Abstract
Genome-wide association studies (GWAS) using family data involve association analyses between hundreds of thousands of markers and a trait for a large number of related individuals. The correlations among relatives bring statistical and computational challenges when performing these large-scale association analyses. Recently, several rapid methods accounting for both within- and between-family variation have been proposed. However, these techniques mostly model the phenotypic similarities in terms of genetic relatedness. The familial resemblances in many family-based studies such as twin studies are not only due to the genetic relatedness, but also derive from shared environmental effects and assortative mating. In this paper, we propose 2 generalized least squares (GLS) models for rapid association analysis of family-based GWAS, which accommodate both genetic and environmental contributions to familial resemblance. In our first model, we estimated the joint genetic and environmental variations. In our second model, we estimated the genetic and environmental components separately. Through simulation studies, we demonstrated that our proposed approaches are more powerful and computationally efficient than a number of existing methods are. We show that estimating the residual variance-covariance matrix in the GLS models without SNP effects does not lead to an appreciable bias in the p values as long as the SNP effect is small (i.e. accounting for no more than 1% of trait variance).
Key Words: Family data, Population-based association analysis, Genome-wide scan, Generalized least squares
Introduction
Genome-wide association studies (GWAS) have been offering increased opportunities for detecting susceptibility genes for complex traits and disease [1, 2, 3]. These studies are performed on a genomic scale involving hundreds of thousands of single-nucleotide polymorphisms (SNPs), and bring new statistical challenges for conducting association analyses using family-based designs. Recently, several family-based GWAS [1, 4, 5, 6, 7, 8] have been conducted and many more are currently being genotyped [9]. Hence there is an increasing need for developing computationally efficient and powerful approaches for conducting association tests on a large group of SNPs using a set of nuclear or large pedigrees.
In family-based designs, a range of association analysis methods are available which fall into 2 major categories: (1) traditional family-based association analyses that use the transmission of alleles within informative families [10], and (2) population-based analysis methods that have been adapted to family data [11, 12, 13, 14, 15]. Traditional family-based association analyses, such as the quantitative trait transmission disequilibrium test (QTDT) [16] and the family-based association test [17], analyze within-family variation and focus on the transmission of alleles from heterozygous parents to their offspring. They are robust to the presence of population stratification in the dataset, at the cost of a loss in power on a per-genotype basis [18, 19]. On the other hand, population-based association analyses [11, 12] provide an overall test of within- and between-family variations, and tend to be more powerful than the family-based association tests [20]. However, they are susceptible to population stratification and are computationally intensive to apply on a genome-wide scale.
The major impediment to rapid computation in population-based approaches adapted to family-based designs involves the need to jointly estimate the fixed SNP effect and the family variance-covariance matrix separately for each SNP. Recently, several approaches have been proposed that aim to maintain the high power of the population-based approaches while substantially reducing computation time. Aulchenko et al. [15] proposed a rapid association analysis (GRAMMAR) for quantitative trait loci (QTL) using a linear mixed model. In their method, familial similarities were modeled by a random polygenic effect. This random effect is assumed multivariate normally distributed with the correlation matrix equal to the kinship matrix. To conduct a rapid genome-wide scan, they proposed to perform a linear regression accommodating this polygenic random effect and fixed effects for covariates using the complete pedigree but ignoring marker data. Regression residuals were then used as a quantitative trait to do single SNP association analyses using a simple linear regression for unrelated individuals. Thus with GRAMMAR, the family variance-covariance matrix is estimated once, as a function of the polygenic effect, and used to residualize the observed quantitative phenotype data for an analysis of the individual SNP effects. p values from the GRAMMAR method were reported to be conservative and a permutation test was suggested [15].
Chen and Yang [14] developed a package for genome-wide association analyses with family data (GWAF) using a method similar to GRAMMAR, but involving joint estimation of the variance of the random polygenic effect and the fixed SNP effect when doing the single SNP analysis. It thus takes much longer for a genome-wide scan than GRAMMAR. Chen and Abecasis [13] proposed another rapid scan scheme using a generalized linear model to conduct variance components analysis. In their method, the quantitative trait is modeled by a multivariate normal distribution where the variance-covariance matrix has 3 variance components: a linked major gene effect, background polygenic effects, and environmental effects. Taking the derivatives of the likelihood function, they constructed a score test for the fixed major gene effect. Like GRAMMAR, they proposed a rapid test by estimating the variance-covariance matrix of the quantitative trait without taking the individual SNP effect into account, i.e. without the linked major gene effect in the variance components and in the fixed effects. This base matrix is then plugged into the score test for computation of the single SNP association analysis.
These rapid methods have provided new opportunities for genome-wide association analyses of family-based design, as the number of typed markers can easily go above a million with advances in genotyping technology. However, approaches like GRAMMAR and that proposed by Chen and Abecasis [13] only model familial resemblance as a function of genetic relatedness. Yet there is a growing list of GWAS that include familial relationships (e.g. between spouses) [1] where phenotypic similarity is likely due to shared environmental rather than genetic factors, and behavioral phenotypes (e.g. personality, obesity) [6, 21] where there are likely environmental as well as genetic contributions to phenotypic similarity amongst relatives. Although these rapid methods provide an efficient way for GWAS analysis when the shared environmental factors are negligible as in some studies of large extended families, they cannot fully express familial similarities when environmental factors are clearly present as in studies of nuclear families [9]. Bravo et al. [22] have demonstrated in their study that it is advantageous to model familial relatedness by including unrelated subjects, such as a spousal relationship, to capture some shared environmental factors.
In this study, we propose an alternative approach: 2 generalized least squares (GLS) models for the rapid association analysis of GWAF data. Like the many population-based approaches discussed above, our methods are linear-regression-based single SNP analyses, model within- and between-family variation to find an efficient estimator of the SNP effect, and achieve computational efficiency by estimating the family variance-covariance matrix once and then using this estimate in every SNP regression. Unlike the methods discussed above, our methods accommodate both genetic and environmental contributions to familial resemblance.
Our first proposed method is a rapid feasible generalized least squares model with unstructured family covariance matrices (RFGLS-UN), and our second proposed method is a rapid feasible generalized least squares model with family covariance matrices modeled as a function of 3 variance components (RFGLS-VC): genetic, shared environmental and non-shared environmental. Because RFGLS-UN places no structure on the family variance-covariance matrix, it can accommodate patterns of familial resemblance due to both genetic and environmental mechanisms. RFGLS-VC also allows for both genetic and environmental contributions to familial resemblance, although it does so in a structured manner separating the genetic and environmental components. Both RFGLS-UN and RFGLS-VC achieve computational efficiency by estimating the family variance-covariance matrix once and then using this estimate in every SNP regression. Conditioning each regression on the same estimated variance-covariance matrix could disturb the distribution of the test statistics, especially if the SNP effect is large. Consequently, we examine the type I error rates and power for these methods for a range of simulated data.
Specifically, we compare these 2 methods with 5 others: an ordinary least squares (OLS) regression model that ignores within-family correlations, a feasible generalized least squares regression model (FGLS) that involves estimating the residual family variance-covariance matrix conditional on each SNP in a genome-wide scan, the GWAF method [14], the QTDT method [16], and a generalized estimating equation (GEE) method. Although our methods can be easily extended to large pedigrees, we focus on their performance in studies of nuclear families and simulates accordingly. Our simulations show that the rapid methods result in appropriate type I error rates and achieve power that approximates that achieved when the real familial similarities were known (i.e. the GLS method). An R package rfgls (http://www.tc.umn.edu/∼lixxx554/rfgls_0.0.tar.gz) was developed to implement the RFGLS-UN/VC and FGLS methods in studies of nuclear families.
Methods
Models
For a set of pedigrees each including one or more related individuals, let yij denote the measured phenotype of individual j in pedigree i (i = 1, …, m, j = 1, …, ni, and Σini = n). Let gij denote the additive genotype score of a SNP with alleles ‘A’ and ‘a’ of individual j in pedigree i, and gij can take values of 0, 1, or 2 depending on the number of minor allele ‘A’ individual i has. Let vectors Yi = {yi1, …, yini}, and Gi = {gi1, …, gini} contain the phenotype and genotype of individuals from pedigree i, respectively. Let Ci be a ni × p matrix that contains p covariates of pedigree i. For a continuous phenotype, we can do a single SNP association test using the following linear regression model:
| (1) |
| (2) |
where α is the population mean, βg is the additive effect of the SNP, βc is a size p vector of the covariate effects, and ∊i is the random residual term which is modeled as
| (3) |
assuming independent pedigrees. The ni × ni matrix Vi is the variance-covariance matrix of pedigree i. To simplify the notation as shown on the right hand side of equ. 1, the observed data on (p + 1) fixed predictors of pedigree i are contained in ni × (p + 2) design matrix Xi, and (p + 2) parameters are contained in vector β.
If Vis are known, the best linear unbiased estimator of β is the GLS estimator [23]:
| (4) |
with variance
| (5) |
The GLS estimator is unbiased and consistent, and achieves the Cramer-Rao lower bound asymptotically [23].
If Vis are unknown, a ‘feasible generalized least squares estimator’ can be used as Vis are estimated from the data, so that the Vi in equ. 4 and 5 can be replaced by its estimator
| (6) |
and
| (7) |
The efficiency of the feasible least squares estimators are often of interest as can take various forms. If is a consistent estimator of Vi, like a maximum likelihood estimator (MLE), then is a consistent estimator of β [23]. If we model Vi by Vi = V(Θ), where Θ is the parameter or a vector of parameters of the covariance matrix, Θ and β can be jointly estimated, usually through iterative methods [24].
If we ignore the within-family correlations and model Vi by Vi = σ2Ini, where Ini, is an ni × ni identity matrix, we obtain the OLS estimator of β:
| (8) |
with variance
| (9) |
where can be easily estimated by
The equality of OLS estimators (equ. 8) and FGLS estimators (equ. 6) modeling within-family correlations has been shown to be satisfied under certain conditions [25], and the efficiency of the 2 estimators depends on their variances. In an FGLS method estimating Vi by its MLE, the jointly estimated SNP effect asymptotically gains efficiency (i.e. the SNP effect estimator achieves smaller variance) over that of an OLS estimator [23, 26, 27]. However, the estimation of the Vis can slow down the analysis and be too computationally demanding on a genomic scale. We hence propose a RFGLS method to increase the computational efficiency and conduct a rapid genome-wide scan. The RFGLS method includes the following steps:
-
(1)Perform a single FGLS analysis using the complete pedigree but ignoring marker data. Suppose there are m families of l different pedigree types in the data. Let Yij contain the phenotype of individuals from family j in pedigree type i (i = 1, …, l, j = 1, …, mi, and Σimi = m). So equ. 1 becomes
where . A MLE of Vi by is obtained for each type of pedigree (see the following descriptions).(10) -
(2)Do a single SNP analysis using the as the variance-covariance matrices for each pedigree. Let V be the block-diagonal matrix with on its diagonal blocks. First, a Cholesky decomposition of V is taken as V = SST. Then a simple linear regression for unrelated observations is carried out using the Cholesky-factor-transformed data:
where X is the n × (p + 2) matrix containing predictor values of all m pedigrees, Y is the size n vector of phenotypes, β = (α, βg, βc) as defined in equ. 2, and the residual term ∊g is distributed as N(0, σ2I).(11) -
(3)
Make association calls based on the F test statistics from the simple linear regressions in step 2. Here we want to test the null hypothesis H0: βg = 0. The F test statistics follows an F(1, s) distribution, where s = n – rank(S−1X).
Here we propose 2 ways to estimate in step 1: (i) the RFGLS-UN method estimating unstructured family blocks. If different pedigree types are present in the data, for example, families with monozygotic (MZ) twins and families with non-twin biological offspring, etc., our model allows separate estimations of the variance-covariance blocks for different pedigree types and models the heteroscedasticity accordingly. Thus if there are l different pedigree types with each of pedigree size ki (i = 1, …, l), there are Σli = 1ki (ki + 1)/2 parameters in the variance-covariance matrix to estimate. (ii) The RFGLS-VC method estimating familial similarities by a combination of ‘genetic relatedness’ and ‘environmental relatedness’. We add a random additive polygenic effect aij in equ. 10:
| (12) |
We let aij follow a multivariate normal distribution N(0, σ2aΦi), where Φi is the relationship matrix of each pedigree in pedigree type i, i.e. twice the kinship matrix, and σ2a is the genetic variance. We also let ∊ij follow a multivariate normal distribution MVN(0, σ2eRi), where σ2e is the environmental variance and Ri is a compound symmetric matrix taking the form (1 – ρi)I + ρiJ, where ρi is the within-family correlation coefficient for each pedigree in pedigree type i, I is the identity matrix and J is the 1's matrix. Hence we have
| (13) |
Our proposed methods address 2 factors that should be considered when evaluating alternative approaches to the analysis of family-based data on a genome-wide scale. The first is computational efficiency, which has been achieved primarily by estimating the family variance-covariance matrix a single time rather than separately for each individual SNP regression. The second concerns modeling familial resemblance, which is modeled in our methods as a function of both genetic and environmental factors. In the following sections, we study the performances of these 2 approaches through simulation studies and compare type I error and power with other (FGLS, OLS, GLS, GWAF, GEE, and QTDT) approaches through a range of simulation studies. We also study the impact of ignoring environmental similarity within each pedigree and show how our method can gain power over the existing population-based approaches. In addition, we compare the computational time to implement these methods to perform a single SNP association analysis.
Simulations
Simulations of 1,800 Families
Simulation I: Single Causal SNP
In order to explore the performance of the alternative approaches to detect association in pedigrees with diverse familial relationships, we modeled our simulations after the family structures that exist within the Minnesota Center for Twin and Family Research (MCTFR), which, in the process of GWAS, is genotyping research participants from families that include MZ twins, dizygotic twins, non-twin biological offspring, and adopted offspring. We considered these types of pedigrees because they justify the need of proposing the RFGLS approach. Each pedigree type showed a very different variance-covariance structure for a set of behavioral phenotypes [9]. There is evidence that the huge variation of phenotypes among these different pedigree types strongly depends on environmental as well as genetic factors [9]. Hence to achieve greater power, it is essential to model the environmental component of the phenotypic variation along with the genetic component.
We simulated 1,800 4-member pedigrees for this simulation study. Each pedigree consisted of 2 parents and 2 siblings. Among the 1,800 pedigrees, there were 600 in which both offspring were MZ twins, 600 in which both offspring were full biological siblings, and 600 in which both offspring were adopted (i.e. not genetically related to each other or their rearing parents). Our RFGLS approach allows us to include all members in twin families and thus would be more powerful compared to approaches that drop one member of each twin pair. Further, we recognize that inclusion of adoptive families in GWAS will never become a common occurrence even if these families are part of the MCTFR GWAS. Nonetheless, inclusion of adoptive families allows us to investigate the performance of the alternative methods with samples that include culturally defined clusters (e.g. classrooms, neighborhoods), where phenotypic similarity owes to shared experiences rather than common genetics. Such culturally defined samples will, arguably, become more common in future GWAS.
The total genotyped population consisted of 7,200 individuals. No ethnic stratification was included. One trait locus was simulated using an additive genetic model, which explained 0.6% of the total trait variation. This was then repeated 1,000 times to get 1,000 quantitative traits and 1,000 corresponding trait loci. The minor allele frequency (MAF) of the 1,000 trait loci varied uniformly from 0.2 to 0.5. To study type I error rates of the different methods in a large-scale analysis, we simulated 10,000 independent SNPs that did not contribute to the phenotype.
In order to span a diverse set of familial phenotypic similarities, family variance-covariance structures were modeled using a quantitative behavioral trait, substance use and abuse, which shows both genetic and environmental contributions to familial resemblance [9]. Specifically, we considered the following 3 variance-covariance models when generating the continuous traits from multivariate normal distributions. The first model (HomoG) is homoscedastic with a polygenic component of 40% additive heritability, and no environmental contribution to familial resemblance. The first model adheres to the assumptions that underlie the GWAF approach. The second model (HomoGE) adds an environmental component to the first model with a within-family correlation of 0.2 that is constant for all family members. The inclusion of an environmental contribution to familial resemblance violates the assumptions of the GWAF method, while it adheres to the assumptions that underlie the RFGLS-VC method. The third model (HetGE) introduces heteroscedasticity by varying phenotypic variance for fathers, mothers and offspring. The phenotypic variance was chosen such that the trait locus explained 0.6% of the total variance for offspring, 0.5% for mothers, and 0.4% for fathers. The environmental correlations were set to be 0.2 within a generation (i.e. between father and mother or between offspring) but 0 between generations (i.e. between parent and offspring). The third model, which closely parallels what is found with substance abuse [9] as well as other behavioral traits, sought to investigate the added effect of phenotypic variance heterogeneity and unequal environmental correlation.
Simulation II: Single Causal SNP – RFGLS-UN versus FGLS(-UN)
We further sought to determine if it is valid to estimate the variance-covariance matrix through a single regression without adjusting for the SNP effect. Specifically, we varied the size of the major gene effect and compared the p values from the RFGLS-UN method and the FGLS method. Let r2 represent the proportion of the total variance explained by the trait locus. We randomly picked 10 trait loci out of the 1,000 trait loci simulated, and varied the r2 of each locus to be at 5 values under the HomoGE scenario, i.e. r2 = 0.1, 0.5, 1, 3 and 5%. Thus there were in total 50 sets of continuous traits simulated, each a composite of a QTL effect, and a polygenic and environmental variance component following the specification used in Simulation I.
Simulation III: Multiple Causal SNPs
To study the performance of our rapid methods when there are multiple trait loci, we simulated a trait with 10 causal SNPs each explaining 0.6% of the total variance (with MAF ranging from 0.2 to 0.5). The trait was simulated using the same family structures (HomoG, HomoGE, and HetGE) as those used in Simulation I. The simulation was repeated 1,000 times.
Simulations of 180 Families
To study the performance of our proposed method at a much smaller sample size, we randomly selected 60 families from each of the 3 pedigree types as described in ‘Simulations of 1,800 Families’ above. Our study consisted of 180 4-member pedigrees. Single causal SNP analysis was carried out with the causal SNP, nonassociated SNPs, and the 3 phenotype models simulated as described in ‘Simulations of 1,800 Families’ above.
The MCTFR GWAS
As a complement to the simulation studies presented above, we analyzed the MCTFR GWAS data for height. Height was measured while participants were standing with their backs straight against a wall and their shoes off. Height results are reported here for 4,711 individuals in 1,817 2-generation pedigrees. This represents the first approximately 60% of MCTFR participants to be genotyped. Genotyping on more than 500,000 SNP markers was completed using Illumina's 660W Quad array. Markers underwent standard quality control filters before being analyzed, resulting in the final set of 527,469 markers used here.
Analysis
Besides the proposed 2 rapid generalized least squares approaches, RFGLS-UN and RFGLS-VC, the data was also analyzed using the following methods:
-
(1)
FGLS(-UN): This is similar to the proposed RFGLS-UN method. Instead of fixing the variance-covariance matrix as in RFGLS-UN, the variance-covariance matrix was jointly estimated with each marker effect by a block-diagonal matrix, using unstructured blocks. As discussed in the Models section, the FGLS method gives a consistent estimator of the SNP effect, and is reported to be more efficient than OLS estimators. Our rapid method RFGLS-UN is a simplification of the FGLS method, and a comparison between the two will be illustrated in the Results section.
-
(2)
OLS: A single SNP association analysis was carried out ignoring family structure. Thus the individuals in all pedigrees were analyzed as independent observations. This is the fastest approach conducting simple linear regressions, and gives consistent estimators.
-
(3)
GLS: The variance-covariance matrix used to simulate the phenotype data was plugged into the association analysis as the known variance-covariance matrix (equ. 4 and 5). The GLS method gives the most efficient estimator of the SNP effect among the unbiased estimators [23], and achieves the Cramer-Rao lower bound asymptotically. This method was used as our ‘gold standard’ in the model comparisons.
-
(4)
GWAF [14]: This method uses the linear mixed effects model implemented in the R package GWAF (http://cran.r-project.org/web/packages/GWAF/index.html). Familial similarity was modeled through a polygenic random effect.
-
(5)
GEE [28]: A GEE model with an exchangeable within-family correlation structure was fit. This method was implemented using the geeglm function in the R package geepack[29, 30, 31].
-
(6)
QTDT: The orthogonal association test under an allelic transmission scoring model in extended pedigrees by Abecasis et al. [16] was carried out using the QTDT software.
In the rest of the paper, we will refer to QTDT as the family-based association analysis, and methods 1–5 as population-based association analyses. The family-based methods analyze within-family variation, while the population-based approaches analyze both within- and between-family variation. The RFGLS-UN/VC, FGLS, OLS, and GLS methods were implemented in our R package rfgls.
Results
Simulations of 1,800 Families
Simulation I
To evaluate the performance of our proposed RFGLS methods and several other available methods, we first checked the type I error rates (obtained by calculating the empirical probability of the SNP effect p values exceeding the prespecified significance level α from the 1,000 replicates of the 10,000 nonassociated SNPs). Table 1 summarizes the type I error rates for each method under the 3 variance-covariance models at α = 1 × 10−4 and α = 5 × 10−6, respectively, with the latter being the Bonferroni adjusted nominal type I error rate. The 4 GLS methods along with the GWAF, GEE, and QTDT methods gave type I error rates at the prespecified α level. When within-family correlations were ignored, as in the OLS method, the type I error rate was clearly inflated due to the underestimation of standard errors. We calculated the genomic inflation factor λ by computing the ratio between the median of the observed test statistics of the simulated independent SNPs and the median of the expected values from the same distribution (table 2). Lambda was close to 1 for all the implemented methods except for OLS which gave inflated λs at 1.2, 1.4 and 1.2 for the HomoG, HomoGE and HetGE simulations, respectively. Therefore, there was no evidence of additional inflation of the test statistic from the methods accounting for familial correlations.
Table 1.
Type I error at two significant levels (1,800 families)
| RFGLS-UN | RFGLS-VC | FGLS | GLS | OLS | GWAF | GEE | QTDT | |
|---|---|---|---|---|---|---|---|---|
| α = 1 × 10−4a | ||||||||
| HomoG | 1.0 | 1.0 | 1.0 | 1.0 | 4.2 | 1.0 | 1.1 | 1.0 |
| HomoGE | 1.0 | 1.0 | 1.0 | 1.0 | 10.0 | 1.0 | 1.1 | 1.0 |
| HetGE | 1.0 | 1.0 | 1.0 | 1.0 | 4.9 | 1.0 | 1.1 | 1.0 |
| α = 5 × 10−6b | ||||||||
| HomoG | 5.0 | 5.8 | 5.0 | 5.0 | 45 | 5.1 | 5.7 | 5.0 |
| HomoGE | 5.0 | 5.0 | 5.0 | 5.0 | 120 | 5.0 | 6.6 | 5.0 |
| HetGE | 4.8 | 6.0 | 4.9 | 5.0 | 48 | 5.1 | 6.2 | 4.9 |
There were 1,000 simulations and 10,000 nonassociated SNPs in each simulation.
Values × 10−4
values × 10−6.
Table 2.
Genomie control parameter X of the null SNPs (1,800 families)
|
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| RFGLS-UN | RFGLS-VC | FGLS | GLS | OLS | GWAF | GEE | QTDT | ||
| HomoG | 0.998 | 0.999 | 0.999 | 0.998 | 1.210 | 0.998 | 0.999 | 0.998 | |
| HomoGE | 0.998 | 0.999 | 0.999 | 0.998 | 1.399 | 0.998 | 0.999 | 0.998 | |
| HetGE | 0.999 | 0.999 | 0.999 | 0.998 | 1.247 | 0.999 | 0.999 | 0.998 | |
λ was calculated using the test statistics from the independent SNPs of the 1,000 simulations each with 10,000 nonassociated SNPs.
We computed the power for each method under the 3 variance-covariance models at α = 1 × 10−6 and α = 5 × 10−8 (obtained by counting the proportion of times the trait SNP was called significant out of the 1,000 simulations). The latter significance level is often recommended as the genome-wide significance level [32, 33]. Table 3 summarizes the empirical power for the methods implemented corrected for any inflation in type I error. Overall, the population-based analysis methods adapted to family data outperformed the family-based association method. Under the first scenario where only genetic factors contributed to familial resemblance (HomoG), the 3 GLS methods (RFGLS-UN, RFGLS-VC, and FGLS) and the mixed model approach (GWAF) gave power comparable to our gold standard, the GLS method. We observed a minor loss of power (4% less than with the GLS method) using the GEE method at the more stringent α level of 1 × 10−6. The power of the OLS method correcting for inflated type I error was lower than that of the ones accounting for within-family correlations. This can be due to the loss of efficiency when familial similarities are ignored. The family-based analysis, the QTDT method, demonstrated very low power. When we introduced an environmental contribution to familial resemblance (HomoGE), the 3 least squares methods (RFGLS-UN, RFGLS-VC and FGLS) continue to approximate the GLS gold standard, although now we observe some loss of power with the mixed model GWAF method, which assumes familial resemblance is due entirely to genetic factors. In addition, a minor loss of power using the GEE method is also observed here. Finally, under the third scenario, which is most realistic for behavioral data, only the RFGLS-UN and FGLS methods provide levels of power comparable to that seen with GLS.
Table 3.
Empirical powera at a given threshold (1,800 families): proportion of times out of 1,000 simulations that the reported p value was more significant than the listed significance level (in percentages)
| RFGLS-UN | RFGLS-VC | FGLS | GLS | OLSb | GWAF | GEE | QTDT | |
|---|---|---|---|---|---|---|---|---|
| α = 5 × 10−6 | ||||||||
| HomoG | 93.2 | 93.5 | 94.1 | 93.7 | 83.3 | 93.7 | 93.1 | 4.5 |
| HomoGE | 95.8 | 96.1 | 96.3 | 96.2 | 85.3 | 94.6 | 95.4 | 5.4 |
| HetGE | 88.5 | 87.8 | 89.2 | 89.2 | 81.6 | 87.4 | 88.3 | 2.4 |
| α = 5 × 10−8 | ||||||||
| HomoG | 77.1 | 77.3 | 78.5 | 78.7 | – | 78.1 | 75.2 | 0.4 |
| HomoGE | 81.0 | 81.9 | 82.3 | 83.0 | – | 77.2 | 79.5 | 0.5 |
| HetGE | 64.7 | 61.1 | 65.5 | 65.4 | – | 60.4 | 60.9 | 0.1 |
Power of detecting a single causal SNP which accounts for 0.6% of the variance.
The power of the OLS method was calculated after the inflated type I error was corrected.
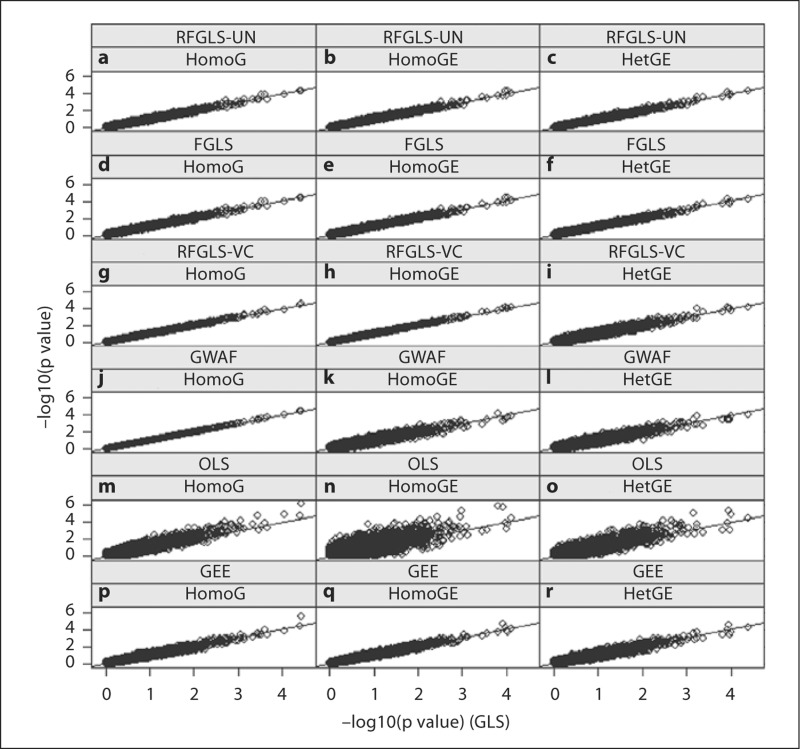
When we plot the p values from the GLS method versus those from the others (fig. 1), we see that the RFGLS-UN approach generally gave good alignment (fig. 1a–c). The variance components approaches, especially GWAF, suffer most when model assumptions are not correct (fig. 1k, l). The OLS method had generally lower p values (fig. 1m–o).
Fig. 1.
−log10(p values) comparing the ‘gold standard’ GLS method and the other methods in HomoG, HomoGE, and HetGE scenarios. There are 10,000 points on each panel. The first descriptions label the 6 methods, which are RFGLS-UN, FGLS, RFGLS-VC, GWAF, OLS, and GEE (from top to bottom). The second descriptions label the 3 simulation scenarios, which are HomoG, HomoGE, and HetGE (from left to right)
Simulation II
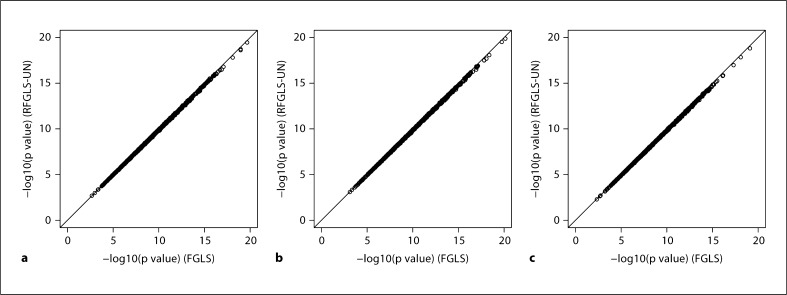
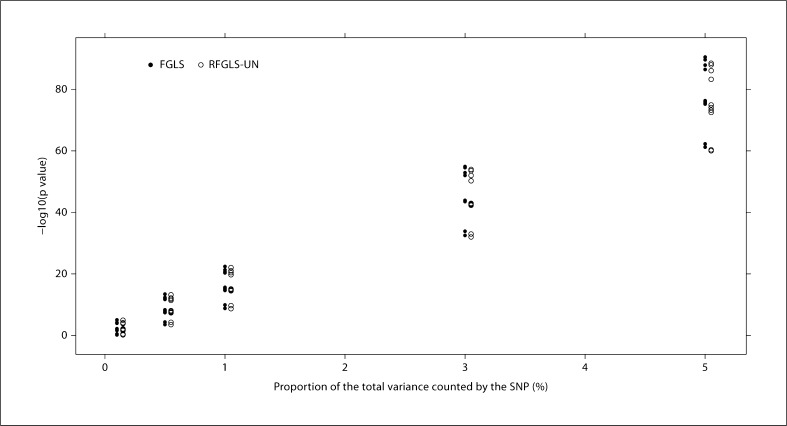
From Simulation I, we plotted the p values (fig. 2) from the rapid method (RFGLS-UN) versus those from the method where variance and fixed genetic effects were jointly estimated (FGLS). The rapid method was shown to be almost perfectly correlated with the latter. In Simulation II, we further sought to determine the effect of different SNP effect sizes on the performance of the RFGLS-UN method. Specifically, we varied the size of the major gene effect and compared the p values from the RFGLS-UN method and the FGLS method. In our simulations, no inflation/deflation of the p values was observed when the variance explained by the trait locus was less than 1% (fig. 3). This shows that estimating the variance-covariance matrix in the GLS models under the null hypothesis (i.e. without conditioning on the SNP effect) does not lead to appreciable bias in the association p values as long as the major gene effect is small. When the SNP effect size gets bigger (i.e. the 3 and 5% levels), some inflated p values were observed in our rapid method which gave larger p values than the FGLS approach. At such a big effect size (greater than 1% of the total variance), however, the p values obtained from both methods have already passed even the most stringent significance level (5 × 10−8), and the minor inflation of the p values in our rapid method will not affect the detection of the SNP.
Fig. 2.
−log10(p values) comparing the rapid FGLS method (RFGLS-UN) and its corresponding full FGLS method. There are 10,000 points on each panel. a HomoG, b HomoGE, c HetGE.
Fig. 3.
Comparison between the RFGLS-UN and the FGLS method by increasing the proportion of the total variance explained by each SNP from 0.1 to 5% (Simulation II). There are 10 SNPs being compared at each vertical line. Solid circles represent the FGLS method, and empty circles represent the RFGLS-UN method.
Simulation III
In our proposed rapid methods, the variance-covariance matrix is estimated without adjusting for the SNP effect. Although one causal SNP may explain only a small proportion of the total variance, a group of causal SNPs may exist that jointly account for a larger proportion of the total variance. In Simulation III, we sought to compare the rapid method with the standard FGLS and GLS methods when there are multiple causal SNPs. In our 10 causal SNP simulations, we computed the power for detecting each SNP at α = 5 × 10−8, and illustrated the 10 power rates (obtained by counting the proportion of times the causal SNP was called significant out of the 1,000 simulations) in a box plot (fig. 4) for each of the method under the 3 scenarios. When familial resemblance is entirely due to genetic factors (HomoG), the 3 GLS methods (RFGLS-UN, RFGLS-VC, and FGLS) and the mixed model approach (GWAF) have similar power, which approximated that for GLS, the gold standard. When there are both genetic and environmental contributions to familial resemblance (HomoGE), there is a notable loss of power with the GWAF method relative to the other methods. Finally, when we introduce heteroscedasticity and a more complex pattern of environmental contributions to familial resemblance (HetGE), relative loss of power is observed with both the GWAF and RFGLS-VC methods. The GEE method suffers some loss of power as compared to the gold standard under all 3 variance-covariance models. It trails all the other methods in the HomoG scenario, and slightly outperforms GWAF in the HomoGE and HetGE scenarios. The minor allele frequency showed no effect on the detecting of the SNPs.
Fig. 4.
Power of the multiple causal SNPs in a boxplot for each method (Simulation III). y-axis is power. x-axis is method. a HomoG, b HomoGE, c HetGE. Each of the 10 causal SNP explains 0.6% of the total variance. α = 5 × 10−8.
Simulations of 180 Families
The performance of the proposed methods and several other methods were also evaluated through type I error rate and power using a much smaller sample size of 180 families. Table 4 summarizes the type I error rates (obtained by calculating the empirical probability of the SNP effect p values exceeding the prespecified significance level α from the 1,000 replicates of the 10,000 nonassociated SNPs) for each method under the 3 variance-covariance models at α = 5 × 10−2 and α = 5 × 10−3, respectively. The 2 proposed GLS methods (RFGLS-UN/VC) along with the GLS and QTDT methods gave type I error rates at the prespecified α level. Similar to the large sample size case presented above, we observed the most inflated type I error rate using the OLS method where within-family correlations were ignored. Under the small sample size, however, we also observed some inflation using the FGLS methods, the GEE method, and the GWAF method. The inflation of the type I error rate was more obvious when a relatively large number of parameters were estimated, as with the FGLS method. There was only a minor inflation in the type I error rate with the GWAF method. The inflation of type I error rate for small sample sizes due to the underestimation of the variance by the FGLS method and the GEE method has also been reported in previous studies [34]. The genomic inflation factors λ for this small sample size study are listed in table 5. Lambda was close to 1 for all implemented methods except for OLS and FGLS.
Table 4.
Type I error at two significant levels (180 families)
| RFGLS-UN | RFGLS-VC | FGLS | GLS | OLS | GWAF | GEE | QTDT | |
|---|---|---|---|---|---|---|---|---|
| α = 5 × 10−2a | ||||||||
| HomoG | 5.0 | 5.0 | 7.2 | 5.0 | 7.6 | 5.1 | 5.4 | 5.0 |
| HomoGE | 5.0 | 5.0 | 7.2 | 5.0 | 9.8 | 5.1 | 5.3 | 5.0 |
| HetGE | 5.0 | 5.0 | 7.2 | 5.0 | 7.9 | 5.1 | 5.4 | 5.0 |
| α = 5 × 10−3b | ||||||||
| HomoG | 5.0 | 5.0 | 10.0 | 5.0 | 11.0 | 5.4 | 6.1 | 5.0 |
| HomoGE | 5.0 | 5.0 | 10.0 | 5.0 | 18.0 | 5.3 | 6.1 | 5.0 |
| HetGE | 5.0 | 5.0 | 10.0 | 5.0 | 11.9 | 5.4 | 6.1 | 5.0 |
There were 1,000 simulations and 10,000 nonassociated SNPs in each simulation.
Values × 10−2
values × 10−3.
Table 5.
Genomie control parameter X of the null SNPs (180 families)
| RFGLS-UN | RFGLS-VC | FGLS | GLS | OLS | GWAF | GEE | QTDT | ||
|---|---|---|---|---|---|---|---|---|---|
| HomoG | 1.001 | 1.002 | 1.186 | 1.001 | 1.218 | 1.009 | 1.016 | 1.001 | |
| HomoGE | 1.000 | 1.002 | 1.183 | 1.002 | 1.405 | 1.005 | 1.013 | 1.000 | |
| HetGE | 1.001 | 1.002 | 1.186 | 1.001 | 1.248 | 1.009 | 1.015 | 1.001 | |
X was calculated using the test statistics from the independent SNPs of the 1,000 simulations each with 10,000 nonassociated SNPs.
We computed the power for each method under the 3 variance-covariance models at α = 5 × 10−2 and α = 5 × 10−3 (obtained by counting the proportion of times the trait SNP was called significant out of the 1,000 simulations). Table 6 summarizes the empirical power for the methods implemented in this small sample size study corrected for any inflation in type I error. Similar to the large sample case, the population-based analysis methods adapted to family data outperformed the family-based association method. The power of the OLS method correcting for inflated type I error was lower than the power achieved by methods that modeled the within-family correlations. The performance of the rapid method, RFGLS-UN, continued to approximate that of FGLS under all 3 simulation models, after adjustment for the inflated type I error rates of FGLS. However, there was a minor loss of power using the RFGLS-UN and FGLS method in the 3 scenarios as compared to using the GLS gold standard. Due to the relatively large number of parameters in the unstructured variance-covariance matrices in RFGLS-UN and FGLS, the loss of power came from the less accurate estimation of the parameters using the small sample size. The performance of the proposed RFGLS-VC method, where fewer parameters are estimated, approximated that of the GLS gold standard in all 3 scenarios. It slightly outperformed GWAF under the HomoGE scenario where there is an environmental contribution to familial resemblance, and the 2 methods were comparable in the HomoG and HetGE scenarios.
Table 6.
Powera at a given threshold (180 families): proportion of times out of 1,000 simulations that the reported p value was more significant than the listed significance level (in percentages)
| RFGLS-UN | RFGLS-VC | FGLSb | GLS | OLSb | GWAFb | GEEb | QTDT | |
|---|---|---|---|---|---|---|---|---|
| α = 5 × 10−2 | ||||||||
| HomoG | 47.2 | 49.2 | 47.4 | 50.4 | 42.4 | 50.0 | 50.3 | 16.9 |
| HomoGE | 49.7 | 51.5 | 49.9 | 51.8 | 44.3 | 50.1 | 51.2 | 17.7 |
| HetGE | 44.6 | 44.8 | 44.8 | 47.1 | 37.6 | 45.5 | 44.6 | 15.0 |
| α = 5 × 10−3 | ||||||||
| HomoG | 16.3 | 19.4 | 16.3 | 19.7 | 13.6 | 19.7 | 18.2 | 2.7 |
| HomoGE | 19.1 | 21.6 | 19.7 | 21.3 | 15.3 | 19.5 | 20.2 | 4.2 |
| HetGE | 14.6 | 15.4 | 14.6 | 16.1 | 11.3 | 15.2 | 15.5 | 2.0 |
Power of detecting a single causal SNP which accounts for 0.6% of the variance.
The power of the FGLS, OLS, GWAF and GEE methods were calculated after the inflated type I error was corrected.
The MCTFR GWAS
The genome-wide scan of the MCTFR height data of 4,711 individuals in 1,817 2-generation pedigrees was carried out using our proposed rapid methods, RFGLS-UN/VC, as well as FGLS, OLS, GWAF and GEE. European Americans made up about 94% of the MCTFR height data. The rest were mostly Asian Americans (3% of the whole sample). To account for ethnic stratification, we used EIGENSOFT [35] and computed the principal components using the genotype data of the founders of the pedigrees. The top ten principal components were used as covariates in the methods implemented. Height was also adjusted for sex and generation.
There were 3 types of pedigrees in the data analyzed, which include MZ twin families, non-twin biological-offspring families, and adopted-offspring families. Individuals in the adopted-offspring families (342 out of 4,711) were treated as independent observations in RFGLS- UN and FGLS. Pedigree kinship coefficients for the 3 types of families were used in RFGLS-VC and GWAF. An exchangeable correlation structure was estimated in GEE.
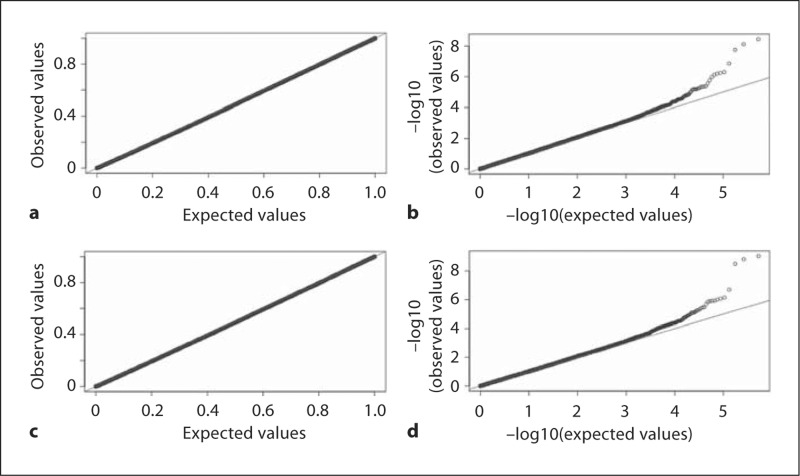
Table 7 presents the genomic inflation factors λ. The λ from the OLS method at 2.94 is much larger than 1. The λs from the other 5 methods correcting for family structures are close to 1. The OLS result is therefore excluded in the following discussions. Figure 5 summarizes the genome scan results. In general, all 5 methods give similar results showing the most significant association with trait at the 3 closely located SNPs rs17034592, rs972583, and rs10516510 on chromosome 4. Two lower peaks on chromosome 3 and chromosome 2 also appear at the same place in the 5 outputs. The Q-Q plots (fig. 6a, c) show that the p values of the 2 proposed methods are distributed uniformly between 0 and 1. The Q-Q plots on a log-scale (fig. 6b, d) reveal outliers in the tail of the distribution. Among all 22 p values ≤10−5, 17 correspond to the 3 peaks on chromosome 4, 3, and 2, and map within 200 kb of each other. Table 8 lists all associations reaching the Bonferroni corrected significance level (nominal p value = 1.9 × 10−6, using an overall α = 0.05) in the 5 methods. RFGLS-VC reported 11 hits, RFGLS-UN reported 10 hits, FGLS and GWAF reported 9 hits, and GEE reported 6 hits. All 5 methods reported the same 3 closely located SNPs on chromosome 4 exceeding the genome-wide significance level at 5 × 10−8, which is more stringent than the Bonferroni corrected one. The genome scan of the MCTFR height data provides additional support that our proposed methods behave correctly.
Table 7.
Genome scan of the MCTFR height data: genomie control parameters X
| RFGLS-UN | RFGLS-VC | FGLS | OLS | GWAF | GEE | |
|---|---|---|---|---|---|---|
| 1.035 | 1.034 | 1.044 | 2.94 | 1.038 | 1.034 | |
Fig. 5.
Manhattan plot of the genome scans for the MCTFR height phenotype using the 5 methods. The x-axes represent the chromosome number. The y-axes represent the −log10 of p values.
Fig. 6.
Q-Q plots and log Q-log Q plots. a, b RFGLS-UN. c, d RFGLS-VC.
Table 8.
Genome scan of the MCTFR height data: SNPs reaching the Bonferroni corrected threshold
| Gene | Chromosome | SNP | Position, bp | p valuesa (×10−6) |
||||
|---|---|---|---|---|---|---|---|---|
| RFGLS-UN | RFGLS-VC | FGLS | GWAF | GEE | ||||
| NCOA1 | 2 | rsl7734306 | 24'575'942 | 0.4972 | 0.7124 | 0.6817 | 1.312 | 1.2393 |
| NCOA1 | 2 | rs2044148 | 24'591'337 | 0.5656 | 1.1625 | 0.7797 | 2.3934 | 1.6555 |
| NCOA1 | 2 | rs6720514 | 24'680'123 | 0.6306 | 0.8414 | 0.8785 | 1.6203 | 1.3927 |
| NCOA1 | 2 | rs2119115 | 24'740'285 | 1.0572 | 1.4161 | 1.4923 | (2.6626) | (2.1101) |
| ZBTB38 | 3 | rs13095453 | 142'557'652 | 1.7148 | 1.2829 | (2.0034) | 0.5056 | (92.0618) |
| ZBTB38 | 3 | rs6763931 | 142'585'523 | 0.7525 | 1.2135 | 0.8521 | 0.5958 | (32.7139) |
| ZBTB38 | 3 | rs6785073 | 142'622'020 | 0.1372 | 0.1982 | 0.1515 | 0.0904 | (4.0754) |
| – | 4 | rs7682418 | 105'647'866 | (6.2730) | 0.9890 | (7.6710) | (2.0177) | (2.2290) |
| – | 4 | rsl0516510 | 105'656'811 | 0.0175 | 0.0031 | 0.0168 | 0.0115 | 0.0003 |
| – | 4 | rs972583 | 105'661'751 | 0.0075 | 0.0015 | 0.0116 | 0.0055 | 0.0002 |
| – | 4 | rs17034592 | 105'674'409 | 0.0035 | 0.0009 | 0.0070 | 0.0025 | 0.0002 |
The Bonferroni corrected threshold is 1.9 × 10−6. p values in brackets are those not reaching the Bonferroni corrected significance level using the method presented, but reaching the threshold using any other method on the list.
Computation Time
Table 9 shows the relative computation time of all implemented methods. We set the computation time for the OLS method to be 1. It takes around 8 min to run an analysis of 7, 200 observations and 10,000 SNPs using a single core on a Dell M605 compute node which has 2 AMD Opteron 431 six-core processors (2.4 GHz with 6 × 512 KB cache), 16 GB RAM and one 146 GB 10,000 RPM SAS drive. Our proposed RFGLS approach was the fastest among all other methods. It is about 50 times faster than the FGLS method, and 10 times faster than the GWAF method. It will take about 20 days for the QTDT or GWAF method to scan the whole genome on a single core, and 2 days for the RFGLS method.
Table 9.
Relative computation time of the different methods completing the 10,000 single SNP analysis
| Relative time | |||||
|---|---|---|---|---|---|
| RFGLS-UN/VC | FGLS | OLS | GWAF | GEE | QTDT |
| 1.8 | 100 | 1 | 19.1 | 3.8 | 11.5 |
The computation time for the OLS method is set to be 1.
Discussion
We proposed 2 rapid methods (RFGLS-UN and RFGLS-VC) for a genome-wide association analysis of QTL in family-based designs. These methods take advantage of the fact that the genetic variance associated with any particular marker in a GWAS usually accounts for a small proportion of the total variance [36]. Consequently, the rapid methods involve first estimating the variance-covariance matrix for the family data ignoring the effects of individual markers, and then carrying out single SNP analyses using this estimated matrix. This proposed rapid method is a simplification of the FGLS estimator, where the variance and the fixed effects are jointly estimated for each marker. In the RFGLS-VC method, family covariance matrices are modeled as a function of 3 variance components: an additive genetic effect (estimated assuming known genetic relationships among family members), a family environmental effect (estimated assuming compound symmetric structure), and a residual effect (estimated assuming common variance across family members). In the RFGLS-UN method, family variance-covariance matrices are estimated assuming no structure and phenotypic variances are allowed to vary. Our simulations showed that the rapid methods resulted in appropriate type I error rates and power that approximated that for the gold standard estimator where the real familial similarities were known (the GLS method).
In the study of a large sample size of 1,800 nuclear families, when we simulated family data in which familial resemblance owed to additive genetic effects only, we found that RFGLS-UN and RFGLS-VC performed similarly to the GWAF method, which assumes that familial resemblance is due to additive genetic factors only, and the FGLS method, in which the residual variance-covariance matrix is estimated for each individual marker. When we simulated family data in which familial resemblance owed to both additive genetic and environmental factors, both RFGLS-UN and RFGLS-VC showed power that was greater than the one in GWAF and comparable to the one in FGLS. Finally, when we simulated family data in which there were both additive genetic and familial environmental effects as well as heterogeneity of phenotypic variance, the RFGLS-UN method had power that exceeded that for the other methods and approximated that for the GLS gold standard. The GEE method that gave robust estimation of the effect size suffered a minor loss of power in all simulated cases. The QTDT showed low power in all simulated cases, no doubt a reflection of the family clusters we considered, in which fully two-thirds of the families (the MZ twin and adoptive families) were not informative for the within-family QTDT test. In the study of a small sample size of 180 nuclear families, we found that RFGLS-UN was slightly underpowered compared to the GLS gold standard due to the many parameters required to be estimated. The performance of RFGLS-VC continued to approximate that of the GLS gold standard. Both RFGLS-UN and RFGLS-VC maintained the correct type I error rate, while FGLS showed inflated type I error rate in this small sample size.
The rapid methods were computationally much more efficient than all alternatives other than OLS, whose speed is achieved by making the invalid assumption of no familial correlation. This dramatic reduction in the computation time for our proposed methods means that these methods could be used to carry out permutation tests, find the null distribution of the F test implemented in this study or any other test to be developed, and get the empirical p values. This can be especially valuable when dealing with no normality in data. Our rapid method also allows to extensively expand the genotyped marker set by imputation, usually by employing HapMap data as a reference. Our method can also be used as the first stage in a 2-stage design where the first stage genome-wide scan is used to select noteworthy SNPs, and a second stage analysis can then be carried out, e.g. with FGLS, so that computation time is a lesser concern.
In general, large genetic association studies are primarily interested in the detection of SNPs associated with a trait, and our approaches have provided a rapid and powerful way to detect an association. In the proposed rapid methods, along with those available such as GRAMMAR, there is bias in the estimation of a genetic effect and it increases with increased effect size. However, the rapid methods are powerful in detecting an association.
Our proposed population-based association analysis was shown to be much more powerful than the family-based association analysis QTDT when ethnic stratification is not present. If there is ethnic stratification, we can use EIGENSOFT and compute the principal components using the founders of the pedigree [35], then select the top principal components and use them as covariates in our RFGLS approach. This was efficiently demonstrated in our analysis of the MCTFR height data. We can also estimate a genomic kinship matrix, instead of the pedigree kin- ship matrix used in this study, to be used in the RFGLS- VC method. The use of a genomic kinship matrix has been advocated by Aulchenko et al. [15] to get a better estimation of the ‘true’ covariance between individual genomes. The impact of estimating the kinship matrix on the power of the RFGLS-VC method is a topic for future study.
In our study, we compared our proposed approaches with others using simulated nuclear family data. It is worth noting that our methods can also be applied to extended families. In our RFGLS-UN method, within-generation or within-household constrains can be put in the estimation of the family variance-covariance matrix. In our RFGLS-VC method, the relationship matrix of an extended family instead of a nuclear family can be easily applied and an environmental correlation of the extended family can be estimated. The performance of our proposed method applied to extended families is a topic for further study.
We used an F test in our proposed method. GWAF and GRAMMAR use a score test. Different tests may have different effects on the p values, although they are asymptotically equivalent at a large sample size. Future study should include the likelihood ratio test and the score test by Chen and Abecasis [13] to measure the depth of the effect. We did not compare the performance of the GRAMMAR approach in our study since the package does not support sparse matrix computation, and is too computationally demanding for the sample size we used. However, we did find that in methods where environmental similarity was not modeled explicitly (e.g. GWAF) this resulted in loss of power when such similarity existed in the data.
To summarize, we developed a fast and powerful approach for family-based GWAS of quantitative traits. Currently available approaches like GWAF and GRAMMAR estimate familial similarities through kinship relationships. Because environmental factors are likely to contribute to familial resemblance for many phenotypes, and especially for behavioral phenotypes, we combined kinship relationships and ‘environmental relatedness’ in our estimation of the familial correlation structures in 2 rapid methods, RFGLS-UN and RFGLS-VC, and achieved comparable power to those from the GLS analysis when the real variance-covariance matrix is known. Among all the alternatives implemented that did not show inflated type I error, our methods were computationally most efficient. Further study will aim at extending our rapid approach to categorical trait analyses using a GEE method.
Acknowledgements
We thank Scott Vrieze and the reviewers for their helpful comments on an earlier version of this paper. Research was supported in part by NIH grant U01 DA024417.
References
- 1.Cupples LA, Arruda H, Benjamin E, D'Agostino R, et al. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8(suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benyamin B, Visscher PM, McRae AF. Family-based genome-wide association studies. Pharmacogenomics. 2009;10:181–190. doi: 10.2217/14622416.10.2.181. [DOI] [PubMed] [Google Scholar]
- 5.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scuteri A, Sanna S, Chen WM, Uda M, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:1200–1210. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham R, Cotsapas C, Davies L, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poduslo SE, Huang R, Huang J, Smith S. Genome screen of late-onset Alzheimer's extended pedigrees identifies TRPC4AP by haplotype analysis. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:50–55. doi: 10.1002/ajmg.b.30767. [DOI] [PubMed] [Google Scholar]
- 9.Hicks B, Schalet B, Malone S, Iacono W, McGue M: Psychometric and genetic architecture of substance use disorder and behavioral disinhibition measures for genetic association studies. In press. [DOI] [PMC free article] [PubMed]
- 10.Ewens WJ, Li M, Spielman RS. A review of family-based tests for linkage disequilibrium between a quantitative trait and a genetic marker. PLoS Genet. 2008;4:e1000180. doi: 10.1371/journal.pgen.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopper JL, Mathews JD. Extensions to multivariate normal models for pedigree analysis. II. Modeling the effect of shared environment in the analysis of variation in blood lead levels. Am J Epidemiol. 1983;117:344–355. doi: 10.1093/oxfordjournals.aje.a113547. [DOI] [PubMed] [Google Scholar]
- 12.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen WM, Abecasis GR. Family-based association tests for genome-wide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aulchenko YS, Koning DJD, Haley C. Genome-wide rapid association using mixed model and regression: a fast and simple method for genome-wide pedigree-based quantitative trait loci association analysis. Nat Genet. 2007;177:577–585. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abecasis GR, Cardon L, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laird N, Horvath S, Xu X. Implementing a unified approach to family based tests of association. Genet Epidemiol. 2000;19(suppl 1):S3–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 18.Fulker D, Cherny S, Sham P, Hewitt J. Combined linkage and association analysis for quantitative traits. Am J Hum Genet. 2003;64:259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardon L, Palmer L. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 20.Havill LM, Dyer TD, Richardson DK, Mahaney MC, Blangero J. The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification. BMC Genet. 2005;6(suppl 1):S91. doi: 10.1186/1471-2156-6-S1-S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terracciano A, Sanna S, Uda M, Deiana B, Usala G, et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. 2010;15:647–656. doi: 10.1038/mp.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bravo HC, Lee KE, Klein BEK, Klein R, Iyengar SK, Wahba G. Examining the relative influence of familial, genetic, and environmental covariate information in flexible risk models. Proc Natl Acad Sci USA. 2009;106:8128–8133. doi: 10.1073/pnas.0902906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baltagi BH. Econometrics, ed 4. Berlin: Springer; 2008. pp. 221–226. [Google Scholar]
- 24.Lindstrom MJ, Bates DM. Newton-Raphson and EM algorithms for linear mixed-effects models for repeated-measures data. J Am Stat Assoc. 1988;83:1014–1022. [Google Scholar]
- 25.Puntanen S, Styan G. The equality of the ordinary least squares estimator and the best linear unbiased estimator. The American Statistician. 1989;43:153–164. [Google Scholar]
- 26.Grubb D, Magee L. A variance comparison of OLS and feasible GLS estimators. Econometric Theory. 1988;4:329–335. [Google Scholar]
- 27.Single RM, Finch SJ. Gain in efficiency from using generalized least squares in the haseman-elston test. Genet Epidemiol. 1995;12:889–894. doi: 10.1002/gepi.1370120660. [DOI] [PubMed] [Google Scholar]
- 28.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 29.Hjsgaard S, Halekoh U, Yan J. The R package geepack for generalized estimating equations. J Stat Soft. 2005;15:1–11. [Google Scholar]
- 30.Yan J, Fine JP. Estimating equations for association structures. Stat Med. 2004;23:859–880. doi: 10.1002/sim.1650. [DOI] [PubMed] [Google Scholar]
- 31.Yan J. geepack: Yet another package for generalized estimating equations. R-News. 2002;2–3:12–14. [Google Scholar]
- 32.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 33.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malzahn D, Schillert A, Müller M, Bickeböller H. The longitudinal nonparametric test as a new tool to explore gene-gene and gene-time effects in cohorts. Genet Epidemiol. 2010;34:469–478. doi: 10.1002/gepi.20500. [DOI] [PubMed] [Google Scholar]
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Manolio TA, Collins FS, Cox NJ, Goldstein DB, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]