Abstract
The neuronal transcription splicing factor, A2BP1, has been implicated in a variety of neurodevelopmental disorders; however, the role of A2BP1 in brain development and gene regulatory function remains to be explicated. Here, we map A2bp1 gene expression, focusing on the developing forebrain of the C57BL6J mouse. Early in forebrain development, A2bp1 expression is highly reminiscent of the expression of genes marking postmitotic GABAergic cells emanating from the ventral telencephalon during migration to the dorsal pallium. Ventral pallial expression remains low after the migratory period. Broader dorsal pallial expression becomes more evident late prenatally and early postnatally. This is paralleled by dense, restricted expression in the ventrobasal dorsal thalamic complex and mid-hypothalamic region. Outside of the forebrain, there is significant expression in motor pathways. These data indicate that A2BP1 mutations may clinically affect very selective forebrain neuron types from early periods of development.
Key Words: C57BL6J mouse, Embryonic forebrain, Neurodevelopment, mRNA splicing regulation, Gene expression, GABA, Interneurons, Thalamus, Neocortex
Introduction
The regulation of gene splicing in neurons (reviewed in Li et al. [1]), particularly during development, appears to be an important mechanism for controlling expression of genes key to histogenic processes. Splicing results in multiple protein products arising from a single gene locus, based on specific exon splicing. Exon usage in the nervous system varies by cell type, developmental stage, and can be regulated by activity [1]. The full dynamic range of alternative splicing mechanisms of genes expressed in neurons is unclear, but there are significant pathophysiological consequences of a disruption in the ability to splice [2].
Ataxin-2-binding protein 1 (A2BP1) is a neuronal splicing regulator necessary for proper exon usage in several mammalian genes with tissue-specific splice variants, such as exon EIIIB inclusion in fibronectin [3,4], calcitonin/calcitonin gene-related peptide [5], CaV1.2 voltage-gated calcium channels [6] and NMDA receptor 1 [7]. Gene disruption of A2BP1 has been associated with a variety of neurodevelopmentally based brain disorders, including autism [8,9,10], mental retardation with epilepsy [11], attention deficit hyperactivity disorder [12] and schizophrenia [13,14].
Mammalian A2bp1 was first identified by Shibata et al. [15] in a yeast two-hybrid screen for interacting partners of ataxin-2, which is disrupted in spinocerebellar ataxia type 2. Three isoforms of A2BP1 were identified in postmortem adult human brain by Northern blot [15]. A 4.4-kb isoform was most predominantly expressed in the cerebellum, while 2 less abundant isoforms (3.4 and 6.2 kb) were expressed in the cerebral cortex and putamen [15]. In mice, this highly conserved RNA-binding protein also shows expression in the brain by Northern blot analysis and immunocytochemistry [16]. As in the human central nervous system, in the mouse brain, there are 3 mRNA isoforms by Northern blot analysis (6.2, 4.0 and 3.4 kb).
A2BP1 has been identified in adult mouse brain by immunohistochemistry. Abundant protein expression in neuronal nuclei exists throughout the adult brain including the hippocampus, cortex, cerebellum and olfactory bulb [4]. There are no detailed in situ hybridization studies or database entries to date of developmental expression of A2bp1, which is somewhat surprising given the functional importance of splicing regulation during brain development, and implications of A2BP1 involvement in neurodevelopmental disorders. In this report, we describe the developmental expression of a transcript from the A2bp1 locus in C57BL6J mouse, characterizing mRNA localization in the forebrain from early stages of neuron production (E11.5) through eye opening (P14).
Materials and Methods
Mouse Care and Use
Timed pregnant C57BL6J mice were bred in-house under protocols approved by the Institutional Animal Care and Use Committee of Vanderbilt University. Mice were maintained on a 12-hour light/12-hour dark cycle and were permitted food and water ad libitum. Noon on the day following a time-delimited overnight pairing was considered E0.5. Pregnant females were deeply anesthetized with isofluorane vapors followed by rapid decapitation in order to harvest fetuses for in situ hybridization. Crown-rump length and external physical features were used to verify fetal ages.
Riboprobe Labeling
An A2bp1 I.M.A.G.E clone (6821627) was obtained from ATCC (Manassas, Va., USA). Informatics analysis indicates that the synthesized probe binds to the large exon in the 3′UTR which is present in two transcript variants. These transcripts encode an A2BP1 protein that terminates in the amino acids FAPY. BLASTing mouse refseq and non-refseq RNA databases with the generated probe did not yield significant homology to other mouse sequences. A transcript from the A2bp1 locus has been mapped in C57BL/6 mouse at one age (E14.5) in the sagittal plane on www.genepaint.org. The probe in those studies only recognizes transcript variant 2. The identity of the I.M.A.G.E. clone was confirmed by sequencing at the Vanderbilt DNA Sequencing Facility. The clone was linearized with Sal I and transcribed using T3 polymerase (Promega) in a 20-μl reaction. Digoxigenin-11-uridine-5′-triphosphate (350 μM; Roche, Indianapolis, Ind., USA) was included in the transcription reaction to allow for non-radioactive colorimetric detection of transcripts. Hydrolyzed and non-hydrolyzed probes yielded the same results. Based on gel electrophoresis of the probe, the overwhelming majority of hydrolyzed products were between 650 and 1,650 nucleotides in length. The images in this report are all from non-hydrolyzed probes.
In situ Hybridization
Fetuses were harvested into cold PBS and whole heads or microdissected brains were immersion fixed for 24 h in 4% formaldehyde made from 4% paraformaldehyde by weight in alkaline water heated to 60°C in 0.156 M NaH2PO4, 0.107 M NaOH, pH 7.12 with HCl. After fixation, brains were cryoprotected in graded 10, 20 and 30% sucrose in PBS followed by embedding in TFM tissue freezing medium (Triangle Biomedical Sciences, Inc., Durham, N.C., USA) over liquid nitrogen vapors. Brains were stored at −80°C until sectioning into 6 series at 20 μM in a cryostat. Cut sections were stored at −80°C until they were fixed, acetylated and dehydrated. After this step, they were returned to −80°C until processed for in situ hybridization. The in situ hybridization was performed on a Tecan Evo 150 (Tecan Group Ltd., Männedorf, Switzerland) following the Allen Brain Atlas (http://www.brain-map.org/) and Genepaint (http://www.genepaint.org/) protocols. After completion of the Tecan-run protocol, BCIP and NBT (Roche) were applied manually. The time in color development was 40 min for embryonic tissue and 3 h for P14. After color development, the slides were rinsed 4 times with double distilled water and then twice with 4% formaldehyde, dehydrated through a series of alcohols and coverslipped with VectaMount (Vector Laboratories, Burlingame, Calif., USA). Two cases per age were tested. A sense control probe generated from the same I.M.A.G.E. clone was tested at E14.5 and P14.
Light Microscopy
Images of the rostrocaudal extent of the prenatal material were obtained on an Ariol SL-50 automated imaging platform at ×10 in the Epithelial Biology Center Imaging Resource Core of Vanderbilt University. Images of the rostrocaudal extent of the P14 material were captured on an Olympus SZX12 microscope. Additional images illustrating higher magnification or comparison across multiple ages were captured on a Zeiss Axioplan with a high-resolution color camera. Any images appearing in the same figure together were captured with a single microscope in the same image capture session, and resulting images underwent the same manipulations for brightness and contrast. Anatomical references were made using an embryonic mouse brain atlas [17].
Western Blotting
Protein was harvested from 5 different ages of wild-type mice on a C57BL6J background. The dorsal telencephalon/cortex was dissected and nuclear and cytoplasmic fractions were prepared. Twenty micrograms of protein was loaded on a Nupage 4–12% Bis/Tris gel (Invitrogen, Carlsbad, Calif., USA) run for 1.5 h at 140 V and transferred to nitrocellulose at 15 V for 40 min. The blot was incubated in 1:100 anti-Fox-1 (sc-135476; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif., USA) in 0.5% casein, 0.05% Tween-20 in PBS. After washes in PBS, the blot was incubated in goat anti-rabbit IgG-HRP (GE Healthcare, Piscataway, N.J., USA) at a final concentration of 1:20,000 and was detected with Super Signal Dura ECL (Pierce/Thermo-Fisher Scientific, Inc., Rockford, Ill., USA).
Results
In situ Hybridization Mapping of Mouse A2bp1
We have characterized A2bp1 expression in prenatal and P14 mouse brain using a probe that recognizes two characterized mRNA transcripts of the A2bp1 locus (fig. 1). At all ages examined prenatally, A2bp1 mRNA detection was evident only in regions containing postmitotic cells. Thus, proliferative zones lining the ventricles throughout the forebrain were unlabeled. Labeling intensity of individual cells was similar at each age. Early pools of postmitotic cells were darkly labeled. Later pools of postmitotic cells (perhaps glia) were unlabeled. This gave the appearance of a lighter label at P14. A sense probe tested at E14.5 and P14 demonstrated the specificity of the anti-sense label (data not shown).
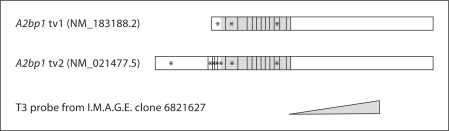
Fig. 1.
Schematic of known mouse A2bp1 transcript variants (tv) and the location of ISH probe used in this study. The I.M.A.G.E. clone is more similar to transcript variant 2, but identical to both transcript variants 1 and 2 in the large 3′ exon, which is the region of the clone that includes the probe that was generated in this study. Although there are other known A2bp1 splice variants, when BLASTing for mRNA hits in NCBI databases with the probe, these are the only two transcripts retrieved. Both of these transcripts encode an A2bp1 isoform which ends in amino acids FAPY. Vertical lines indicate exon boundaries. Asterisks denote transcript-specific exon usage. The shaded region represents the coding region.
A2bp1 mRNA was detected by in situ hybridization in a small number of cells at E11.5, the earliest time point that we assessed (fig. 2). In particular, the pattern of expression resembled the distribution of postmitotic cells at the cortical plate-mantle region at the early corticostriatal boundary. Labeled cells are evident in the geniculate pretectum and hypothalamus. At this age, there were no labeled cells in the dorsal pallium. Outside of the forebrain, caudally, there was A2bp1 signal in the medullary ventral motor column as well as the ventral spinal cord.
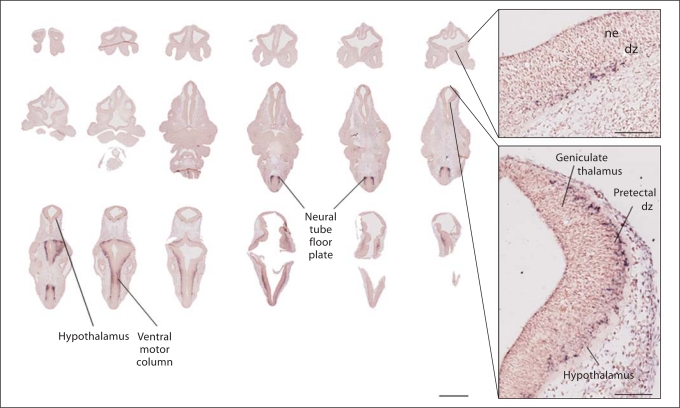
Fig. 2.
Serial sections through E11.5 embryo demonstrates caudal to rostral emergence of forebrain A2bp1 expression. Labeling is light in the forebrain at this age, and more abundant in more caudal sections. Labeling is evident in pallidal and striatal differentiating zones, usually marked by tyrosine hydroxylase, calbindin and calretinin-positive cells. Labeling is very dense in the floor plate of the neural tube. ne = Neuroepithelium; dz = differentiating zone. Scale bar for whole slices = 1 mm, inset scale bar = 100 μm.
By E12.5, the expression of A2bp1 was much more robust (fig. 3) and tracked closely with the known emergence of interneurons arising from the lateral ganglionic eminence (LGE). Transcript-expressing cells were not seen emerging from the medial ganglionic eminence, which gives rise to the majority of parvalbumin cortical interneurons, or within the developing striatum, which contains long-projecting GABAergic neurons. Rather, at this early age, more ventral and posterior areas contained A2bp1-expressing cells, including the preoptic differentiating zone, the anterobasal nucleus and the lateral hypothalamus. Outside of the forebrain, in the rhombencephalon, there was labeling in the region generating the facial motor nucleus, the ventral pontine differentiating zone and the medullary differentiating zone, the principal sensory trigeminal nucleus and the pontine reticular formation. Outside of the central nervous system, there was emerging labeling in the trigeminal ganglion V and vestibulocochlear ganglion VIII (not depicted). Although dorsal pallial neurons are being produced by this age, and there is a transition from preplate to cortical plate, this dorsal forebrain region remains A2bp1 negative.
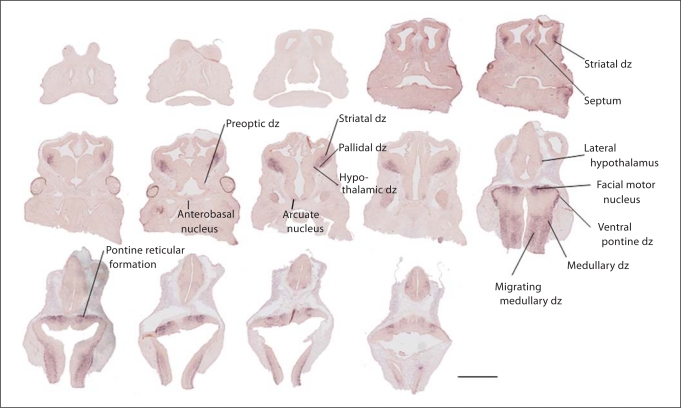
Fig. 3.
At age E12.5, there is a significant increase in A2bp1-positive cells. The expression is restricted to regions that are enriched for postmitotic cells and there is no labeling in neuroepithelial zones. Robust expression is evident in the differentiating zones of the striatum, pallidum and hypothalamus, including the preoptic dz, anterobasal nucleus, arcuate nucleus and lateral hypothalamus. The presumptive pons and medulla also show robust labeling. There is no evidence of A2bp1 expression in the neocortical neuroepithelium at this age. Scale bar = 1 mm. dz = Differentiating zone.
By E13.5 (fig. 4), there continued to be robust A2bp1 expression in cells differentiating in the same areas evident at earlier ages, including those that have migrated from the LGE, hypothalamic and brain stem nuclei. There also was the first appearance of A2bp1-positive cells in the most lateral regions of the dorsal pallium. This timing is coincident with the major increase in neurons populating the developing cortical plate. At this age, there was an abundance of A2bp1 cells residing in the lateral migratory stream from the ganglionic eminence, coincident with the increase in cortical interneuron migration [18]. Labeled neurons in the lateral septum, bed nucleus of the stria terminalis, amygdala and the preoptic and ventromedial hypothalamus also emerged at this age. Labeling was also seen in the dorsal thalamus.
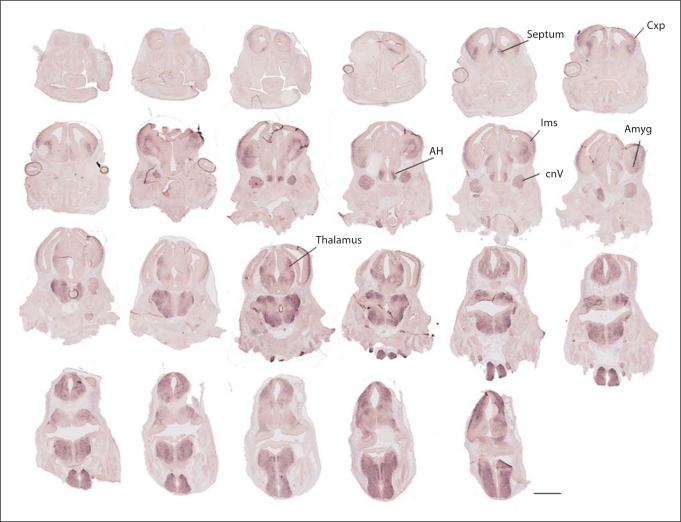
Fig. 4.
Forebrain progression of A2bp1 label at E13.5 resembles expression seen at E12.5 with the addition of labeling in the cortical plate. The A2bp1 expression in the neocortex presumptively includes both locally generated cells, as well as interneurons from the LGE which are tangentially migrating in the intermediate zone. The majority of the labeling in the cortex at this age is in the emerging cortical plate. CxP = Cortical plate; lms = lateral migratory stream; AH = anterior hypothalamus; cnV = cranial nerve V; Amyg = amygdala. Scale bar = 1 mm.
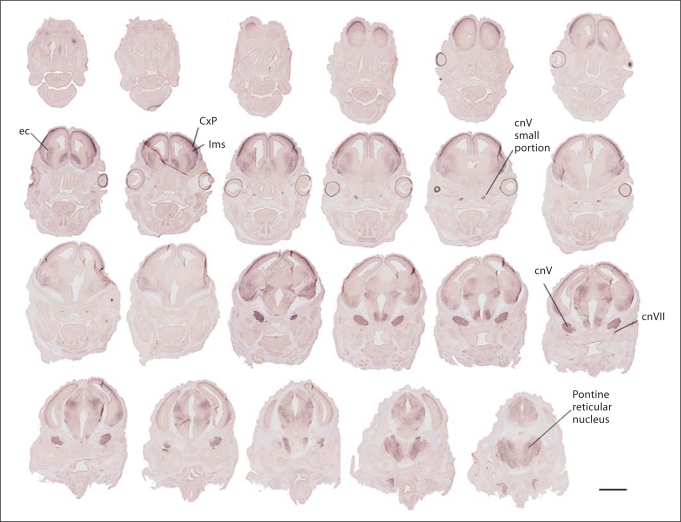
By E14.5 (fig. 5), all lateromedial aspects of the dorsal pallium contained neurons expressing A2bp1 in the cortical plate. Transcript-expressing cells were evident in the striatum distal to the ganglionic eminence. Labeled cells also accumulated in lateral regions of the basal forebrain that include the nascent amygdala. At E15.5–E18.5 (see online supplementary figures 1–4, www. karger.com/doi/10.1159/000323732), expression continued to be excluded from progenitor zones, while labeled cells were abundant in several postmitotic differentiated telencephalic, diencephalic, mesencephalic and rhombencephalic areas.
Fig. 5.
Forebrain progression of A2bp1 label from E14.5 demonstrates the rapid rise in A2bp1 expression in the cortex and persistent expression in postmitotic cells of the basal forebrain. Medial to the external capsule (ec), ample expression is evident in the striatum, while lateral to the ec, there is expression in the lateral migratory stream (lms), which is relatively light compared to the darker labeling in the cortical plate (CxP). Labeling is still apparent in cranial nerves (cnV and cnVII). Scale bar = 1 mm.
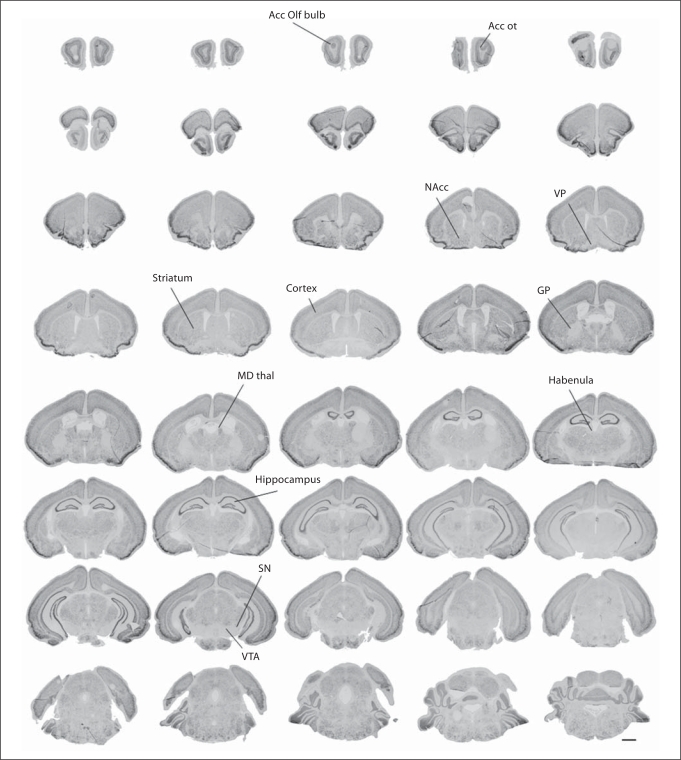
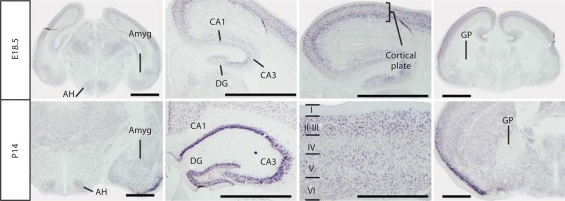
By the end of the second postnatal week, the expression of A2bp1 became easier to detect in discrete areas (fig. 6). There were many brain areas that retained high levels of expression including the olfactory bulb, nucleus accumbens, thalamus, cortex, hippocampus and striatum to indicate a few. At this age, there were several brain areas that emerged with no signal for this transcript including the ventral pallidum, globus pallidus, habenula, and the ventral tegmentum, which includes the ventral tegmental area and the substantia nigra. At low magnification, A2bp1 expression appeared to be lighter at P14 than at the late prenatal ages (online suppl. figures 1–4). This age difference was likely related to the postnatal emergence of non-neuronal cells that remain unlabeled and does not indicate a decrease of expression in postnatal development. Indeed, at higher magnification (fig. 7), the distribution and intensity of expression across multiple brain regions looked very similar between P14 and E18.5. At both ages, there was expression in a wide variety of brain regions, including the anterior hypothalamus, the amygdala, the hippocampus and the cortical plate/neocortex. The two ages also shared a noticeable lack of labeling in some brain regions such as the globus pallidus and ventral pallidum. The main difference between these two ages was that there were more cells at P14 that do not contain A2bp1. These were potentially glia as they develop later and specifically do not express A2bp1.
Fig. 6.
Postnatal expression of A2bp1 at P14. As the brain matures, A2bp1 signal becomes easier to detect in discrete areas. There is ample signal in a wide variety of areas, including the accessory olfactory bulb (Acc Olf bulb) and the accessory olfactory tract (Acc ot), nucleus accumbens (NAcc), mediodorsal thalamus (MD thal), cerebral cortex, hippocampus, and striatum. However, expression is noticeably absent in the ventral pallidum (VP), globus pallidus (GP), habenula, ventral tegmental area (VTA) and substantia nigra (SN). Scale bar = 1 mm.
Fig. 7.
Comparison of expression at E18.5 and P14. The amygdala (Amyg) and anterior hypothalamus (AH) are labeled at both E18.5 and P14. Within the hippocampus, the major subdivisions [CA1, CA3, dentate gyrus (DG)] show similar patterns of expression at embryonic and postnatal ages. Within the cortical plate at E18.5 and the more mature laminated cortex at P14, there is dense expression of A2bp1. The globus pallidus (GP) is one of only a few landmark brain areas with no evident expression of the A2bp1 probe target at any age examined. Scale bar = 1 mm.
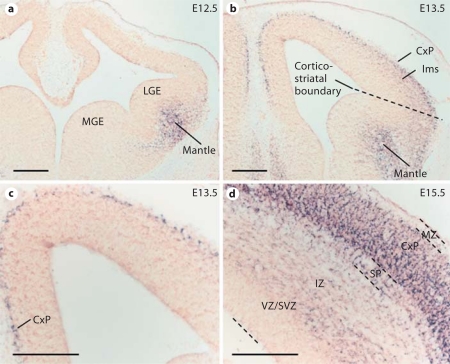
At all of the examined ages, the A2bp1 expression was excluded from proliferative zones and was highly restricted to regions that are known to posses differentiated cells (fig. 8). We observed no expression in the ventricular zone of the dorsal pallium in the medial or LGE. A2bp1-positive cells were not yet evident in the cortical plate at E12.5 (fig. 8a), but were easily detected in the cortical plate at E13.5 (fig. 8b, c). At E13.5, A2bp1-positive cells were also evident in the lateral migratory stream. Labeling at E15.5 (fig. 8d) was highly consistent with labeling in postmitotic cells. There was no labeling detected in the proliferating cells of the ventricular zone, and modest labeling in the subventricular zone. The majority of the label at this age was in the cortical plate. Based on this labeling pattern at these ages, it is possible that A2bp1 marks populations of postmitotic tangentially migrating interneurons as well as radially migrating projection neurons.
Fig. 8.
A2bp1 is exclusively expressed in nonproliferating postmitotic zones. a A2bp1 is not expressed in the ganglionic eminence [medial (MGE), lateral (LGE)], but rather in the mantle at E12.5. b A2bp1-positive cells are evident in the mantle, the corticostriatal boundary, the lateral migratory stream (lms) and the cortical plate (CxP) at E13.5, which suggests that A2bp1 can be found in a variety of postmitotic cell types including striatal cells, tangentially migrating interneurons as well as cortical projection neurons. c A higher magnification view of the cortex at E13.5 shows A2bp1-positive cells along the medial surface of the cortex, at an age slightly before interneurons reach this area. d The presumptive projection neurons as well as some tangentially migrating interneurons in the dorsal pallium are darkly labeled as shown here at E15.5. The ventricular zone (VZ) contains undifferentiated A2bp1-negative cells, the subventricular zone (SVZ) contains A2bp1-positive cells that could be radially or tangentially migrating cells, the intermediate zone (IZ) contains some A2bp1-positive cells suggesting migrating interneurons and the subplate (SP) and cortical plate (CxP) contain A2bp1-positive cells, with the CxP containing the densest population of A2bp1-positive cells. The marginal zone (MZ) contains some A2bp1-positive cells (out of view). Scale bar = 200 μm.
At several pre- and postnatal ages, A2BP1 protein in the dorsal telencephalon/neocortex was detectable by Western blotting (fig. 9). A distinct band at approximately 43 kDa was apparent at all ages examined and it was evident in both nuclear and cytoplasmic fractions.
Fig. 9.
a Western blot with selective antibody against A2bp1 detecting the presence of a band at approximately 43 kDa at prenatal and postnatal ages in both the cytoplasm and the nucleus. b Ponceau S stain of the nitrocellulose membrane prior to Western blotting showing relatively equal loading of protein across samples.
Discussion
In this report, we describe the prenatal and early postnatal expression profile of A2bp1. The data demonstrate that the gene, which serves as a transcriptional splicing factor, is expressed in a developmentally restricted pattern along the neuroaxis that excludes mitotically active cells. A2BP1 was also evident in the forebrain as detected by Western blotting, demonstrating that the mRNA labeled in this study is likely of functional relevance. The clinical implications of mutations in A2bp1 increasing risk for neurodevelopmental disorders including autism, attention deficit hyperactivity disorder, epilepsy and schizophrenia are consistent with the expression patterns in specific regions of the basal forebrain, hippocampus and neocortex. While the expression of A2bp1 became more evident in dorsal pallial regions later in fetal development, the early expression of A2bp1 is consistent with a role in both postmitotic projection neuron and interneuron development.
The mapping of A2bp1 in the fetal brain establishes an important dataset for deciphering the potential regulation of downstream target genes. Remarkably, there have been a number of recent analyses addressing potential target genes of A2bp1 regulation with little knowledge of spatiotemporal expression patterns during brain development. For example, Zhang et al. [19] used a bioinformatic approach to identify all of the potential targets of A2BP1 that may contain the conserved splicing motif UGCAUG. Of these genes, several known genes in interneuron specification were listed, including Cutl1 – a gene that includes two known protein products: CASP and Cut1. Other targets of A2BP1 of neurodevelopmental relevance are the activity-dependent alternative splicing of CaV1.2 L-type calcium channels [6] and NMDA receptor 1 [7]. In addition to regulating the alternative splicing of other transcripts, A2bp1 appears to have the potential to self-regulate. Zhang et al. [19] also report that the OMIM database is enriched for genes that contain the UGCAUG recognition motif. Many of the genes in OMIM that contain this recognition motif are associated with epilepsy and a variety of neurodevelopmental disorders including autism, Asperger's syndrome, Rett syndrome, X-linked mental retardation, lissencephaly and schizophrenia. It is important to note that the analysis by Zhang et al. [19] does not distinguish between A2BP1 (FOX1)-specific targets and RBM9 (FOX2)-specific targets, which will be necessary to determine precisely the gene-regulatory targets in A2bp1-expressing cells during fetal brain development.
An additional informatics report, based on a meta-analysis combining GWAS data, functional studies from animal models, and tissue markers from postmortem samples [20], ranked A2BP1 as the fifth of 41 top candidates to pursue in bipolar disorder. Given the expression pattern of A2bp1, its role in neuronal splicing (and perhaps even sex-steroid-dependent splicing) and its chromosomal position in a highly replicated locus for ASD genome association studies, A2BP1 is a promising candidate for future studies of the molecular etiology of a broad range of neurodevelopmental disorders. Given the large number of UGCAUG sites in the genome and the robust expression from the A2bp1 locus evident in selective neuronal populations in the developing fetal mouse brain, experimental disruption of A2BP1 activity may reveal cell type-specific target genes that lead to clinically relevant dysregulated phenotypes.
Supplementary Material
Supplementary figures
Acknowledgements
We would like to acknowledge the technical support of Eric Yetter, Christine Svitek, Frank Liu and Deborah Gregory. We would especially like to acknowledge Phil Ebert, PhD, for facilitating the implementation of the high throughput in situ hybridization protocols and Joseph Roland, PhD, in Vanderbilt University's Epithelial Biology Center Imaging Resource Core for expert help with high throughput image acquisition. We also acknowledge funding from the Vanderbilt Kennedy Center P30 grant from the NIH (HD015052), Vanderbilt University Program in Neurogenomics (NIH T32 MH065215) and Vanderbilt Kennedy Center Biobehavioral Interventions Training Program (NIH T32 MH075883). We are grateful for the helpful advice of the anonymous reviewers.
References
- 1.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 2.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Underwood JG, et al. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang ZZ, et al. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnby G, et al. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin CL, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 10.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhalla K, et al. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- 12.Elia J, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 14.Hamshere ML, et al. Genetic utility of broadly defined bipolar schizoaffective disorder as a diagnostic concept. Br J Psychiatry. 2009;195:23–29. doi: 10.1192/bjp.bp.108.061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 16.Kiehl TR, et al. Identification and expression of a mouse ortholog of A2BP1. Mamm Genome. 2001;12:595–601. doi: 10.1007/s00335-001-2056-4. [DOI] [PubMed] [Google Scholar]
- 17.Jacobowitz DM, Abbott LC. Chemoarchitectonic Atlas of the Developing Mouse Brain. Boca Raton: CRC Press; 1997. [Google Scholar]
- 18.Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le-Niculescu H, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures