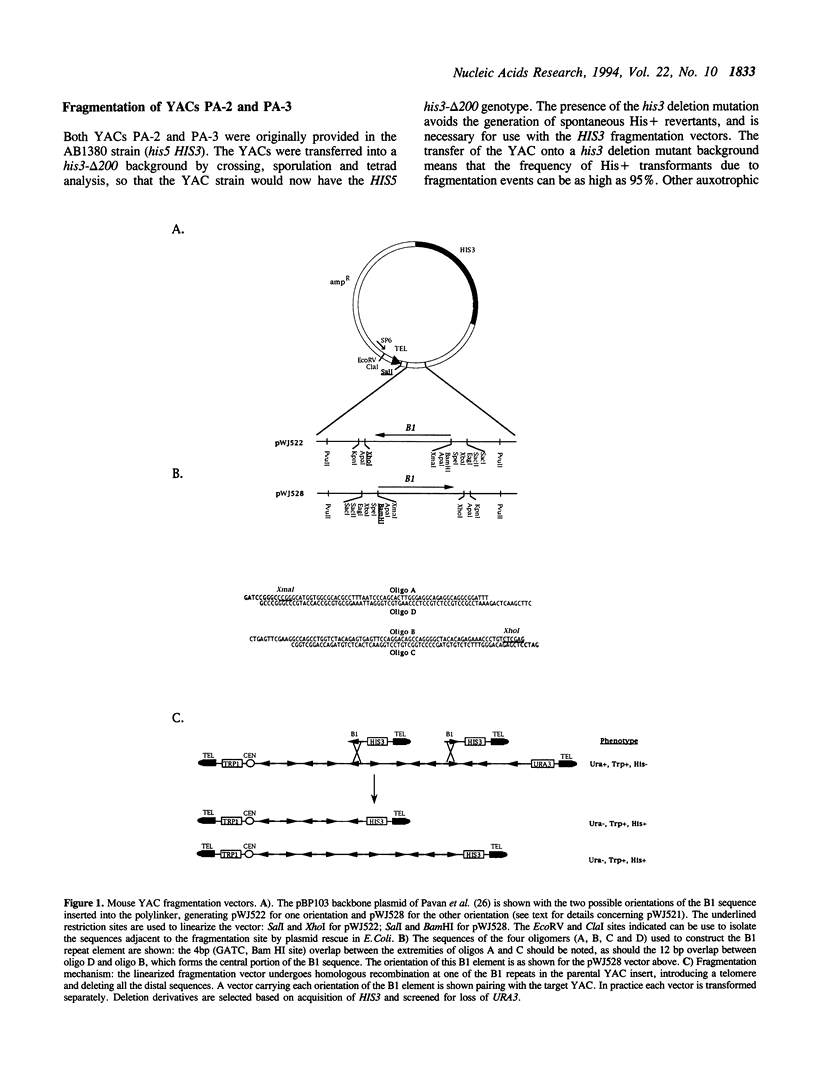
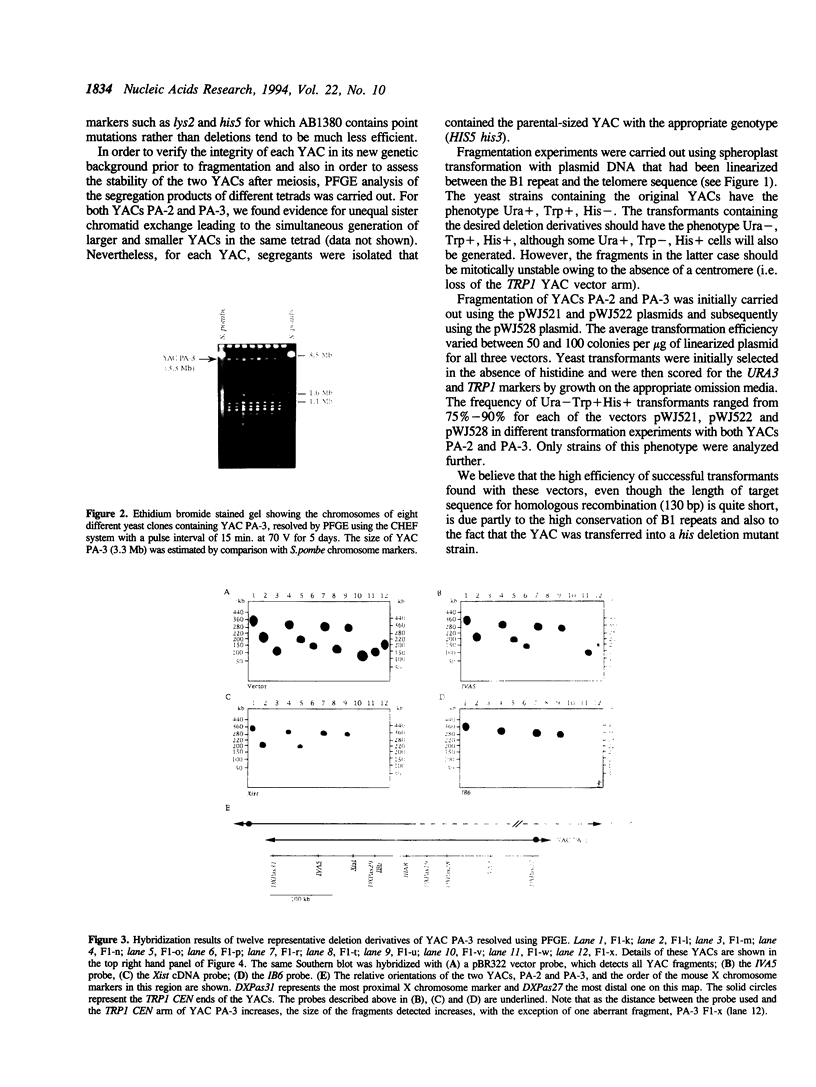
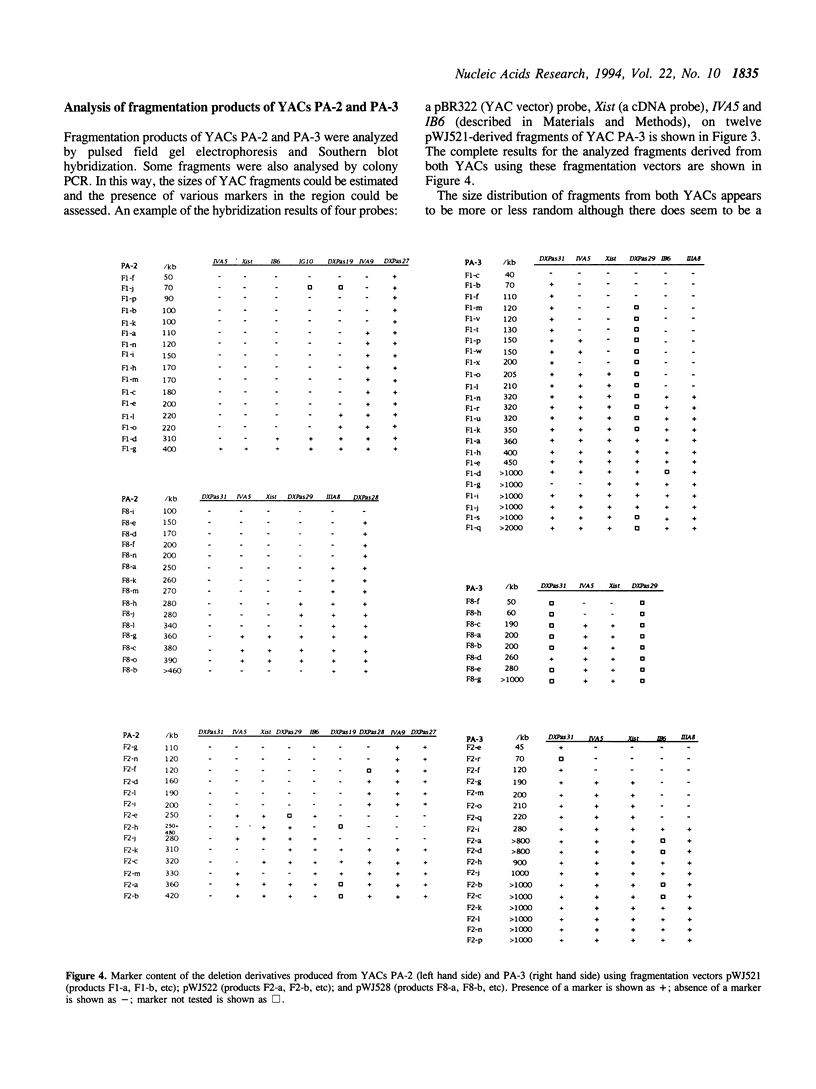
Abstract
Two mouse YACs, PA-2 and PA-3, contain the Xist gene and are 460 kb and 3.3 Mb long respectively. While PA-2 is non-chimeric, PA-3 contains a substantial proportion of non-contiguous DNA. As a prerequisite to functional studies of the role of this region in X inactivation, we have created a deletion series of YACs that are spaced at approximately 50 kb intervals and were able to eliminate the unwanted chimeric sequences in YAC PA-3. For this purpose, we have constructed mouse B1 fragmentation vectors based on those described for human Alu fragmentation. Having created this series of YAC deletion derivatives, we were able to eliminate efficiently the 10-15% aberrant YACs that arise during the course of a fragmentation experiment by assessing their marker content. The overlap and the opposite orientation of the two YAC inserts permitted the creation of deletions on both sides of the 500 kb region around Xist. The use of this series of YACs in a biological assay will help us define the extent of the sequences necessary to bring about X chromosome inactivation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett K. L., Hill R. E., Pietras D. F., Woodworth-Gutai M., Kane-Haas C., Houston J. M., Heath J. K., Hastie N. D. Most highly repeated dispersed DNA families in the mouse genome. Mol Cell Biol. 1984 Aug;4(8):1561–1571. doi: 10.1128/mcb.4.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G., Tonlorenzi R., Simmler M. C., Dandolo L., Arnaud D., Capra V., Grompe M., Pizzuti A., Muzny D., Lawrence C. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991 May 23;351(6324):325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S., McCabe V. M., Norris D. P., Penny G. D., Patel D., Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991 May 23;351(6324):329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., McCabe V. M., Norris D. P., Cooper P. J., Swift S., Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992 Oct 30;71(3):515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Hendrich B. D., Rupert J. L., Lafrenière R. G., Xing Y., Lawrence J., Willard H. F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992 Oct 30;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Lafreniere R. G., Powers V. E., Sebastio G., Ballabio A., Pettigrew A. L., Ledbetter D. H., Levy E., Craig I. W., Willard H. F. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991 Jan 3;349(6304):82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- Brownstein B. H., Silverman G. A., Little R. D., Burke D. T., Korsmeyer S. J., Schlessinger D., Olson M. V. Isolation of single-copy human genes from a library of yeast artificial chromosome clones. Science. 1989 Jun 16;244(4910):1348–1351. doi: 10.1126/science.2544027. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Percival K. J. Transformation of yeast spheroplasts without cell fusion. Anal Biochem. 1987 Jun;163(2):391–397. doi: 10.1016/0003-2697(87)90240-5. [DOI] [PubMed] [Google Scholar]
- Capecchi M. R. Mouse genetics. YACs to the rescue. Nature. 1993 Mar 18;362(6417):205–206. doi: 10.1038/362205a0. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach B. M., Pollard C. E., Perez J. N. Controlling elements in the mouse X-chromosome. I. Interaction with the X-linked genes. Genet Res. 1969 Dec;14(3):223–235. doi: 10.1017/s0016672300002068. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., Rasberry C., Evans E. P., Dandolo L., Simmler M. C., Avner P. Genetic and molecular evidence of an X-chromosome deletion spanning the tabby (Ta) and testicular feminization (Tfm) loci in the mouse. Cytogenet Cell Genet. 1991;56(3-4):137–143. doi: 10.1159/000133070. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Colleaux L., Rougeulle C., Avner P., Dujon B. Rapid physical mapping of YAC inserts by random integration of I-Sce I sites. Hum Mol Genet. 1993 Mar;2(3):265–271. doi: 10.1093/hmg/2.3.265. [DOI] [PubMed] [Google Scholar]
- Forget B. G. YAC transgenes: bigger is probably better. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):7909–7911. doi: 10.1073/pnas.90.17.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A., Huxley C., Peterson K., Olson M. V. Microinjection of intact 200- to 500-kb fragments of YAC DNA into mammalian cells. Genomics. 1993 Mar;15(3):659–667. doi: 10.1006/geno.1993.1121. [DOI] [PubMed] [Google Scholar]
- Heard E., Simmler M. C., Larin Z., Rougeulle C., Courtier B., Lehrach H., Avner P. Physical mapping and YAC contig analysis of the region surrounding Xist on the mouse X chromosome. Genomics. 1993 Mar;15(3):559–569. doi: 10.1006/geno.1993.1108. [DOI] [PubMed] [Google Scholar]
- Huxley C., Gnirke A. Transfer of yeast artificial chromosomes from yeast to mammalian cells. Bioessays. 1991 Oct;13(10):545–550. doi: 10.1002/bies.950131009. [DOI] [PubMed] [Google Scholar]
- Kay G. F., Penny G. D., Patel D., Ashworth A., Brockdorff N., Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993 Jan 29;72(2):171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Markusheva T. V., Kramerov D. A., Ryskov A. P., Skryabin K. G., Bayev A. A., Georgiev G. P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982 Dec 11;10(23):7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larin Z., Monaco A. P., Lehrach H. Yeast artificial chromosome libraries containing large inserts from mouse and human DNA. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4123–4127. doi: 10.1073/pnas.88.10.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Schultes N. P., Szostak J. W. Chromosome length controls mitotic chromosome segregation in yeast. Cell. 1986 May 23;45(4):529–536. doi: 10.1016/0092-8674(86)90284-9. [DOI] [PubMed] [Google Scholar]
- Pachnis V., Pevny L., Rothstein R., Costantini F. Transfer of a yeast artificial chromosome carrying human DNA from Saccharomyces cerevisiae into mammalian cells. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5109–5113. doi: 10.1073/pnas.87.13.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan W. J., Hieter P., Reeves R. H. Generation of deletion derivatives by targeted transformation of human-derived yeast artificial chromosomes. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1300–1304. doi: 10.1073/pnas.87.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan W. J., Hieter P., Sears D., Burkhoff A., Reeves R. H. High-efficiency yeast artificial chromosome fragmentation vectors. Gene. 1991 Sep 30;106(1):125–127. doi: 10.1016/0378-1119(91)90576-w. [DOI] [PubMed] [Google Scholar]
- Quentin Y. Successive waves of fixation of B1 variants in rodent lineage history. J Mol Evol. 1989 Apr;28(4):299–305. doi: 10.1007/BF02103425. [DOI] [PubMed] [Google Scholar]
- Reeves R. H., Pavan W. J., Hieter P. Yeast artificial chromosome modification and manipulation. Methods Enzymol. 1992;216:584–603. doi: 10.1016/0076-6879(92)16051-k. [DOI] [PubMed] [Google Scholar]
- Simmler M. C., Cattanach B. M., Rasberry C., Rougeulle C., Avner P. Mapping the murine Xce locus with (CA)n repeats. Mamm Genome. 1993 Sep;4(9):523–530. doi: 10.1007/BF00364788. [DOI] [PubMed] [Google Scholar]
- Vollrath D., Davis R. W., Connelly C., Hieter P. Physical mapping of large DNA by chromosome fragmentation. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6027–6031. doi: 10.1073/pnas.85.16.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]