Abstract
Next-generation sequencing technologies are making a substantial impact on many areas of biology, including the analysis of genetic diversity in populations. However, genome-scale population genetic studies have been accessible only to well-funded model systems. Restriction-site associated DNA sequencing, a method that samples at reduced complexity across target genomes, promises to deliver high resolution population genomic data—thousands of sequenced markers across many individuals—for any organism at reasonable costs. It has found application in wild populations and non-traditional study species, and promises to become an important technology for ecological population genomics.
Keywords: RADSeq, population genetics, next-generation sequencing, genetic marker discovery, SNP discovery
INTRODUCTION
The ability to produce gigabases of DNA sequence in a short time and at minimal cost using platforms such as Illumina [1], Roche 454 [2] and AB SOLiD [3] means that complete genomes can now be sequenced from scratch within the limits of a normal research grant. Many other applications are being developed for these platforms, such as transcriptome sequencing, gene expression profiling and small RNA characterization. However, the standard methods for these platforms are not ideally suited to population genetic studies, where the discovery and use of genetic markers across many individuals is paramount. Genome-wide marker analysis in pedigrees and populations is essential for evaluation of patterns and processes in evolutionary change, and for the investigation of the genetic architecture underpinning quantitative and other phenotypic traits. A dense linkage map is also a near-essential requirement for completing the assembly of newly-sequenced large genomes, as de novo assembly from raw next-generation data remains a significant problem [4].
The development and deployment of genetic marker toolkits for new species has typically involved marker discovery, assay development for each marker, and proving of the assays in a screening population before full deployment across large populations. Common marker types include microsatellites (microsats), single nucleotide polymorphisms (SNPs) and insertion–deletion polymorphisms (indels). This process is costly (in time and research funding) and usually results in generation of very few (tens) of working markers. Given the availability of ultra-high-throughput sequencing, one strategy might be to just sequence completely the genomes of several of the target organisms, and identify SNPs by comparing these total data sets. However, this is still not feasible for eukaryotes with genomes many megabases (or gigabases) in size, and is less informative for poorly-assembled new genomes (where linkage between the sequenced fragments is unknown).
Several recent papers by Cresko and colleagues [5–7] describe a method called restriction site-associated DNA sequencing (RADSeq) that can identify and score thousands of genetic markers, randomly distributed across the target genome, from a group of individuals using Illumina technology. RADSeq can be used to carry out population genetic studies on species with no, or limited, existing sequence data, and has several advantages over previous methods for marker discovery. It is akin to analyses using restriction fragment length polymorphisms (RFLPs) and amplified fragment length polymorphisms (AFLPs) in that it reduces the complexity of the genome by subsampling only at specific sites defined by restriction enzymes. RADSeq surpasses these methods in its ability to identify, verify and score markers simultaneously (rather than requiring an extensive development process) and to robustly identify which markers derive from each site. RADSeq can be used on crosses of any design, and in wild populations, enabling not only genotyping and SNP discovery, but also more complex analyses such as quantitative genetic and phylogeographic studies, as shown below.
THE RADSeq METHOD: FINDING AND SCORING POLYMORPHISMS ACROSS THE GENOME
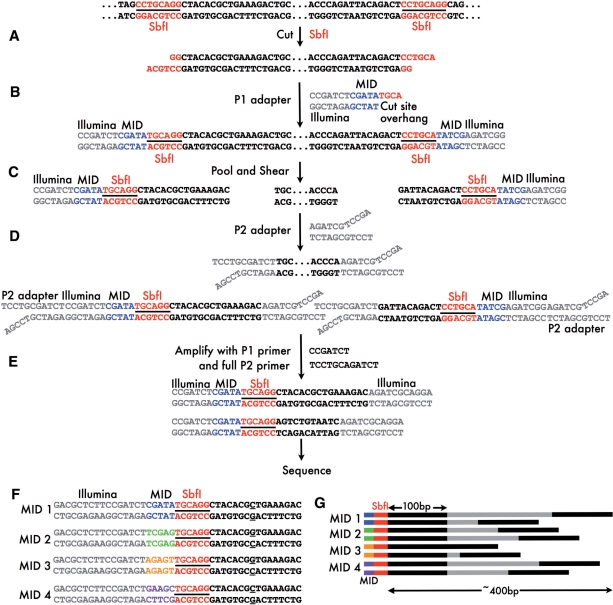
RADSeq combines two simple molecular biology techniques with Illumina sequencing: the use of restriction enzymes to cut DNA into fragments (as for RFLPs and AFLPs), and the use of molecular identifiers (MID) to associate sequence reads to particular individuals (Figure 1 and Table 1). DNA from an individual is cut with the chosen restriction enzyme, producing a set of sticky-ended fragments (Figure 1A). To be sequenced on an Illumina machine, these fragments must be ligated to adapters that will bind to an Illumina flow cell. RADSeq uses modified Illumina adapters that enable the binding and amplification of restriction site fragments only. The sticky-end fragments are ligated to a P1 adapter that contains a matching sticky-end (for example, TGCA for SbfI—Table 1) and a MID (Molecular Identifier), a short sequence that will uniquely identify the individual (Figure 1B). Individuals can also be pooled according to any desirable criteria before restriction digestion and P1 ligation (an approach akin to bulk segregant analysis). The tagged restriction fragments from a number of individuals are pooled, and then sheared randomly to generate fragments with a mean length of a few hundred base pairs (Figure 1C). The sheared fragments are ligated to a second, P2 adapter (Figure 1D) and PCR amplified using P1 and P2 primers (Figure 1E). The P2 adapter has a divergent ‘Y’ structure that will not bind to the P2 primer unless it has been completed by amplification by the P1 adapter. This ensures that all amplified fragments have the P1 adapter and MID, the partial restriction site, a few hundred bases of flanking sequence, and a P2 adapter. These sheared, sequencer-ready fragments are then size selected (approximately 200–500 base fragments are isolated) and this RADSeq library sequenced on the Illumina platform. Sequence is generated from the MID in the P1 adapter and across the restriction enzyme site, generating a data set of RAD tags (sequences downstream of restriction sites) that derive from a much-reduced part of the original genome (Table 1). If the restriction site is symmetric, then two RAD tags will be produced from each site. The Illumina platform currently permits sequencing out to 150 bases, and thus approximately 300 bases flanking each restriction site can be screened for polymorphisms.
Figure 1:
The process of RADSeq. (A) Genomic DNA is sheared with a restriction enzyme of choice (SbfI in this example). (B) P1 adapter is ligated to SbfI-cut fragments. The P1 adapter is adapted from the Illumina sequencing adapter (full sequence not shown here), with a molecular identifier (MID; CGATA in this example) and a cut site overhang at the end (TGCA in this example). (C) Samples from multiple individuals are pooled together and all fragments are randomly sheared. Only a subset of the resulting fragments contains restriction sites and P1 adapters. (D) P2 adapter is ligated to all fragments. The P2 adapter has a divergent end. (E) PCR amplification with P1 and P2 primers. The P2 adapter will be completed only in the fragments ligated with P1 adapter, and so only these fragments will be fully amplified. (F) Pooled samples with different MIDs are separated bioinformatically and SNPs called (C/G SNP underlined). (G) As fragments are sheared randomly, paired end sequences from each sequenced fragment will cover a 300–400 bp region downstream of the restriction site.
Table 1:
Enzymes and adapters for RADSeq
| Adapter set overhang | Enzyme | Site | Predicted numbers of sites (assuming a Poisson distribution) |
Actual numbers of sites in Caenorhabditis elegans reference genome | |
|---|---|---|---|---|---|
| Per Mega base (50% GC) | Caenorhabditis elegans (100.2 Mb, 36% GC) | ||||
| TGCA 3′ | SbfI | CCTGCA*GG | 15 | 348 | 323 |
| PstI | CTGCA*G | 244 | 10 750 | 13 548 | |
| NsiI | ATGCA*T | 244 | 33 974 | 24 216 | |
| GGCC 5′ | NotI | GC*GGCCGC | 15 | 110 | 395 |
| EagI | C*GGCCG | 244 | 3401 | 6298 | |
| EaeI | Y*GGCCR | 977 | 26 244 | 14 305 | |
| GATC 5′ | BamHI | G*GATCC | 244 | 10 750 | 11 749 |
| BclI | T*GATCA | 244 | 33 974 | 30 831 | |
| BglII | A*GATCT | 244 | 33 974 | 21 992 | |
| BstYI | R*GATCY | 977 | 36 864 | 36 741 | |
Examples of restriction enzymes suitable for RADSeq, including enzymes with ambiguous recognition sites. Restriction enzymes with the same core recognition site can be amplified with the same adapter set. The choice of enzyme has a considerable impact on number of fragments produced. The number of fragments can be roughly predicted by assuming sites are Poisson distributed, but some enzymes depart considerably from this expectation (e.g. EagI).
In wild populations where the expected diversity is of the order of 0.1%, ∼20–30% of all restriction sites should be flanked by a polymorphism in the flanking 200–300 bases of sequence. The sequences from each individual are separated after sequencing using the MID. RADSeq can be used to detect restriction site presence-absence polymorphisms (by identifying a marker that is present in one set of individuals but absent in another, indicating a variation in the restriction site) or SNPs and indels in the sequence flanking the restriction site. If a reference genome is available, raw sequence reads can be aligned to the reference sequence, and SNPs and indels identified using existing next-generation sequencing bioinformatics tools such as Bowtie [8], BWA [9] and SAMtools [10]. Mapping to a reference genome automatically corrects for the low level of sequencing error in the reads.
If a reference sequence is not available, RAD tags can be analysed de novo. Identical reads are aggregated into unique sequences and treated as candidate alleles. By clustering together unique sequences that have only a small number of mismatches between them, SNPs and indels can be called between alleles at the same locus, and errors corrected by comparing counts of each base at each position. Real homozygous or heterozygous alleles will have relatively high read counts, whereas errors will have low counts.
RADSeq OF ECOLOGICALLY SIGNIFICANT PHENOTYPIC TRAITS IN THREESPINE STICKLEBACK IN A CONTROLLED CROSS
RADSeq was first applied to investigation of the genetics of an important ecological trait in the threespine stickleback (Gasterosteus aculeatus), the genome of which has been sequenced. The lateral plate armor of the stickleback is present in oceanic populations but has been lost independently in many derived freshwater populations. This phenotype displays quantitative genetic variation, with the complete-plate phenotype generally dominant to plate-loss, but modified by other loci. A locus of major effect had previously been mapped to the ectodysplasin gene (Eda) on stickleback linkage group (LG) IV [11]. Baird et al. [5] demonstrated that RADSeq could be used to independently identify markers linked to plate loss at the Eda locus and several other loci on linkage group IV.
The study used an F2 mapping cross, with one parent from a lake where the stickleback population has the complete-plate phenotype and the other parent from a population with the plate-loss phenotype. Parental genomic DNA was cut with the eight-base recognition site enzyme SbfI (Table 1) and RAD tags generated using 36 base, single-end Illumina reads. The parents were sequenced to greater depth than the F2 offspring, so as to define the possible set of RAD tags with some certainty. The parental tags were mapped to the 460 Mb stickleback genome, identifying 41 622 RAD tags, distributed evenly over the genome.
DNA from F2 individuals was then pooled by lateral plate phenotype, and two RADSeq libraries generated and sequenced. The resulting sequences were mapped to the RAD tags identified in the parents. Of the RAD tags that were polymorphic across the F2 offspring, 1136 were only found in the complete-plate individuals and 1097 were only found in the plate-loss individuals. Sixty percent of the polymorphic markers contained SNPs and 40% had disruptions to the SbfI cut site. Markers completely associated with the plate-loss phenotype were found not only as expected around the Eda locus but also in two further regions on LGIV.
SbfI is expected to cut roughly every 65 kb. To map the LGIV loci more closely, Baird et al. [5] also RAD sequenced the F2 population using the 6-base recognition site enzyme EcoRI, which cuts every 3.6 kb. Instead of pooling by phenotype, a different MID was used for each of 96 fish, and the pooling was done bioinformatically. Ninety-one fish were used for the mapping analysis, 60 of which had the complete-plate phenotype, and 31 had the plate-loss phenotype. An astounding 148 390 EcoRI RAD tags were identified, of which 2311 were plate-loss-specific and 4530 were complete-plate-specific. A region of complete linkage to the plate-loss phenotype <1.5 Mb in size surrounding the known Eda locus was defined.
Because the F2 fish were uniquely identified, the genetics of other phenotypes could be analysed using the same RADSeq data. Fish without lateral plate armor often also have a reduced pelvic structure compared to fish with complete lateral plates. The 91 F2 individuals were resorted based on pelvic structure, with 60 fish possessing full pelvic structure and 29 with reduced pelvic structure. Using these new pools, a locus controlling the pelvic trait was identified on the distal tip of LGVII.
This study thus established RADSeq as a method for generating and assaying polymorphism across the entirety of a genome in non-traditional model species. Using the genome sequence as a mapping reference, hundreds of thousands of potential marker sites can be surveyed, and the mapping precision of the method is limited only by the standing diversity in the parents and the numbers of crossovers present in the progeny (as in all laboratory cross situations).
RADSeq OF ECOLOGICALLY SIGNIFICANT PHENOTYPIC TRAITS IN THREESPINE STICKLEBACK IN WILD POPULATIONS
Wild populations offer a major challenge to population genetic analyses, as development of high-density markers is a burden that has effectively excluded most species from consideration for study. Because many populations will have private alleles, the discovery stage of the process of developing traditional markers has to include screening of large numbers of individuals in advance of the main study. In cases where multiple sets of populations must be studied, for example when examining the repeatability or predictability of evolution, this issue becomes insurmountable. RADSeq, because it discovers, proves and assays markers simultaneously, overcomes this problem, and delivers data that can be used for sophisticated genome-wide population genetics. Hohenlohe et al. [6] investigated whether evolutionary trajectories of threespine stickleback were repeatable by comparing three independently derived freshwater populations and two oceanic populations from south Alaska, near Anchorage. Twenty fish from each of three freshwater populations and two different oceanic populations were processed for RADSeq, using SbfI, and the RAD tags were mapped to the reference stickleback genome. SNPs (45 789) were identified (2.5% of the 2092 294 nt covered by the RAD tags), with each RAD tag sequenced 5–10 times in each fish.
Standard population genetics statistics [mean nucleotide diversity (π) and heterozygosity (H), population differentiation (FST), Tajima’s D and private allele density (p)] were then calculated in sliding windows across the whole genome for the five populations. Regions where these statistics significantly deviated from background were easily identified. For example, FST values between the two oceanic populations, which are separated by ∼150 km as the crow flies, but ∼600 km as the fish swims as they are isolated on either side of the Kenai peninsula, did not depart from background levels at any point across the genome, indicating ongoing genetic exchange between these widely-separated populations. However, when the oceanic populations were compared to the freshwater populations, nine regions of substantial population differentiation were identified (despite the lake populations being only 20–50 km from the northerly oceanic one) across six different linkage groups, including the previously identified lateral plate phenotype QTL mapping to the Eda locus on LGIV [11] and two other regions on LGIV. These nine regions, covering 12.2 Mb of the genome, contained 590 annotated genes, of which 31 candidate genes had previously been linked in the literature to skeletal or osmotic traits, two phenotypes known to be under selection in freshwater stickleback populations.
By examining statistics in combination, signatures of selection could be identified. For example, a 1.3 Mb region at the end of LGIII has markedly high nucleotide diversity and heterozygosity across all five populations, but substantially reduced population differentiation (FST), indicating that genetic variation is being maintained under balancing selection. This region contains orthologues of genes implicated in pathogen resistance (ZEB1, APOL), inflammation pathways (LTB4R, CEBPD) and innate immune response (TRIM14, TRIM35), all systems likely to be under balancing selection by Red Queen dynamics [12, 13]. These results clearly show the power of RADSeq for population genomics.
RADSeq FOR PHYLOGEOGRAPHIC ANALYSIS OF A MOSQUITO THAT DOES NOT HAVE A SEQUENCED GENOME
Generation of RAD tags from a species with a sequenced genome allows the researcher to use the advanced read-mapping tools available for next-generation sequencing to identify sites, deal with sequencing error, and define SNPs and indels. When working with a species for which there is no genome sequence, RAD tags must be assembled de novo, sequence error dealt with efficiently, tags deriving from repetitive elements identified, and allelism inferred. These informatic tasks are complex but not impossible. Emerson et al. [7] used RADSeq to resolve the post-glacial phylogeographic relationships of populations of the non-model organism Wyeomyia smithii, a pitcher plant mosquito.
Wyeomyia smithii is geographically separated into two, fully interfertile northern and southern groups across eastern North America. Use of a standard marker gene, mitochondrial cytochrome oxidase 1, to analyse the relationships of 21 geographically distinct populations yielded support only for the north–south divide. Emerson et al. [7] thus turned to RADSeq to screen the 21 populations for polymorphisms to resolve their relationships. DNA from six individuals per population (126 individuals in total) was digested with SbfI and RAD sequenced. Identical reads were aligned together into RAD stacks (a group of reads aligned to a RAD tag), and stacks with at most 1 nt mismatch between them were assumed to be allelic. Sequences from repeat regions were removed by discarding any RAD stacks with mean read depth >2 SDs from the mean. RADSeq defined 13 627 non-repeat loci, containing 3741 SNPs. Analysis of these 3741 SNPs revealed that the 21 populations fall into four major clades across the two groups, with all but four nodes of the tree resolved with high confidence. These data reveal that the Appalachian population is sister to all the other ‘northern’ group populations, and that continental populations from the Great Lakes and central Canada have been derived from more easterly ones found spanning the St Lawrence corridor.
Thus even without a reference genome, by stringent filtering of reads and definition of allelic sequences, RADSeq can deliver huge numbers of SNPs for analysis. Importantly, the technology used to find the polymorphic loci is one and the same as the technology used to score those SNPs in the study populations. There are obvious improvements and extensions to this approach. For example, Emerson et al. [7] accepted as allelic only tags that differed by a maximum of one base: if tags differing by two or three bases could be defined as alleles, many, many additional SNPs could be mined.
FUTURE PROSPECTS: MAXIMIZING VARIATION DETECTION WITH RADSeq
These three publications are the current published state of the art in RADSeq analysis. In the busy months since these studies were performed, the RADSeq technology has evolved significantly, most importantly with the improvement in Illumina read lengths (now up to 150 bases) and in the use of paired end sequencing (generating 300 bases per sequenced fragment). RADSeq yields two kinds of markers: presence–absence markers resulting from polymorphism in the restriction enzyme cut site, and substitutional (SNP, indel) markers in the tag sequences. In populations with a hypothetical expected level of between-haploid variation of one difference in 1000 bases, short reads (such as the 36 base reads used by Baird et al. [5]) would be predicted to identify one SNP per 28 RAD tags. Illumina 150 base reads would yield one SNP per seven RAD tags.
Paired-end sequencing can also be performed from RADSeq libraries. Because fragments are randomly sheared, the paired sequences associated with each RAD tag will begin at different positions downstream of the restriction site. These RADSeq pair tags can be assembled to produce extended (200–300 base) contigs linked to each RAD site (Figure 1F). The ‘target’ for identification of SNPs and indels is thus four times the length of the RAD tag. The paired contigs can also be used for the development of PCR-based assays for higher throughput analyses.
RADSeq has several advantages compared to other SNP genotyping approaches such as AFLPs and oligonucleotide SNP chips. First, rather than just detecting changes in 16-base (AFLP) or 20-base (oligonucleotide hybridization) targets, the full sequences of the RAD tag and its paired contig can be screened for SNPs and indels. There is no variant discovery and assay design step, and there are no hybridization optimization issues. For example, acquiring additional sets of markers to increase density simply requires the use of different restriction enzymes, rather than an extended discovery and design process. The initial analysis of RAD tag sequences is also arguably simpler than the interpretation of AFLP gels or oligoarray images. Obviously, like all genetic association experiments, the resolution of RADSeq in identification of the loci underpinning traits of interest depends sensitively on the number of independent markers assayed, their levels of variability, and the numbers of crossovers that have occurred in the mapping population. By increasing the numbers of markers (resampling the same genomes with additional restriction enzymes) and the numbers of individual cross progeny or population representatives, the accuracy of RADSeq mapping can be improved.
The simple bioinformatics steps outlined above provide a framework for RADSeq analysis, but this is just the start [14]. Simple approaches to SNP calling and error correction may yield thousands of markers, but will leave much of the data unmined. Genomes differ by complex patterns of substitution and indels, and the error rate of the Illumina platform may obscure some true alleles. RAD tags that appear to be ‘repetitive’ paralogue clouds when just the RAD tag is considered may be revealed as sets of alleles when paired contigs are analysed. Much more comprehensive approaches are possible, particularly for de novo RAD, where no reference genome is available.
Hohenlohe et al. [6] established a maximum likelihood framework for calculating the likelihood of each homozygote or heterozygote genotype at each locus given the bases called at the locus and the sequencing error at the locus. The error model used treats the error rate as varying across a single read, as opposed to being identical at all bases across all reads. More complex error models could be used, taking into account issues such as chimeras and copying errors induced during PCR and the GC bias in Illumina sequencing data [15]. The GC bias issue makes it difficult to separate heterozygotes, homozygotes, repeats and paralogues by read count alone. Further normalization of raw count data will be required to take full advantage of de novo RAD sequencing for population genetics studies.
In addition, the availability of tens of thousands of markers across hundreds of individuals will considerably deepen the possibilities for population genomic analyses. It will be possible to detect subtle effects in multiple markers across the genome that were previously unobservable, not least because it will be possible to calculate a high confidence, genome-wide average for any chosen statistic simultaneously with the scoring of outliers, enabling simple identification of divergence from neutrality wherever it occurs. Signatures of positive, balancing, divergent and background selection can be identified separately within and across populations.
RADSeq: POISED TO CHANGE THE GENETIC ANALYSIS LANDSCAPE
RADSeq is a very versatile method; it is expected to work with any restriction enzyme on any species. This means it can be used on any number of individuals at any depth of sequencing, depending on available resources. Current sequencing costs make sequencing of tens of individuals realistic, but it is expected that with new instruments, such as the HiSeq 2000 [16], it will be possible to sequence hundreds of individuals at substantial depth per sequencer run. This means RADSeq can be applied to many research problems, from identifying a handful of markers for large-scale population genotyping purposes, to creating complete linkage maps for mapping of Mendelian or quantitative trait loci. RADSeq will be a powerful tool for generation of dense linkage maps for scaffolding of newly sequenced genomes.
While the cost of sequencing is plummeting, sampling a defined, restricted portion of a genome is always going to be cheaper than sequencing the whole genome. As third generation sequencing technologies come on line, it is likely to become possible to sequence RAD tags kilobases in length. Therefore RADSeq opens up rich prospects for analysis of genetic markers, both in the detailed information that can be gained from single markers and from the complex interactions between thousands of markers across the genome. RADSeq brings population genetic analysis of essentially every sexual organism firmly into the next-generation sequencing age.
Key Points.
RADSeq is an important new method for the discovery of thousands of sequenced markers in any organism of choice.
RADSeq makes possible population genetics studies of unprecedented depth and complexity.
RADSeq is feasible for genomes of any size and does not require a reference genome, enabling studies of non-model organisms and wild populations.
FUNDING
Biotechnology and Biological Sciences Research Council in the UK (grants G00661X and F021135 to J.W.D. and M.L.B.); Medical Research Council (grant MRC G0900740 to M.L.B. and the GenePool Genomics Facility); Natural Environmental Research Council in the UK (NERC R8/H10/56).
Acknowledgements
The authors would like to thank Karim Gharbi, Marian Thomson and others at the GenePool Genomics Facility for many useful discussions on RADSeq.
Biographies
John W. Davey is a postdoctoral bioinformatician in the Institute of Evolutionary Biology in Edinburgh. He is developing tools for the analysis of RADSeq data.
Mark L. Blaxter is a Professor of Evolutionary Biology in the University of Edinburgh and Director of the GenePool Genomics Facility, Edinburgh. His research focuses on the genomics of non-vertebrate animals.
References
- 1.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothberg J, Leamon J. The development and impact of 454 sequencing. Nat Biotechnol. 2008;26:1117–24. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- 3.Pandey V, Nutter RC, Prediger E. Applied biosystems SOLID system: ligation-based sequencing. In: Janitz M, editor. Next Generation Genome Sequencing: Towards Personalized Medicine. Germany: Wiley-VCH, Weinheim; 2008. pp. 431–44. [Google Scholar]
- 4.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–27. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baird N, Etter P, Atwood T, et al. Rapid SNP Discovery and Genetic Mapping Using Sequenced RAD Markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohenlohe PA, Bassham S, Etter PD, et al. Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD Tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson KJ, Merz CR, Catchen JM, et al. Resolving postglacial phylogeography using high-throughput sequencing. Proc Natl Acad Sci USA. 2010;107:16196–200. doi: 10.1073/pnas.1006538107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colosimo PF, Hosemann KE, Balahbadra S, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–33. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 12.Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2(4):e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer-Admetlla A, Bosch E, Sikora M, et al. Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 2008;181:1315–22. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- 14.Davey J. UK RAD Sequencing Wiki. https://www.wiki.ed.ac.uk/display/RADSequencing/Home (18 October 2010, date last accessed)
- 15.Harismendy O, Ng PC, Strausberg RL, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Illumina Inc. HiSeq 2000. http://www.illumina.com/systems/hiseq_2000.ilmn (18 October 2010, date last accessed) [Google Scholar]