) | DEK–NUP214 | <1 | Adverse | M2/M4 |
| t(1;22)(p13;q13) | RBM15–MKL1 | <1 | Intermediate | M7 |
| t(8;16)(p11;p13) | MYST3–CREBBP | <1 | Adverse | M4/M5 |
| t(7;11)(p15;p15) | NUP98–HOXA9 | <1 | Intermediate | M2/M4 |
| t(12;22)(p12;q11) | MN1–TEL | <1 | Variable | M4/M7 |
| inv(3)(q21;q26) | RPN1–EVI1 | <1 | Adverse | M1/M2/M4/M6/M7 |
| t(16;21)(p11;q22) | FUS–ERG | <1 | Adverse | M1/M2/M4/M5/M7 |
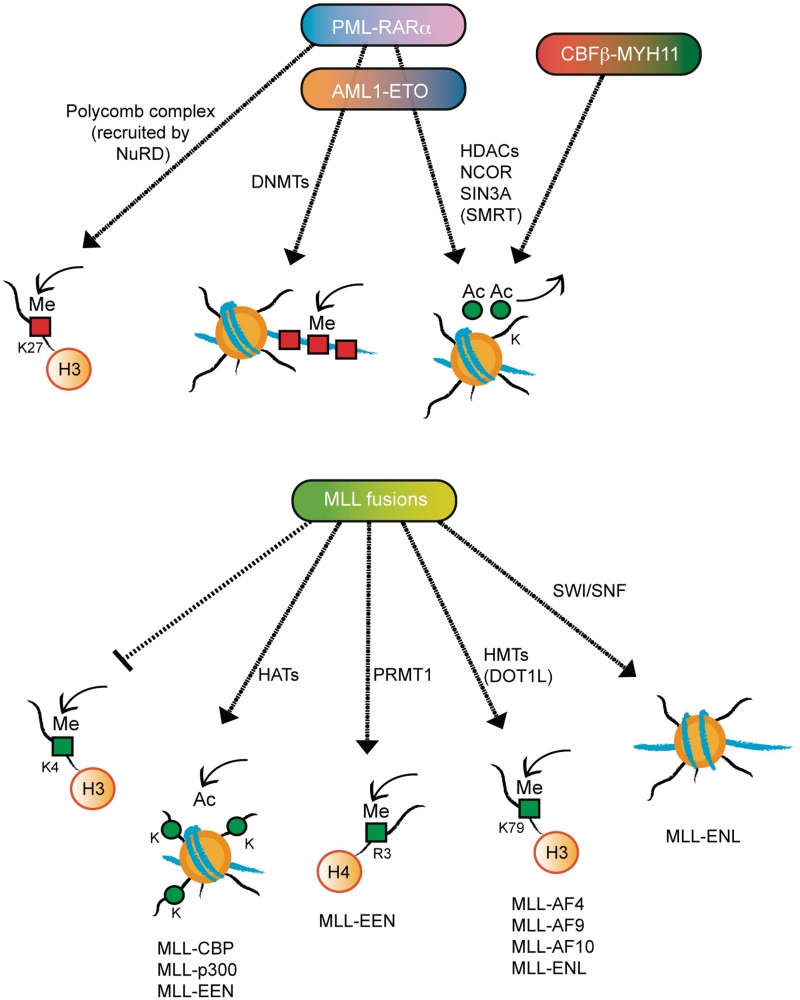
The aberrant recruitment of the epigenetic machinery by the oncofusion proteins to the promoters of key differentiation genes plays a central role in disrupting normal gene transcription and thus triggering AML. As a result, the normal patterns of histone epigenetic marks and DNA methylation are altered in leukemic blasts. Recently, genome-wide promoter DNA methylation patterns have been shown to correlate with unique AML subgroups and to be able to predict clinical outcome [41]. Interestingly, although all oncofusion proteins aberrantly recruit epigenetic modulators, the specific mechanisms involved are different depending on the oncoprotein formed. In the next sections we will review the current knowledge about the aberrant gene expression found in the most frequently occurring AMLs (Figure 2).
Figure 2:
Main epigenetic mechanisms of transcription deregulation by AML fusion proteins. The aberrant recruitment of epigenetic machineries to the promoters of key genes in differentiation by fusion proteins is a central step to disrupt normal gene transcription regulation and thus trigger AML. The mechanisms to deregulate transcription are specific for each oncofusion protein, being determined by the characteristics of its moieties.
PML–RARα FUSION PROTEIN AND ACUTE PROMYELOCYTIC LEUKEMIA
Acute promyelocytic leukemia (APL) is an AML subtype characterized by an aberrant expansion of immature myeloid precursors that are arrested at the promyelocytic stage. The chromosomal translocations associated with APL involve the retinoic acid receptor alpha (RARα) gene in chromosome 17 with one of five different partner genes (PML, PLZF, NUMA, NPM or STAT5b), with the PML–RARα fusion present in >95% of all APL cases [14, 42]. PML–RARα contains the cystein and coiled-coil domains of PML fused to the B-F domains of RARα which contain the DNA binding, heterodimerization, ligand binding and corepressor/coactivator interaction domains [43].
RARα plays a key role in myeloid terminal differentiation as end point of the retinoic acid (RA) signaling pathway [44]. RARα binds to specific DNA sequences called RA-responsive elements (RARE) in the promoter regions of target genes in the form of a heterodimer with the Retinoid X receptor (RXR). In the absence of RA, the heterodimer binds to corepressor complexes (N-CoR, SMRT, Sin3A) and to HDACs, leading to gene silencing [45, 46]. Binding of the RA ligand triggers the dissociation of these complexes and promotes the interaction of RAR/RXR with coactivators, such as p300/CBP, and with the chromatin remodeler Swi/Snf, which works to relax the chromatin structure to allow transcription [47, 48].
The PML–RARα oncofusion protein interacts with, and disrupts the normal functions of, both RARα and PML expressed by wild-type alleles from the same cell. PML–RARα is required but not sufficient to induce frank leukemia (as is the AML1–ETO oncofusion expression) [49]. Other genetic events are required to fully develop APL. The PML–RARα interaction with nuclear corepressors is stronger than that of the wild-type RARα. Additionally, it recruits DNMTs (DNMT1 and DNMT3A) [50], thus imposing an aberrant silencing through intensive histone deacetylation and DNA hypermethylation at RARα target promoters [14]. The nucleosome remodelling and deacetylase corepressor complex is also aberrantly recruited by PML–RARα to target genes and facilitates PRC2 binding, which catalyzes the methylation of H3K27, contributing to gene silencing [51, 52]. Furthermore, PML–RARα homodimerizes through the coiled-coil domain of PML moiety, thus forming large protein complexes with high affinity for corepressors. Once established, PML–RARα-induced epigenetic modifications are maintained throughout the cell cycle.
In PML–RARα-expressing cells, RARα target genes no longer are activated by physiological doses of RA (10−9 to 10−7 M) but rather require pharmacological doses of the ligand (10−6 M). Pharmacological doses of RA can partially overcome the dominant repression of PML–RARα by inducing its degradation, consequently enabling recruitment of coactivators and normal granulopoiesis by the wild-type copy of RARα [53].
Importantly, PML–RARα has been shown to bind to non-canonical RAREs, thus contributing to a widespread transcriptional deregulation [54]. PML–RARα enhances the expansion of leukemic cells by repressing key genes involved in myeloid differentiation as well as in DNA repair processes, while activating growth-promoting genes from the Wnt/Catenin and Jagged/Notch pathways [55, 56]. In contrast to repression, gene activation seems to be indirect, most probably due to the sequestration of corepressors by the fusion proteins. It has also been proposed that the reciprocal RARα–PML fusion protein can cooperate in the deregulation of additional non-RA typical targets, such as cyclin A1 and C/EBPα, which additionally play an important role in controlling myeloid differentiation [57, 58].
Although all fusion proteins retain the RARα moiety, not all are responsive to RA ligand binding. In leukemic cells expressing PLZF-RARα, the corepressor complex remains attached to the oncofusion protein even in the presence of large pharmacological doses of 10−5 M RA [59]. The wild-type PLZF moiety is a transcriptional repressor that can itself interact directly with corepressor [60–64]. These interactions are maintained in the PLZF–RARα fusion protein and are insensitive to RA-induced gene activation. Combinatory treatments of RA with an HDAC inhibitor (such as trichostatin A, TSA) are effective in blocking the recruitment of corepressors by both RARα and PLZF moieties, respectively [65]. This suggests that the moiety fused to RARα plays a critical role in the pathogenesis of the disease and needs to be taken into account in the treatment strategy.
AML1–ETO AND CBFβ–MYH11 FUSIONS
The core binding factor (CBF) complex is a key regulator of definitive hematopoiesis. The CBF family is composed of four proteins: the three alpha subunits AML1 (also called RUNX1 or Cbfα2), RUNX2 (Cbfα1) and RUNX3 (Cbfα3), and the single beta subunit CBFβ [66, 67]. The alpha subunit is the DNA-binding element, while the beta subunit stabilizes the DNA binding but without direct contact with DNA [68]. Mutations and chromosomal translocations involving this complex are frequently implicated in leukemogenesis.
The most frequent recurrent chromosomal abnormality involving AML1 is the translocation t(8;21), in which the first five exons of the AML1 gene, containing the DNA-binding domain (RHD), are fused to almost the entire ETO gene (also called MTG8 or RUNX1T1). Although wild-type AML1 functions as a transcriptional activator, its normal role in initiating hematopoietic differentiation is impaired when fused to ETO. The ETO transcription factor interacts with HDACs and corepressors, such as the nuclear receptor corepressor complex (N-CoR) and Sin3A, causing transcriptional repression by deacetylating histones and creating repressive chromatin structures [69–72]. In addition, AML1–ETO can also recruit DNMT1 [73], which causes promoter DNA hyper-methylation at target genes. Similar to PML–RARα, ETO forms oligomers that increase the stability of corepressor recruitment at target promoters.
Another frequent chromosomal translocation in AML patients fuses the CBFβ subunit to MYH11 (also known as SMMHC). CBFβ–MYH11, similar to AML1–ETO, functions by dominantly repressing normal CBF complex activity. MYH11 is able to recruit corepressors (Sin3A and HDAC8) to the AML1-regulated genes [74, 75]. Although the members of the CBFα family are predominantly nuclear, wild-type CBFβ remains in the cytoplasm and is recruited to the nucleus upon heterodimerization with the alpha subunit. CBFβ–MYH11 is both nuclear and cytoplasmic. In vitro studies indicate that CBFβ–MYH11 has a higher affinity for AML1 than does endogenous CBFβ, and it has been proposed that CBFβ–MYH11 can also repress AML1 transactivation by sequestering it in the cytoplasm [76].
Despite the general view that the AML1–ETO and CBFβ–MYH11 fusion proteins primarily act by aberrantly repressing CBF target genes, recent findings point toward additional CBF repression-independent activities of the fusions that may also contribute to leukemogenesis. These functions (reviewed extensively in ref. [77]) might lead to the development of new therapies to treat leukemias caused by CBF fusion oncoproteins.
LINEAGE LEUKEMIA FUSIONS
The mixed lineage leukemia (MLL) protein is a transcriptional activator that, once it binds either directly or indirectly to DNA, can modify H3K4 through the HMT activity of its C-terminal SET domain [78, 79]. MLL coordinates cell fate and cell cycle regulation, and thus plays an essential role in hematopoiesis. As the mammalian counterpart of D. melanogaster trithorax (trx) [80], MLL works antagonistic to PcG proteins in maintaining proper Hox gene expression through chromatin modifications. Because Hox genes are essential for regulating hematopoiesis, it is critical to tightly regulate their expression in lineage- and stage-specific combinations. Indeed, mis-regulation of Hox genes has been shown to be a common target of most of the oncofusion proteins produced by chromosomal translocations, and thus to be directly linked to leukemogenesis.
The gene region encoding the N-terminus domain of MLL is involved in chromosomal translocations with more than 60 different partner genes, from nuclear factors to cytoplasmic proteins. In ∼80% of the cases, MLL is fused with AF4, AF9, ENL, AF10 and ELL [81]. The individual fusion partners determine the phenotypes of the resulting leukemia, which often is AML, or acute lymphoblastic leukemia. Evidence suggests that MLL fusions can aberrantly activate genes at inappropriate times by recruiting enzymes involved in epigenetic regulation, as discussed below. Transcriptional alteration through deregulated histone modifications appears to play an important mechanism in MLL fusion-based leukemogenesis.
The SET domain, responsible for the H3K4 methyltransferase activity, is consistently lost in MLL fusions [82]. This loss of transcriptional regulation by MLL is exploited by the fusion partners to cause aberrant enhanced activation of the target genes through epigenetic modifications. Indeed, the loss of the SET domain is often compensated by the interaction with alternative HAT/HMT enzymes’ activity through partner proteins. For example, the CBP and p300 fusion partners are HATs, the ENL fusion partner is a subunit of SWI/SNF complex [83] and the MLL–EEN fusion protein recruits CBP and the protein arginine methyltransferase PRMT1 (H4R3) to MLL target genes [84]. Moreover, it has been reported that MLL fusion partners such as AF4, AF9, AF10 and ENL associate with the H3K79 histone methyltransferase DOT1L [85–88]. H3K79 methylation typically marks actively transcribed chromatin regions [89] and has been shown to play a relevant role in transformation caused by different MLL fusion partners. AF4 is a positive regulator of Pol II transcription elongation factor b (P-TEFb) and, together with AF9/ENL and AF10, acts as a mediator of histone H3K79 methylation by the recruitment of DOT1L to elongating Pol II [88]. In addition, it has been recently identified a super elongation complex (SEC) aberrantly associated with MLL fused to AF4, AF9, ENL and ELL [90]. The macromolecular SEC complex consists of several of the known Pol II elongation factors including ELL, the components of P-TEFb and AFF4, which itself is a rare translocation partner of MLL. Being AFF4 the responsible for SEC assembly and activity, it can mediate enhanced Hox gene expression and leukemogenesis by the above-mentioned MLL fusions [90]. This knowledge, combined with multiple studies on the epigenetic state in several human cancers, suggests that the development of many types of tumors (and/or cancer stem cells) is influenced by epigenetic mechanisms. MLL rearrangements usually predict early relapse with a very poor prognosis, as the current therapeutics are poorly effective, indicating the need for more successful and specific treatments. Although it is not easy to block oncogenic transcription factors with drugs, targeting the modification of H3K79me via DOT1L could be a potentially effective therapeutic strategy in MLL-dependent leukemias [91]. Similarly, other enzymatic interactors of oncofusion proteins are possible target for therapeutic intervention [92] as well as the AFF4 component of the newly identified SEC complex [90].
LEUKEMIC STEM CELLS
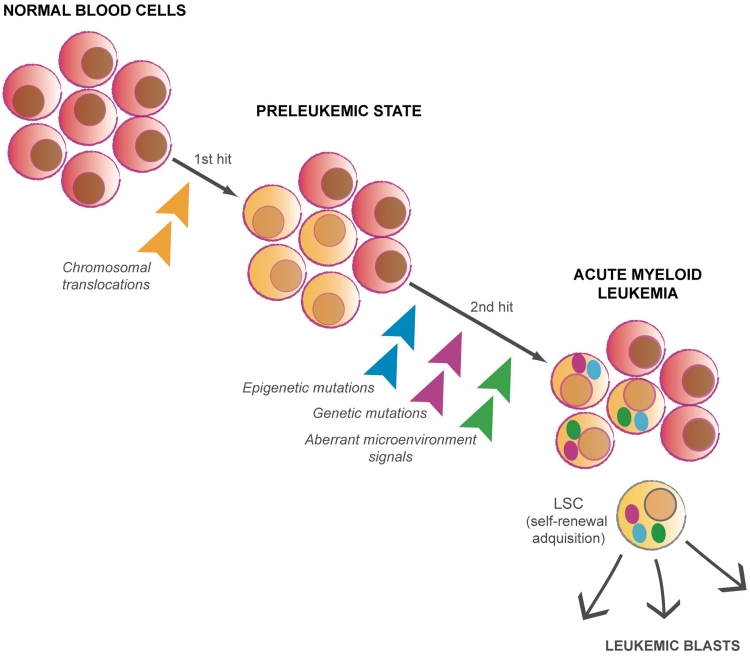
Current evidence shows that fusion proteins found in AML induce a pre-leukemic state in which further genetic and epigenetic mutations are necessary for progression to leukemia [93]. All events that are necessary and sufficient for leukemogenesis to occur are still not clear, and this seems to differ depending on the expressed oncoprotein [35]. However, for all cases within the AML cell population that carry the mutations implicated in the pathogenesis, leukemogenesis appears to require a functional heterogeneity. Specifically, it has been suggested that there is a subpopulation of cells with self-renewal capacity that are able to maintain and propagate the AML phenotype, namely, the leukemic stem cells (LSCs) [94, 95]. The described role of epigenetic regulators such as the PcG protein Bmi-1 in the activity of LSC [96] has underlined the importance of epigenetic modifications in the maintenance of LSC (Figure 3). The existence of cancer stem cells has also been attributed to other types of cancer in addition to leukemia, and its origin is now being extensively investigated. Although it has been estimated that only one LSC is found for every million AML cells, it is not always applicable. Frequent LSCs have been identified in a mouse model of human MLL-AF9 AML, showing that LSCs are not necessarily rare and only located within the stem compartment [97]. In accordance with several studies in mouse models of APL, the identified LSCs in MLL-AF9 AML are phenotypically myeloid cells that have aberrantly acquired self-renewal capacity rather than undifferentiated stem cells [97–99]. Taken together, the current data suggest that LSCs, which establish a leukemia cell hierarchy, may arise from mutations occurring in HSCs and also in committed progenitors [100, 101]. In addition to the chromosomal rearrangements, recent data suggest that aberrant bone marrow microenvironment signaling might also lead to leukemia [102, 103], adding another layer of complexity to elucidating the entirety of the causative factors in leukemogenesis.
Figure 3:
AML development as a multistage process. Chromosomal translocations in either HSCs or committed progenitors induce a pre-leukemic state in which further mutations will lead to AML development. Within the AML cell population carrying the mutations implicated in the pathogenesis, the LSCs subpopulation will maintain and propagate the AML phenotype.
A more in-depth understanding of the origin and maintenance of LSCs, as well as of the genetic and cellular differences between LSCs and HSCs, will be crucial to better comprehend leukemogenesis. Moreover, given the failure of many of the current anti-leukemic therapies, specific targeting of LSCs could be a potential successful approach [104].
CONCLUSIONS
DNA and histone posttranslational modifications provide a basis for regulating gene transcription through the chromatin template. Because these modifications are dynamic in nature, they can occur in highly orchestrated manner, leading to fine-tuning of gene expression. The mis-regulation of this process that is frequently characterized in AMLs is often effected by oncofusion proteins derived from balanced chromosomal aberrations. Developing effective treatments for AMLs will be a priority in the future, because the current prognosis of AMLs is dismal. A more detailed understanding of the mechanistic repercussions of epigenetic mis-regulation will be critical for addressing this. Indeed, DNMT and HDAC inhibitors have been successfully used for treating different type of cancers, including leukemia. Transcriptional therapy with a combination of agents that can also modulate HMTs and HDMs is thus an exciting possibility. Several biotechnology and pharmaceutical companies are investing huge effort in identifying such epigenetics drugs which will be of paramount importance for treating human diseases.
Key Points.
Chromatin modifications at both histones and DNA are key regulators of gene transcription.
Deregulation of the epigenetic marks is often found in leukemia.
Chromosomal translocations are frequent in acute myeloid leukemia (AML), and lead to fusion proteins that aberrantly regulate gene transcription.
Fusion proteins occurring in AML often recruit epigenetic machineries to target genes.
Since epigenetic modifications are reversible, they represent new drug targets for various diseases, including cancer.
FUNDING
This work was supported by the Spanish ‘Ministerio de Educación y Ciencia’, by the AGAUR and Consolider (to L.D.C.); I.U. has been supported by a FPI fellowship.
Acknowledgements
This work was supported by the Spanish ‘Ministerio de Educación y Ciencia’, by the AGAUR and Consolider (to L.D.C.); I.U. has been supported by a FPI fellowship.
Biographies
Iris Uribesalgo is a PhD student at the Center for Genomic Regulation with interests in epigenetics and human blood system function, in both health and disease.
Luciano Di Croce is an ICREA Professor and Group Leader at CRG with long-stand interest in the connection between epigenetics and cancer.
References
- 1.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bednar J, Horowitz RA, Grigoryev SA, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–8. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–59. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laherty CD, Yang WM, Sun JM, et al. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–56. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Iratni R, Erdjument-Bromage H, et al. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–64. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 9.Heinzel T, Lavinsky RM, Mullen TM, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–8. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 10.Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J Cell Physiol. 2007;213:306–15. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- 11.Vakoc CR, Mandat SA, Olenchock BA, et al. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–91. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trowbridge JJ, Snow JW, Kim J, et al. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–9. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villa R, De Santis F, Gutierrez A, et al. Epigenetic gene silencing in acute promyelocytic leukemia. Biochem Pharmacol. 2004;68:1247–54. doi: 10.1016/j.bcp.2004.05.041. [DOI] [PubMed] [Google Scholar]
- 15.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–43. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 16.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 17.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 18.Kokura K, Kaul SC, Wadhwa R, et al. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem. 2001;276:34115–21. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- 19.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 21.Czermin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 22.Muller J, Hart CM, Francis NJ, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 23.Nekrasov M, Klymenko T, Fraterman S, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–88. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarma K, Margueron R, Ivanov A, et al. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–31. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasini D, Hansen KH, Christensen J, et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–55. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng JC, Valouev A, Swigut T, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X, Kim W, Fujiwara Y, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–14. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landeira D, Sauer S, Poot R, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–24. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 31.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 32.Cheshier SH, Morrison SJ, Liao X, et al. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci USA. 1999;96:3120–5. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa M. Differentiation and proliferation of hematopoietic stem cells. Blood. 1993;81:2844–53. [PubMed] [Google Scholar]
- 34.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–64. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 35.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–96. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33:451–8. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 37.Deschler B, Lubbert M. Acute myeloid leukemia: epidemiology and etiology. Cancer. 2006;107:2099–107. doi: 10.1002/cncr.22233. [DOI] [PubMed] [Google Scholar]
- 38.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 39.Di Croce L. Chromatin modifying activity of leukaemia associated fusion proteins. Hum Mol Genet. 2005;14(Spec No 1):R77–84. doi: 10.1093/hmg/ddi109. [DOI] [PubMed] [Google Scholar]
- 40.Martens JH, Stunnenberg HG. The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS Lett. 2010;584:2662–9. doi: 10.1016/j.febslet.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de The H, Chomienne C, Lanotte M, et al. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347:558–61. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 43.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–95. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gratas C, Menot ML, Dresch C, et al. Retinoid acid supports granulocytic but not erythroid differentiation of myeloid progenitors in normal bone marrow cells. Leukemia. 1993;7:1156–62. [PubMed] [Google Scholar]
- 45.Horlein AJ, Naar AM, Heinzel T, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 46.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 47.Kamei Y, Xu L, Heinzel T, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–14. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 48.Chakravarti D, LaMorte VJ, Nelson MC, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 49.Insinga A, Pelicci PG, Inucci S. Leukemia-associated fusion proteins. Multiple mechanisms of action to drive cell transformation. Cell Cycle. 2005;4:67–9. doi: 10.4161/cc.4.1.1400. [DOI] [PubMed] [Google Scholar]
- 50.Di Croce L, Raker VA, Corsaro M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 51.Villa R, Pasini D, Gutierrez A, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–25. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Morey L, Brenner C, Fazi F, et al. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol Cell Biol. 2008;28:5912–23. doi: 10.1128/MCB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu J, Gianni M, Kopf E, et al. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci USA. 1999;96:14807–12. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, Peres L, Honore N, et al. Dimerization-induced corepressor binding and relaxed DNA-binding specificity are critical for PML/RARA-induced immortalization. Proc Natl Acad Sci USA. 2006;103:9238–43. doi: 10.1073/pnas.0603324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alcalay M, Meani N, Gelmetti V, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112:1751–61. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller-Tidow C, Steffen B, Cauvet T, et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol Cell Biol. 2004;24:2890–904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pabst T, Mueller BU, Zhang P, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–70. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 58.Muller C, Yang R, Park DJ, et al. The aberrant fusion proteins PML-RAR alpha and PLZF-RAR alpha contribute to the overexpression of cyclin A1 in acute promyelocytic leukemia. Blood. 2000;96:3894–9. [PubMed] [Google Scholar]
- 59.Licht JD, Chomienne C, Goy A, et al. Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17) Blood. 1995;85:1083–94. [PubMed] [Google Scholar]
- 60.Hong SH, David G, Wong CW, et al. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–33. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin RJ, Nagy L, Inoue S, et al. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–4. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 62.David G, Alland L, Hong SH, et al. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–56. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 63.Melnick AM, Westendorf JJ, Polinger A, et al. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol Cell Biol. 2000;20:2075–86. doi: 10.1128/mcb.20.6.2075-2086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barna M, Merghoub T, Costoya JA, et al. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 65.He LZ, Guidez F, Tribioli C, et al. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18:126–35. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 66.Ogawa E, Maruyama M, Kagoshima H, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–63. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogawa E, Inuzuka M, Maruyama M, et al. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993;194:314–31. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 68.Wang S, Wang Q, Crute BE, et al. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–39. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gelmetti V, Zhang J, Fanelli M, et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–91. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J, Hoshino T, Redner RL, et al. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–5. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lutterbach B, Westendorf JJ, Linggi B, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–84. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amann JM, Nip J, Strom DK, et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol. 2001;21:6470–83. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu S, Shen T, Huynh L, et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res. 2005;65:1277–84. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 74.Lutterbach B, Hou Y, Durst KL, et al. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci USA. 1999;96:12822–7. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Durst KL, Lutterbach B, Kummalue T, et al. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol Cell Biol. 2003;23:607–19. doi: 10.1128/MCB.23.2.607-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adya N, Stacy T, Speck NA, et al. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18:7432–43. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hyde RK, Liu PP. RUNX1 repression-independent mechanisms of leukemogenesis by fusion genes CBFB-MYH11 and AML1-ETO (RUNX1-RUNX1T1) J Cell Biochem. 2010;110:1039–45. doi: 10.1002/jcb.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 79.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–28. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 80.Djabali M, Selleri L, Parry P, et al. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Genet. 1992;2:113–8. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 81.Meyer C, Kowarz E, Hofmann J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–9. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 82.Ernst P, Wang J, Huang M, et al. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–58. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nie Z, Yan Z, Chen EH, et al. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol Cell Biol. 2003;23:294–52. doi: 10.1128/MCB.23.8.2942-2952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheung N, Chan LC, Thompson A, et al. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–15. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 85.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 86.Zhang W, Xia X, Reisenauer MR, et al. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–68. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mueller D, Bach C, Zeisig D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–54. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 89.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 90.Lin C, Smith ER, Takahashi H, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–37. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–68. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeisig BB, Cheung N, Yeung J, et al. Reconstructing the disease model and epigenetic networks for MLL-AF4 leukemia. Cancer Cell. 2008;14:345–7. doi: 10.1016/j.ccr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 93.Yuan Y, Zhou L, Miyamoto T, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA. 2001;98:10398–403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 95.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 96.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–60. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 97.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–68. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 98.Wojiski S, Guibal FC, Kindler T, et al. PML-RARalpha initiates leukemia by conferring properties of self-renewal to committed promyelocytic progenitors. Leukemia. 2009;23:1462–71. doi: 10.1038/leu.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guibal FC, Alberich-Jorda M, Hirai H, et al. Identification of a myeloid committed progenitor as the cancer-initiating cell in acute promyelocytic leukemia. Blood. 2009;114:5415–25. doi: 10.1182/blood-2008-10-182071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cozzio A, Passegue E, Ayton PM, et al. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–35. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.So CW, Karsunky H, Passegue E, et al. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–71. doi: 10.1016/s1535-6108(03)00019-9. [DOI] [PubMed] [Google Scholar]
- 102.Walkley CR, Olsen GH, Dworkin S, et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129:1097–110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walkley CR, Shea JM, Sims NA, et al. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129:1081–95. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25:1315–21. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]