Abstract
Specific binding of transcription factors (TFs) determines in a large part the connectivity of gene regulatory networks as well as the quantitative level of gene expression. A multiplicity of both experimental and computational methods is currently used to discover and characterize the underlying TF–DNA interactions. Experimental methods can be further subdivided into in vitro- and in vivo-based approaches, each accenting different aspects of TF-binding events. In this review we summarize the flexibility and performance of a selection of both types of experimental methods. In conclusion, we argue that a serial combination of methods with different throughput and data type constitutes an optimal experimental strategy.
Keywords: transcription factor, DNA binding, binding affinity, ChIP, MITOMI, PBM, SELEX
INTRODUCTION
The coordinated expression of genes drives a majority of cellular processes. This coordination is in part regulated by interactions between proteins, called transcription factors (TFs) and sequence-specific DNA elements, called TF-binding sites (TFBS). Transcriptional regulation is not an isolated process, but is rather embedded in a highly interconnected gene regulatory network (GRN) consisting of hundreds of TFs, their target promoters and co-regulators (up to 10% of the human ORF-coding genome codes for TFs) [1]. TF binding and function is regulated on several levels. The first and most fundamental order of regulation is achieved by the preferential binding of a TF to specific DNA sequences [2]. Higher orders of regulation are accomplished by post-translational modifications of TF domains or binding of co-regulators. These modifications in turn can modulate the activity and/or cellular location of a TF [3, 4].
It is the specific binding of TFs that determines in large part the connectivity of GRNs as well as the quantitative level of gene expression [5]. Genetic variations in TFBS are frequently associated with differences in transcription among individuals, highlighting the necessity of precise characterization [6]. Thus, in-depth characterization of TF–TFBS interactions on a genome-wide level is pivotal to our understanding of transcriptional regulation. Any comprehensive characterization of GRNs must include TF–DNA-binding specificities as well as the higher-order modes of regulation such as protein modifications and protein-protein interactions [7].
Numerous methods, both experimental and computational, exist that allow one to discover and comprehensively characterize the specificity by which TFs interact with cognate DNA elements. In this review we cover a selection of experimental methods primarily focusing on the flexibility and performance of methods for determining TF-DNA specificities. Within the field of experimental TF biology, two fundamentally different kinds of approaches are used to characterize TF interactions: in vitro- and in vivo-based methods.
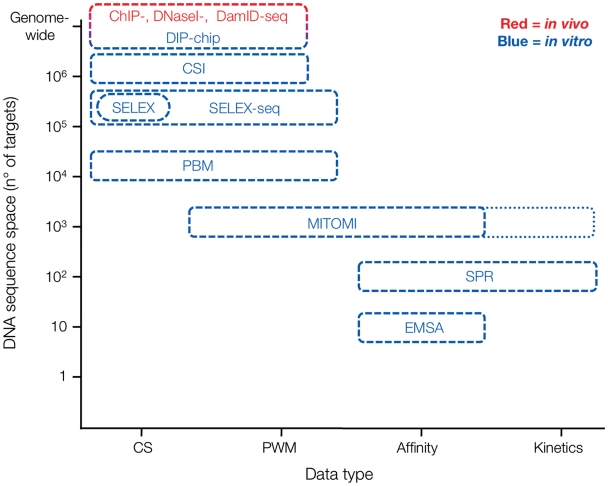
In vitro methods generally aim to identify either TF consensus binding sites [8], binding energy landscapes [9] or the biophysical parameters governing these binding events [10]. In vivo-based methods recover information on TF consensus binding sites, sequence specificity, as well as the biological context of sequence-specific interactions. Experimental methods can be further subdivided into methods that provide qualitative or quantitative data, with a majority of methods falling in the former category (Figure 1 and Table 1). To express these differences more explicit we refer to data type as a qualifier to distinguish qualitative, semi-quantitative, quantitative, and kinetic data of TF–DNA interactions (Figure 1 and Table 1, indicated as ‘+’ to ‘++++’, respectively).
Figure 1:
Comparison of target DNA throughput and obtained data type of selected methods. In vivo-based methods (highlighted in red), offer tremendous throughput but no quantitative data on TF–DNA interaction. In vitro-based methods, while decreasing throughput, capture the quantitative nature of TF–DNA binding events. MITOMI can in principle be extended to collect kinetic information, as indicated by the dotted extension.
Table 1:
Comparison of in vitro- and in vivo-based methods to characterize TF binding specificities
| Method | Synonyms | Throughput (DNA sequence space) | Materials neededc | Data type | Resolution | References | |
|---|---|---|---|---|---|---|---|
| In vitro approaches | |||||||
| Selection of target | SELEX, CASTing | >200 000 sites | mg of P | + | Consensus site | few high affinity binding sites | 35, 36 |
| Selection of target coupled to NGS | HT-SELEX, Bind-n-Seq | >200 000 sites | mg of P | ++ | PWMa, relative KD | Nucleotide resolution feasible | 26, 27 |
| Protein binding microarray | PBM, CSI | up to 1 milion sites | mg of P | ++ | PWMa, relative KD | Nucleotide resolution feasible | 30, 32 |
| DNA immunoprecipitation | DIP-chip | all genomic sites | µg of P | + | PWMa | between 100 and 500 bp | 75 |
| Mechanical trapping | MITOMI | 1000 to 100 sites | ng of P | +++(+) | Absolute KD (kon, koff) | Nucleotide resolution | 9 |
| Gel shift | EMSA | around 10 sites | mg of P | ++++ | Absolute KD, kon, koff | few binding sites only | 17 |
| Surface plasma resonance | BIAcore | up to 100 site | µg of P | ++++ | Absolute KD, kon, koff | few binding sites only | 40 |
| In vivo approaches | |||||||
| ChIP coupled to microarray | ChIP–chip | all genomic sites | ng of D | + | PWMa,b | between 100 and 500 bp | 21 |
| ChIP coupled to NGS | ChIP-seq | all genomic sites | ng of D | + | PWMa,b | between 100 and 500 bp | 22 |
| TF mediated DNA methylation profiling | DamID | all genomic sites | ng of D | + | PWMa | between 100 and 500 bp | 69 |
| Reverse ChIP | PICh | one genomic site | * | – | 71 | ||
| DNaseI sensitivity profiling coupled to NGS | DNaseI-seq | all genomic sites | ng of D | + | PWMa | Nucleotide resolution feasible | 72 |
aqualitative to semi-quantitative.
bno distinction between direct/indirect interaction.
cprotein (P); genomic DNA (D).
*picomole of each protein; MS detection limit.
We refer the reader to excellent reviews [11–14] for a comprehensive overview of in silico methods, which generally rely on conservation of TFBS, either amongst a set of known co-regulated genes, or within homologous promoters of closely related species. The corresponding TF is generally inferred from a priori information if it isn’t known already. We also refer the readers who are interested in the use and performance of ‘one-hybrid’ screens to the following in-depth reviews [15, 16].
We argue that a combination of several in vitro and in vivo methods is currently indispensable to our understanding of transcriptional regulation. Significant advancement in quantitative characterization of genome-wide protein–DNA interactions in space and time is required before it will be possible to accomplish a major goal in transcriptional regulation: the quantitative prediction of GRNs.
METHODS TO ELUCIDATE TF–DNA INTERACTIONS
Traditionally, TFBS have been mapped and characterized in vitro and in vivo using electrophoretic mobility shift assays (EMSA) [17, 18] and promoter deletion analysis coupled to a reporter assay (e.g. β-galactosidase) [19], respectively. In many cases these classical approaches don’t meet the throughput required for a systematic characterization of TF–DNA interactions. The genome-wide characterization of TF-binding profiles only became feasible with the advent of microarray-based methods [20, 21]. To date, several high-throughput approaches have been developed, including in vivo-based ChIP-chip and ChIP-seq methods [20–25] and in vitro methods based on binding site enrichment [26, 27], DNA microarrays [28–32], and microfluidic devices [9, 10, 33, 34].
IN VITRO METHODS
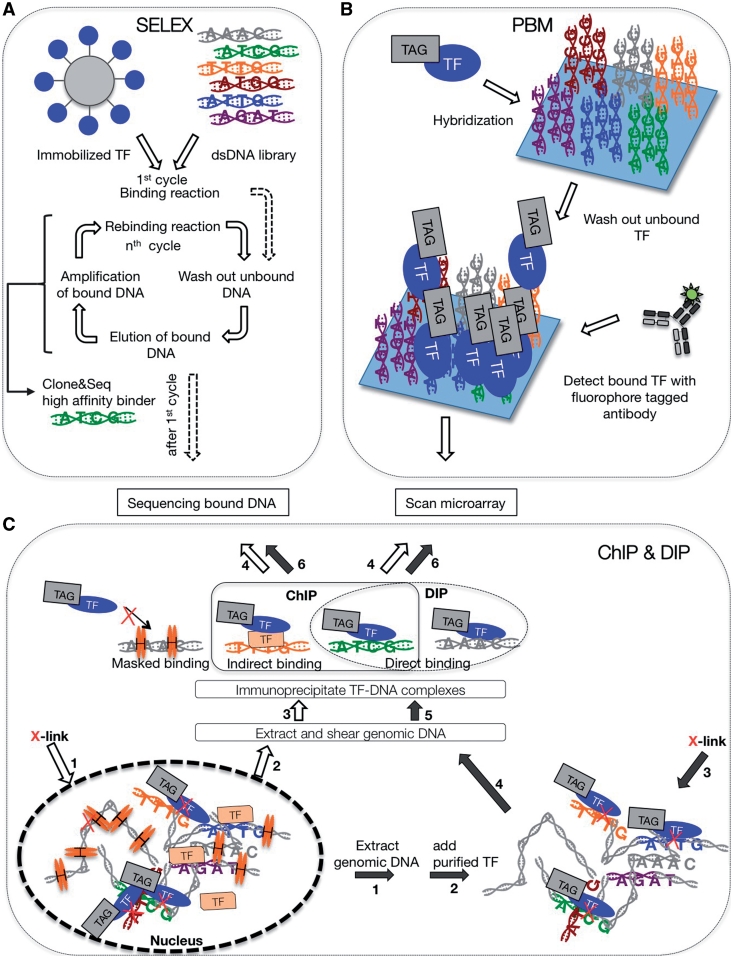
The first in vitro implementation to determine de novo TF-binding sites was developed more than two decades ago. Systematic evolution of ligands by exponential enrichment (SELEX) is based on incubating a purified TF with a pool of random DNA oligos. TF bound oligos are selected, amplified by PCR, and re-incubated with the TF so that repeated rounds of selection identifies high-affinity binders, or the TF’s consensus TFBS [8, 35, 36]. SELEX was one of the first approaches that could determine the consensus binding site of a TF without prior information. Yet the ability to accurately determine the consensus binding site is simultaneously a drawback of SELEX in that only few high-affinity binding sites are selected and amplified, which is insufficient to accurately and comprehensively capture the non-linear relationship between sequence composition and binding affinity of TFBS (Figure 2A) [27, 37].
Figure 2:
Experimental flow chart of TF–DNA characterization. (A) In vitro selection of TF binding sites consists of several rounds of binding and amplification of captured dsDNA targets. Captured targets are either analyzed individually by cloning and sequencing or in bulk by deep sequencing approaches. (B) Protein binding microarray consist of micro-arrayed dsDNA oligos. A binding reaction is performed by adding TF to these microarrays. Following a wash step, bound TFs are immunodetected by a TF specific, fluorescent antibody. (C) Immunoprecipitation based approaches consist of cross-linking TFs to genomic loci in vivo (ChIP) or in vitro (DIP), followed by shearing of DNA and precipitation with a TF specific antibody. Enriched DNA fragments are analyzed after reversal of cross-linking by microarray or deep sequencing. Lined and solid arrows highlight key steps for ChIP and DIP approaches, respectively and the numbers in the arrows indicate the sequence of experimental steps. Note the difference between ChIP and DIP approaches with regard to masked, direct, and indirect binding events.
To overcome this limitation, in vitro selection was recently coupled to massive parallel sequencing approaches [26, 27]. Instead of multiple rounds of binding and amplification, one round of selection is sufficient to capture relative binding affinities as fold enrichments of sequenced DNA fragments. TF throughput of SELEX based methods currently remains limited as sufficient protein needs to be purified and the handling steps have not yet been adapted to high-throughput (Table 1). Nevertheless SELEX-seq may prove to be a more cost-effective, comprehensive and higher-throughput alternative to protein-binding microarrays (PBMs) in the near future [38].
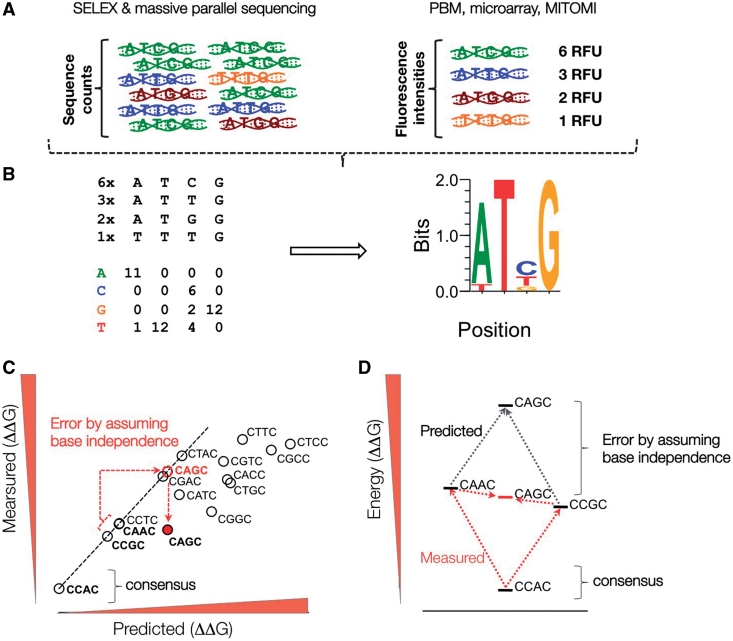
With the availability of DNA microarray chips, binding reactions can be performed on immobilized double-stranded DNA oligonucleotide arrays (Figure 2B) [28–32]. In short, a protein of interest is allowed to bind to a PBM. Following stringent washing steps, binding events are quantified by immuno-detection using protein specific, fluorophore coupled antibodies. Signal intensities are analyzed and interpreted as differential binding profiles. Recent advances in the field of microarray technology allow the fabrication of high-density arrays, harboring practically all permutations of a 10-mer sequence. On a 44 000 feature array all ∼1 000 000 features of a 10-mer space are represented as a nested De Bruijn sequence [28]. Cognate Site Identifier arrays (CSI) based on single stranded oligos that fold over to form dsDNA hairpins have up to 1 000 000 unique features and therefore do not need to rely on De Brujin sequences [31, 32]. Using such ‘universal’ PBMs not only increased the resolution by which binding motifs are detected, but also enabled the use of a single microarray design to examine a broad range of TFs [28]. The information obtained from PBMs is generally significantly reduced into the form of position weight matrices based on the additivity assumption, which posits that bases contribute independently to the binding (PWMs; Figure 4A and B). Recently, Carlson et al. have proposed a visualization method to omit this information reduction. The display of all the information obtained from PBMs highlights the context dependencies of TF binding [31].
Figure 4:
Summary of TF binding site representation. (A) TF binding site preferences are detected as enrichment of TF-bound DNA fragments by massive parallel sequencing or microarray approaches. (B) Sequence counts or microarray-based relative fluorescence units (RFU) are transformed into position-specific weight matrix (PWM) by counting base frequencies of selected DNA sequences. PWMs are commonly represented as sequence logos. (C and D) PWM predicted binding affinities of sequences with multiple base deviations relative to consensus binding site commonly overestimate affinity changes due to assumption of base independence.
DNA immunoprecipitation (DIP-chip) [39] is another in vitro approach, conceptually intermediate between in vitro selection (SELEX) and immunoprecipitation of in vivo cross-linked chromatin (ChIP, see following section; Figure 2C). Instead of synthesized DNA oligos, purified chromosomal DNA is used in binding reactions. Binding complexes are fixed by cross-linking with formaldehyde, sheared into shorter fragments between 100 and 500 bp, and immunoprecipitated with a protein-specific antibody. After reversal of the cross-links, enriched DNA fragments are analyzed by microarrays. Binding site discovery is limited by the inherently low experimental resolution due to sheared fragment size.
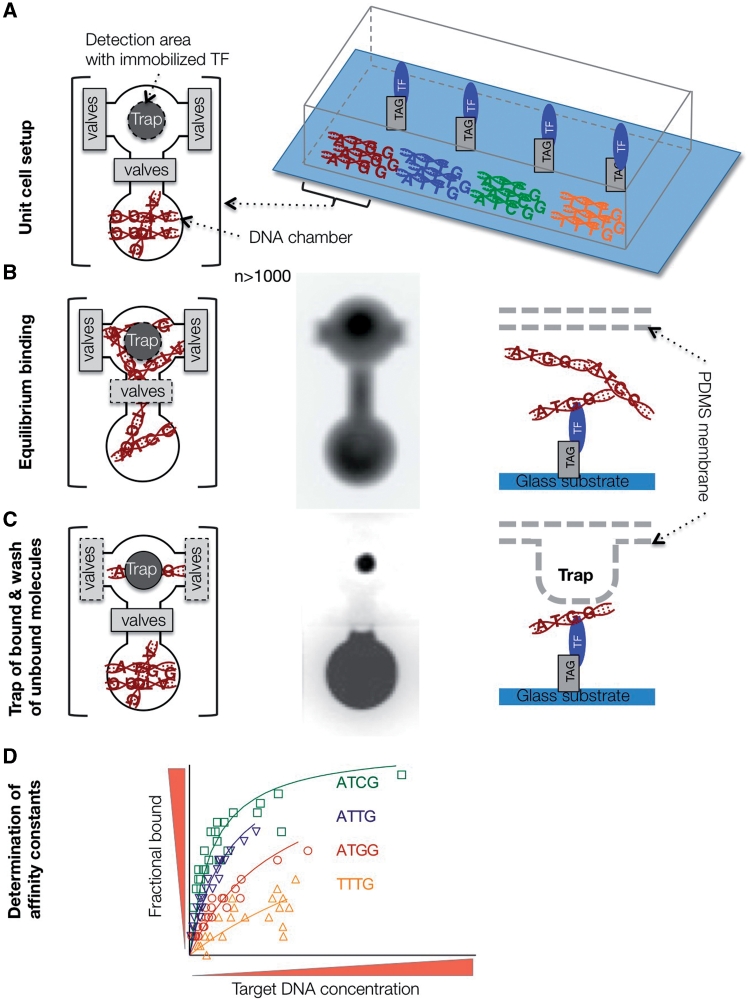
Until recently only methods with relatively low-throughput were available to measure the quantitative parameters of protein–DNA interactions, namely surface plasma resonance platforms, like BIAcore [40, 41], and classical gel shift assays (EMSA) [17, 18]. This experimental gap has recently been filled by the development of a high-throughput microfluidics platform, which employs a novel detection method based on the mechanically induced trapping of molecular interactions (MITOMI; Figure 3) [9]. MITOMI devices, as well as detailed information on how to set up a valve control interface can be obtained from the Stanford Microfluidics Foundry (http://thebigone.stanford.edu/foundry/) and the Caltech Foundry (http://kni.caltech.edu/foundry/). In short, microfluidic chips are fabricated by multilayer soft lithography and aligned to an epoxy-coated glass slide containing thousands of micro-arrayed DNA spots using standard DNA microarray printing instrumentation [9, 42, 43]. Here, each spot codes for a different DNA sequence or concentration, separated and controlled in pL-sized reaction chambers. The concentration-dependent binding to an immobilized TF across the whole chip, and thus hundreds of variable DNA sequences, enables the measurement of thousands of interactions in a single experiment. MITOMI can detect transient and low affinity interactions that are usually missed by other techniques due to the need of stringent wash steps. Indeed mechanical trapping of the interacting molecules completely eliminates loss of molecules and the consequent skew of the apparent affinity before the measurement. Therefore, combined with DNA concentration standards, absolute binding affinities (dissociation constants Kd) can readily be obtained in the nanomolar to micromolar range. Using MITOMI it was shown that the additivity assumption (inherent to PWMs) is not accurate for bHLH TFs (Figure 4C and D) [9].
Figure 3:
Experimental flow chart of MITOMI. (A) Device setup. Target DNAs are spotted on a glass substrate and aligned to DNA chambers of the PDMS chip. One valve separates the DNA chamber from the detection area. TFs are immobilized by selective pull-down underneath the trap. Flanking valves separate unit cells. (B and C) A binding reaction is initiated after opening of DNA chamber valves, and diffusion of target DNA to detection area. Equilibrium bound fraction is separated from unbound fraction by mechanical trap and washing step. From left to right: Top view of unit cell, fluorescence image of diffused fluorescence tagged target DNA, and side view of detection area. (D) Binding affinity constants are determined by non-linear regression fitting of the saturation-binding curve obtained from the measurements.
The same strategy can be used in a reverse configuration by programming reaction chambers with linear templates for cell-free in vitro expression of hundreds of TFs [10, 34]. In this scenario one can either test a promoter fragment of interest for binding to hundreds of TFs, or one can search for interactions between TFs and co-regulators. As previously mentioned, the precise and quantitative characterization of TF–DNA, and TF–co-regulator interactions is fundamental to our understanding of transcriptional regulation, since quantitative modeling of transcriptional regulation relies on quantitative data [44].
So far, most TF–DNA binding studies focused on measuring binding affinities of a given TF to a range of DNA sequences. Only a few studies considered the reverse direction by designing or selecting TFs with altered DNA-recognition properties [45–50]. Yet these types of studies promise to provide us with a better understanding of how TFs recognize DNA and how this recognition could have evolved. Combining on-chip protein synthesis and MITOMI affinity measurement have recently made such permutation studies feasible. The DNA-binding repertoire of 95 TF mutants of a member of the basic Helix–Loop–Helix family [2] has been characterized using such an approach [10]. In this study each of the 19 possible aa point substitutions of five residues known to form DNA base-specific contacts have been tested for binding against 64 DNA sequences. Another, yet related strategy is the comprehensive characterization of related TF families [7, 51–53]. Together the systematic characterization of the functional significance of individual residues, as well as the comprehensive characterization of TF families can help build an understanding of how TF diversity arose.
In vitro measurements of TFs are well suited for discovering consensus sites and binding preferences, as well as for providing a quantitative foundation of TF function. In combination with complete genome sequences, in vitro characterization of TF-binding preferences enables us to map the genome-wide distribution of TFBS and discover candidate target genes in silico [9, 54, 55]. However, a given TF might not always regulate all targeted genes at the same time, or in all cell types, due to cell line-specific modulation of TF activity by co-regulators. In such cases relating in vitro determined binding preferences with in vivo measured protein occupancy profiles is indispensable.
Most in vitro and in vivo experimental approaches rely on a computational framework to detect binding sites [56]. Amongst those MEME, AlignACE, and MDscan are the most commonly used programs to find sequence elements conserved in a set of DNA sequences [57–59]. Although computational techniques for binding site detection have greatly improved over the past years, the underlying assumptions often oversimplify TF–DNA interactions, which commonly results in a high rate of false-positive predictions [11, 13, 56, 60, 61]. Finally, the quantitative modeling of GRN not only relies on estimated TF-binding affinities, but also on the assumption that TFs bind their targets under equilibrium conditions in vivo [12, 62, 63]. Considering the time scale of transcriptional responses under induced stress, this assumption is likely an oversimplification. To circumvent this, one needs to consider the kinetics of binding events. To date only low-throughput approaches, like the SPR BIAcore platform, allow the reliable measurement of TF-binding kinetics. Recently it has been shown that, in principle, MITOMI can be utilized to measure binding kinetics in high-throughput [64].
IN VIVO METHODS
The most commonly used in vivo method to probe for genome-wide TF binding is based on chromatin immunoprecipitation (ChIP; Figure 2C) [65] integrated with either DNA microarray technology (ChIP-chip) [20, 21, 24] or more recently with massive parallel sequencing (ChIP-seq) [22, 25]. Similar to the previously mentioned DIP-chip experiments, TF–DNA complexes are fixed in situ by cross-linking with formaldehyde, sheared into pieces with average length of 100–500 bp, and precipitated from solution using a TF-specific antibody. The enriched DNA is quantified after reversal of the cross-links by either hybridization to DNA microarrays or deep sequencing. In general, both ChIP-chip and ChIP-seq offer a tremendous throughput in profiling genome-wide protein occupancies (Figure 1); while in direct comparison to ChIP–chip, ChIP-seq has improved resolution, lower noise levels, and a higher dynamic range [25]. However, throughput for both ChIP-based methods is limited by the fact that experimental feasibility is strongly dependent on: (i) protein abundance, (ii) cross-linking efficiency, and (iii) antibody availability and specificity [25]. Finally, to identify potential binding sites raw ChIP data needs to be processed with computational techniques, which might remain unsatisfactory as the distinction of direct and indirect protein–DNA interactions are problematic (Figure 2C, see e.g. comparison between ChIP and DIP) [66].
Recently, the use of ChIP-seq experiments across several humans elucidated the impact of genetic variation in TFBS between individuals on TF occupancy [6]. Interestingly, the same study could show that genomic loci with strong ChIP-seq signal, and thus high TF occupancy, are also more frequently occupied in chimpanzee than weaker signals, pointing towards a divergence of weaker, low-affinity binding sites. The importance of low-affinity TF binding in coordinating transcriptional regulation has already been proposed in previous studies [67]. In direct agreement the quantitative variation of TFBS occupancy between closely related Drosophila species have been attributed to modest levels of sequence divergence of otherwise highly conserved binding motifs [68]. However, it remains to be evaluated to what extent these variations translate into alternative transcriptional and developmental programs [68]. Both, the widespread functionality of weak TFBS [67], and the apparent evolutionary divergence of quantitative TFBS traits [68] point towards the necessity to capture minute differences amongst binding sites across a broad affinity regime.
Instead of cross-linking and immunoprecipitation of proteins with DNA, the protein of interest can be fused to Escherichia coli DNA adenine methyltransferase (DamID) [69]. Upon binding to DNA, nucleotides in close vicinity of TF binding are methylated. The methylated DNA is then immunoprecipitated and analyzed by either microarrays or sequencing approaches. Since methylation is restricted to adenine in GATC sites, the resolution of binding site mapping is limited by the distance between two consecutive such sites. This bias in resolution is omitted in approaches that use micrococcal nuclease fusion proteins. In chromatin endogenous cleavage (ChEC) TF-tagged with micrococcal nuclease is activated in vivo by rising levels of Ca2+ [70]. Binding events are detected by mapping of induced double-strand DNA breaks. So far ChEC has not been integrated to high-throughput readouts by deep sequencing or microarray approaches.
A different approach is reverse ChIP or proteomics of isolated chromatin segments (PICh), which is an alternative method for identifying TFs bound to a given locus. Briefly, following a cross-linking step, a desthiobiotin conjugated DNA probe is used to hybridize to a specific genomic locus, and associated proteins are isolated and analyzed by mass spectrometry [71]. This approach alone does not discriminate whether identified proteins are TFs that bind directly or indirectly to DNA. Also, the general applicability of PICh remains to be evaluated, not least because probe design and mass spectrometric analysis will need refinement to adapt to a high-throughput setting.
None of the above-mentioned methods is unbiased with regard to the TF or DNA segment under investigation. DNaseI hypersensitivity assays offer an unbiased, genome-wide mapping of protein binding in vivo when integrated with microarray or massive parallel sequencing [72–74]. The degree of chromatin DNaseI sensitivity allows for the distinction between nucleosome bound and unbound genomic loci. An alternative approach is the use of ChIP based methods to profile genome-wide histone modifications. In this case the detection of alternative chromatin structure can be used to profile genomic regions accessible to TFs [75]. Whether unbound genomic regions are due to the binding of regulatory proteins remains to be validated experimentally or analyzed computationally by considering known TF-binding preferences.
One of the first ChIP–chip experiments revealed that many in silico predicted binding sites are not occupied in vivo [23]. Thus, in addition to the precision of in vitro approaches in determining TF-binding preferences, an in vivo viewpoint is necessary to distinguish between biologically functional and non-functional sites. The prediction of in vivo binding sites from in vitro derived TF-binding preferences is still far from being accurate. One reason is the lack of detailed knowledge of the combinatorial interaction between TFs, cofactor proteins, and chromatin modifiers [76, 77]. Ravasi et al. have recently addressed this issue by combining mammalian two-hybrid screens with gene expression studies [77]. Their analysis highlighted the importance of TF–TF interactions to establish precise transcriptional programs during developmental processes. Solely considering TF–DNA interactions would have missed this regulatory network. On the other hand the identification of TF-binding sites by in vivo experimental methods suffers from drawbacks in (i) the resolution by which binding site can be identified, (ii) the lack to distinguish between direct and indirect interactions, (iii) that the observed interactions are context dependent, and (iv) the fact that only qualitative, or at best semi-quantitative, data can be obtained.
CONSOLIDATION OF IN VIVO AND IN VITRO APPROACHES
Recent technological advances in both, in vitro as well as in vivo methods, have greatly improved our ability to study TF–DNA-binding specificity on a comprehensive level. However, no single approach provides sufficient information to reliably predict the quantitative behavior of gene regulation. While in vitro methods are indispensable for the biophysical characterization and quantification of protein–protein and protein–DNA interactions, it is still difficult to translate this information into actual in vivo function. In many cases only a fraction of high-affinity TFBS are occupied in vivo [23], pointing to secondary effects like the masking of binding sites by competing TFs, nucleosomes [78, 79] or the existence of cooperative binding events frequently missed by in vitro approaches [79]. It will be an exciting endeavor to evaluate the extent to which the consolidation of multiple in vitro data sets, including reconstituted nucleosome occupancy maps and TF–TF interactions, will reduce the discrepancy between in vitro and in vivo results [62, 80]. On the other hand, in vivo methods have the advantage of profiling biologically relevant protein–DNA interactions (e.g. ChIP-seq), albeit, at the expense of being inconclusive with regard to the underlying binding causalities (direct versus indirect binding). A significant fraction of ChIP signals often cannot be correlated to a corresponding TFBS, even in cases where a TF is known to directly bind to DNA [81]. In the near term only a consolidated view of both, in vivo as well as in vitro results promises the unambiguous identification and characterization of TF–DNA interactions.
While in vivo approaches differ with regard to resolution, ‘ChIP-chip versus ChIP-seq’, and experimental bias, ‘TF-centered versus unbiased DNaseI sensitivity’, in vitro approaches greatly differ with regard to throughput and data type (Figure 1 and Table 1). Even without a priori knowledge of possible sequence specificity, in vitro selection and PBM approaches offer de novo TFBS identification. Throughput is limited in principle by the fact that proteins need to be purified to sufficient grade and amounts (Table 1). However, both methods suffer from the inability to account for sequence specific TF dissociation rates. Prior to readout, bound fractions need to be selected and thus washed under stringent conditions. This results in sequence-specific dissociation of TF–DNA complexes, the rates of which are non-linear, unknown and probably vary with sequence (in fact it is probably the dissociation rate that dominates affinity). This results in overestimation of high-affinity binders and thus returns skewed binding profiles. Neither in vitro selection nor PBM approaches can provide quantitative information on affinity and kinetics of the interactions. MITOMI based methods, on the other hand, allow absolute binding affinity measurement at medium throughput but are not yet suited for de novo identification of TFBS. The best experimental strategy would be a serial combination of methods with different throughput and data type (Figure 1 and Table 1). Initial consensus and PWM discovery is optimally done with ChIP, HT-SELEX, MITOMI or PBM approaches. This initial discovery-oriented approach can then be followed up with a MITOMI analysis to arrive at quantitative binding information and a controlled environment for higher-order interaction measurements. Indeed as the catalogues of TF consensus sites and PWMs is growing [52, 82, 83], quantitative measurements and their integration with in vivo data are becoming more and more important.
TF characterization has come a long way in the last decade, with the advent of a multitude of powerful and for the most part mutually complementary methods. Consensus site and binding preferences can now be routinely measured both in vivo and in vitro and precise quantitative measurements can be performed using new methods based on microfluidics, interrogating both the DNA sequence space as well as the protein space. Yet, the challenge remains the same: developing a quantitative understanding of GRNs. Ultimately, the most universal model would only rely on biophysical measurements of TFs and co-factors as these are context-independent and therefore need only be measured once but may be applied universally. Developing hybrid solutions, which take into account in vivo and in vitro measurements are more within our reach. Indeed, any model must be validated with comprehensive in vivo measurements, including ChIP-based binding profiles integrated with expression and proteomic data.
Key Points.
The connectivity of GRNs as well as the quantitative level of gene expression is determined by the specific binding of TFs to cognate binding sites. Moderate genetic variations in these binding sites can lead to quantitative differences in TF occupancies and potential differences in transcriptional output. It is pivotal to our understanding of transcriptional regulation to precisely characterize the quantitative nature of TF–DNA interactions.
TF–DNA interactions can be experimentally characterized by two fundamentally different approaches: in vivo- and in vitro-based methods. In vivo-based methods recover information on the TF consensus binding sites as well as the biological context of DNA specific interactions. In vitro methods aim to identify TF consensus binding sites, binding energy landscapes or the biophysical parameters governing these binding events, and thus can be further subdivided into methods that provide qualitative or quantitative data.
A serial combination of methods with different throughput and information content generally constitutes the best experimental strategy to study TF–DNA interactions: (i) ChIP, HT-SELEX, or PBM approaches to derive consensus sequences and PWM. (ii) MITOMI analysis to substantiate PWM data with quantitative binding information. (iii) Cross-comparison of in vitro binding data with in vivo binding profiles derived from ChIP experiments.
FUNDING
This project was financed with a grant from the Swiss SystemsX.ch initiative, evaluated by the Swiss National Science Foundation.
Acknowledgements
The authors thank David Shore for reading the manuscript and valuable suggestions.
Biographies
Marcel Geertz is a post-doctoral fellow at the University of Geneva. His research is focused on biophysics of TF–DNA interactions.
Sebastian J. Maerkl obtained his PhD from the program of Biochemistry and Molecular Biophysics at the California Institute of Technology in 2007. In his graduate work he developed highly integrated microfluidic devices and applied them to protein biochemistry. Since 2008 he holds a tenure track position in the School of Engineering and the Institute of Bioengineering at the Ecole Polytechnique Federale de Lausanne (EPFL) in Switzerland, where he continues to apply microfluidic large-scale integration to biology.
References
- 1.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 2.Luscombe NM, Austin SE, Berman HM, et al. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1:REVIEWS001. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994;369:758–61. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 4.Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–80. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 5.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–8. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasowski M, Grubert F, Heffelfinger C, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–5. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grove CA, De Masi F, Barrasa MI, et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–27. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djordjevic M. SELEX experiments: new prospects, applications and data analysis in inferring regulatory pathways. Biomol Eng. 2007;24:179–89. doi: 10.1016/j.bioeng.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 10.Maerkl SJ, Quake SR. Experimental determination of the evolvability of a transcription factor. Proc Natl Acad Sci USA. 2009;106:18650–5. doi: 10.1073/pnas.0907688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bussemaker HJ, Foat BC, Ward LD. Predictive modeling of genome-wide mRNA expression: from modules to molecules. Ann Rev Biophys Biomol Struct. 2007;36:329–47. doi: 10.1146/annurev.biophys.36.040306.132725. [DOI] [PubMed] [Google Scholar]
- 12.Kim HD, Shay T, O'Shea EK, et al. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325:429–32. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das MK, Dai H-K. A survey of DNA motif finding algorithms. BMC Bioinformatics. 2007;8(Suppl 7):S21. doi: 10.1186/1471-2105-8-S7-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–98. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 15.Arda HE, Walhout AJM. Gene-centered regulatory networks. Brief Funct Genomics. 2010;9:4–12. doi: 10.1093/bfgp/elp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walhout AJM. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–54. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- 17.Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–25. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–60. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem. 1995;270:11181–9. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 20.Horak CE, Snyder M. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Meth Enzymol. 2002;350:469–83. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- 21.Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 22.Johnson DS, Mortazavi A, Myers RM, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 23.Lee TI, Rinaldi NJ, Robert F, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 24.Hanlon SE, Lieb JD. Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr Opin Genet Dev. 2004;14:697–705. doi: 10.1016/j.gde.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Comput Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger MF, Philippakis AA, Qureshi AM, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–35. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulyk ML, Huang X, Choo Y, et al. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci USA. 2001;98:7158–63. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee S, Berger MF, Jona G, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–9. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson CD, Warren CL, Hauschild KE, et al. Specificity landscapes of DNA binding molecules elucidate biological function. Proc Natl Acad Sci USA. 2010;107:4544–9. doi: 10.1073/pnas.0914023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren CL, Kratochvil NCS, Hauschild KE, et al. Defining the sequence-recognition profile of DNA-binding molecules. Proc Natl Acad Sci USA. 2006;103:867–72. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Einav S, Gerber D, Bryson PD, et al. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol. 2008;26:1019–27. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerber D, Maerkl SJ, Quake SR. An in vitro microfluidic approach to generating protein-interaction networks. Nat Methods. 2009;6:71–4. doi: 10.1038/nmeth.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 36.Wright WE, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–10. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerland U, Moroz JD, Hwa T. Physical constraints and functional characteristics of transcription factor-DNA interaction. Proc Natl Acad Sci USA. 2002;99:12015–20. doi: 10.1073/pnas.192693599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jolma A, Kivioja T, Toivonen J, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–73. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Noll DM, Lieb JD, et al. DIP-chip: rapid and accurate determination of DNA-binding specificity. Genome Res. 2005;15:421–7. doi: 10.1101/gr.3256505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fägerstam LG, Frostell-Karlsson A, Karlsson R, et al. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J Chromatogr. 1992;597:397–410. doi: 10.1016/0021-9673(92)80137-j. [DOI] [PubMed] [Google Scholar]
- 41.Majka J, Speck C. Analysis of protein-DNA interactions using surface plasmon resonance. Adv Biochem Eng Biotechnol. 2007;104:13–36. [PubMed] [Google Scholar]
- 42.Duffy D, Cooper McDonald J, Schueller O, et al. Rapid prototyping of microfluidic systems in Poly(dimethylsiloxane) Anal Chem. 1998;70:4974–84. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 43.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–4. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 44.Bintu L, Buchler NE, Garcia HG, et al. Transcriptional regulation by the numbers: models. Curr Opin Genet Dev. 2005;15:116–24. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pabo CO. Specificity by design. Nat Biotechnol. 2006;24:954–5. doi: 10.1038/nbt0806-954. [DOI] [PubMed] [Google Scholar]
- 46.Blancafort P, Beltran AS. Rational design, selection and specificity of artificial transcription factors (ATFs): the influence of chromatin in target gene regulation. Comb Chem High Throughput Screen. 2008;11:146–58. doi: 10.2174/138620708783744453. [DOI] [PubMed] [Google Scholar]
- 47.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–31. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 48.Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev. 2009;61:513–26. doi: 10.1016/j.addr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Meng X, Thibodeau-Beganny S, Jiang T, et al. Profiling the DNA-binding specificities of engineered Cys2His2 zinc finger domains using a rapid cell-based method. Nucleic Acids Res. 2007;35:e81. doi: 10.1093/nar/gkm385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nilsson MTI, Widersten M. Repertoire selection of variant single-chain Cro: toward directed DNA-binding specificity of helix-turn-helix proteins. Biochemistry. 2004;43:12038–47. doi: 10.1021/bi049122k. [DOI] [PubMed] [Google Scholar]
- 51.Noyes MB, Christensen RG, Wakabayashi A, et al. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–89. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Badis G, Berger MF, Philippakis AA, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–3. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu S, Xie Z, Onishi A, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–22. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–16. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharon E, Lubliner S, Segal E. A feature-based approach to modeling protein-DNA interactions. PLoS Comput Biol. 2008;4:e1000154. doi: 10.1371/journal.pcbi.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elnitski L, Jin VX, Farnham PJ, et al. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–64. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- 57.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20:835–9. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 58.Roth FP, Hughes JD, Estep PW, et al. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–45. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- 59.Bailey TL, Gribskov M. Score distributions for simultaneous matching to multiple motifs. J Comput Biol. 1997;4:45–59. doi: 10.1089/cmb.1997.4.45. [DOI] [PubMed] [Google Scholar]
- 60.Roulet E, Fisch I, Junier T, et al. Evaluation of computer tools for the prediction of transcription factor binding sites on genomic DNA. In Silico Biol (Gedrukt) 1998;1:21–8. [PubMed] [Google Scholar]
- 61.Tompa M, Li N, Bailey TL, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23:137–44. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 62.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–56. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinitz J, Hou S, Sharp DH. Transcriptional Control in Drosophila. Complexus. 2003;1:54–64. [Google Scholar]
- 64.Bates SR, Quake SR. Highly parallel measurements of interaction kinetic constants with a microfabricated optomechanical device. Appl Phys Lett. 2009;95:73705. doi: 10.1063/1.3211382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 66.Gordân R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009;19:2090–100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–72. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bradley RK, Li X-Y, Trapnell C, et al. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 2010;8:e1000343. doi: 10.1371/journal.pbio.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–8. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 70.Schmid M, Durussel T, Laemmli UK. ChIC and ChEC; genomic mapping of chromatin proteins. Mol Cell. 2004;16:147–57. doi: 10.1016/j.molcel.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 71.Déjardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–86. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hesselberth JR, Chen X, Zhang Z, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–9. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sabo PJ, Kuehn MS, Thurman R, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–8. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 74.Crawford GE, Davis S, Scacheri PC, et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods. 2006;3:503–9. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu CL, Kaplan T, Kim M, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fedorova E, Zink D. Nuclear architecture and gene regulation. Biochim Biophys Acta. 2008;1783:2174–84. doi: 10.1016/j.bbamcr.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 77.Ravasi T, Suzuki H, Cannistraci CV, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–72. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–40. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–71. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Massie CE, Mills IG. ChIPping away at gene regulation. EMBO Rep. 2008;9:337–43. doi: 10.1038/embor.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Badis G, Chan ET, van Bakel H, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–87. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wingender E, Dietze P, Karas H, et al. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–41. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]