Abstract
Cellular senescence is a program of irreversible cell cycle arrest that normal cells undergo in response to progressive shortening of telomeres, changes in telomeric structure, oncogene activation or oxidative stress. The underlying signalling pathways, of major clinicopathological relevance, are unknown. We combined genome-wide expression profiling with genetic complementation to identify genes that are differentially expressed when conditionally immortalised human fibroblasts undergo senescence upon activation of the p16-pRB and p53-p21 tumour suppressor pathways. This identified 816 up- and 961 down-regulated genes whose expression was reversed when senescence was bypassed. Overlay of this data set with the meta-signatures of genes up-regulated in cancer showed that nearly 50% of them were down-regulated upon senescence showing that even though overcoming senescence may only be one of the events required for malignant transformation, nearly half of the genes upregulated in cancer are related to it. Moreover 65 of the up- and 26 of the down-regulated genes are known downstream targets of NF-κB suggesting that senescence was associated with activation of the NF-κB pathway. Direct perturbation of this pathway bypasses growth arrest indicating that activation of NF-κB signalling has a causal role in promoting senescence.
Keywords: cellular senescence, conditional immortalisation, senescence bypass, differential expression, NF-κB signalling
Introduction
Cellular senescence is a program of irreversible cell cycle arrest that normal cells undergo after a finite number of divisions, the Hayflick limit (Hayflick and Moorhead, 1961). It is triggered in response to a variety of intrinsic and extrinsic stimuli including progressive shortening of telomeres or changes in telomeric structure or other forms of genotoxic stress such as oncogene activation, DNA damage or oxidative stress resulting in a DNA damage response and growth arrest via activation of the p53 tumour suppressor (Ben-Porath and Weinberg, 2005; Campisi and d’Adda di Fagagna, 2007; Adams, 2009). Activation of p53 induces p21CIP1 which prevents cyclin/cdk complexes from phosphorylating the RB family of proteins, thereby activating the pRB tumour suppressor pathway (Ben-Porath and Weinberg, 2005; Campisi and d’Adda di Fagagna, 2007; Adams, 2009). Under other circumstances, the pRB pathway can be activated independently of p53, through upregulation of p16INK4A, an inhibitor of the cyclinD/cdk4,6 kinases that also phosphorylate the RB family of proteins (Ben-Porath and Weinberg, 2005; Campisi and d’Adda di Fagagna, 2007; Adams, 2009).
Senescence can compromise tissue repair and regeneration and contribute to tissue and organismal ageing due to depletion of stem/progenitor cell compartments. It could also lead to removal of defective and potentially cancerous cells from the proliferating pool thereby limiting tumour development (Campisi and d’Adda di Fagagna, 2007). Acquisition of a limitless replicative potential has been proposed to be one of the key events required for malignant transformation (Hanahan and Weinberg, 2000). The underlying mechanisms that control cellular senescence, the signal transduction pathways involved and how the diverse signals that result in senescence are all integrated, remain poorly defined.
The main stumbling block in studying senescence has been the absence of suitable model systems because it is stochastic and occurs asynchronously in heterogeneous cell populations that are typically used for serial sub-cultivation. Studies with human cells are further complicated by the genetic, epigenetic and proliferative variation that can exist between different donors, as well as phenotypic differences between cells within the cultures. To simplify this process many investigators study oncogene-induced senescence due to expression of activated oncogenes such as RASV12, RAF or BRAF E600 or stress or irradiation induced senescence where it occurs prematurely and can be induced acutely in a variety of cell types (Collado et al., 2007).
We have taken a different approach by making use of the finding that reconstitution of telomerase activity by the catalytic subunit of human telomerase (hTERT) alone was incapable of immortalising all human somatic cells, but inactivation of the p16-pRB and p53-p21 pathways were required in addition (Kiyono et al., 1998; Counter et al., 1998). The ability of SV40 large T (LT) antigen to inactivate the p16-pRB and p53-p21 pathways enabled the use of a thermolabile mutant (U19tsA58) of LT antigen, in conjunction with hTERT, to develop conditionally immortalised human mammary (HMF3A) fibroblasts, that remain stringently temperature sensitive and show no sign of transformation in >300 population doublings (O’hare et al., 2001). These cells are immortal if grown at 34°C but upon inactivation of the thermolabile LT antigen they undergo an irreversible growth arrest within 7 days, induce senescence-associated (SA)-β-galactosidase and have morphological features and express genes in common with senescent cells (Hardy et al., 2005).
Previously we used cDNA microarrays to identify genes that were differentially expressed when these conditionally immortal HMF3A fibroblasts underwent irreversible growth arrest upon activation of the p16-pRB and p53-p21 pathways (Hardy et al., 2005). Although many of the identified changes had previously been found to occur upon replicative senescence (Shelton et al., 1999), we identified a number of genes not previously implicated in senescence and in silico promoter analysis coupled with electrophoretic mobility shift assays suggested that nuclear factor-kappa B (NF-κB) and C/EBP transcription factors may be activated upon senescence (Hardy et al., 2005). Subsequently NF-κB activity was shown to be continually required to enforce many features of ageing in a tissue specific manner (Adler et al., 2007) and be constitutively activated upon ageing (Kriete et al., 2008; Salminen et al., 2008; Kriete and Mayo, 2009). Association of senescence with secretion of senescence-associated-secretory-phenotype (SASP) proteins further suggested a role for NF-κB activation in inducing and reinforcing senescence (Kuilman and Peeper, 2009). In contrast, others have obtained contradictory results (Dimri and Campisi, 1994; Aggarwal et al., 1995; Batsi et al., 2009) and even implicated NF-κB in promoting tumourigenesis (Pikarsky et al., 2004; Karin, 2010). Therefore there is no clear consensus as to whether activated NF-κB signalling promotes growth arrest and ageing or promotes cell growth and cancer.
To further characterise whether NF-κB signalling promotes or abrogates senescence, we carried out a systematic analysis of the conditionally immortal HMF3A cells by genetic complementation and genome wide expression profiling. The results show that senescence is predominantly induced by the p53-p21 pathway and that it is associated with changes in gene expression that are reversed when it is bypassed. Moreover they show that NF-κB signalling is activated and has a causal role in promoting senescence.
Results
Senescence is predominantly induced by the p53-p21 pathway
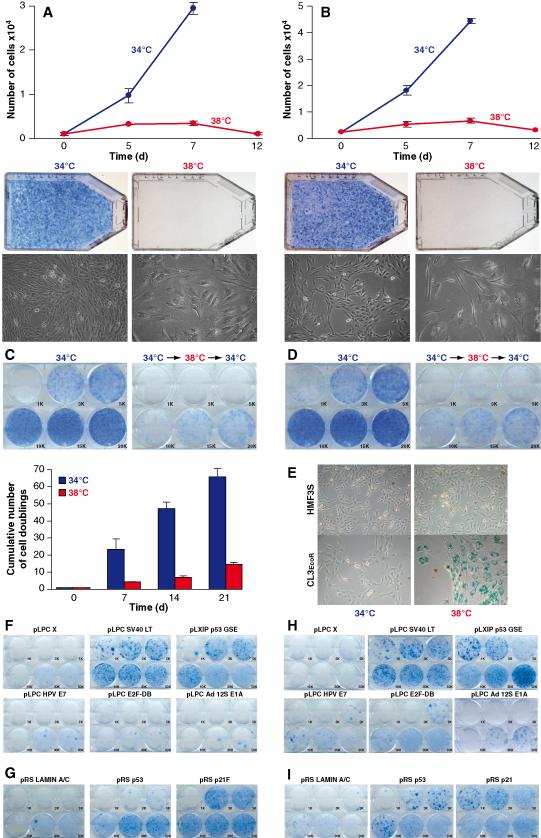
To define the relative contributions of the p16-pRB and p53-p21 pathways towards senescence, each of these pathways was abrogated by ectopic expression or RNAi. To facilitate efficient retroviral infection, HMF3A cells were transduced with the full length murine ecotropic retroviral receptor (receptor expressing cells are designated as EcoR) and 24 single cell clones analysed for temperature dependent growth and infectibility with ecotropic retroviruses. Although all clones exhibited temperature dependent growth, clone 3 (CL3EcoR) cells most closely mirrored the parental cells in their temperature dependent growth characteristics (Fig.1A-D). They undergo an irreversible growth arrest upon shift to 38°C and exhibit the same changes in morphology as the HMF3A cells and express SA-β-galactosidase (Fig.1E).
Figure 1: Characterisation of HMF3A cells.
Growth and morphology of HMF3AEcoR (A) and CL3EcoR (B) cells: Proliferative potential was determined by growing cells at 34°C or 38°C and determining cell numbers or by staining with methylene blue.
Irreversibility HMF3AEcoR (C) and CL3EcoR (D) cells: Irreversibility was tested by incubating cells at 34°C or 38°C for 7 days and then shifting them back to 34°C.
Induction of SA-β galactosidase (E): Cells were stained for SA-β galactosidase activity after 7 days.
Complementation HMF3AEcoR (F) and CL3EcoR (H) cells by ectopic expression: Cells stably transduced with the indicated retroviruses were incubated at 38°C for 21 days before staining. Constructs able to bypass growth arrest yield dark blue colonies of densely growing cells.
Complementation HMF3AEcoR (G) and CL3EcoR (I) cells by RNAi: Cells stably infected with retroviruses that transduce the indicated shRNAs were assayed for bypassing growth arrest.
Complementation analysis indicated that the temperature dependent growth defect was readily overcome with wild type SV40 LT antigen. Different numbers of stably transduced cells were seeded and cultured at 38°C for 3 weeks; densely growing clones were observed after plating 1,000 stably transduced HMF3AEcoR cells (Fig.1F). Growth arrest was also very efficiently overcome upon inactivation of the p53-p21 pathway with p53GSE, that inactivates p53 (Ossovskaya et al., 1996) or shRNAs that target p53 (Berns et al., 2004) or p21CIP1 (Mansfield, 2006; Fig.1F&G). This abrogation was very efficient since clones of densely growing cells were detected after plating <3,000 cells. In contrast, growth arrest was much less efficiently overcome upon inactivation of the p16-pRB pathway (Fig.1F); growing cells were only obtained after plating 30-50,000 cells, stably transduced with Adenovirus (Ad) 5 E1A 12s, Human Papilloma Virus (HPV) 16 E7 or the dominant negative E2F-DB protein (Rowland et al., 2002). Silencing of p21 expression by p53shRNA, p53GSE and p21shRNA is shown in Fig. S1; expression of E1A and E7 are shown in Fig. S2. Growth arrest of CL3EcoR cells was also readily bypassed by SV40 LT antigen, p53GSE, p53shRNA or p21shRNA (Fig.1H&I). However they were rescued more efficiently by HPV16 E7 and E2F-DB than the parental HMF3AEcoR cells. Growing colonies were obtained upon replating 5,000 CL3EcoR cells whereas about 30,000 cells needed to be plated with HMF3AEcoR cells. Growth arrest was also overcome upon transduction with Ad5 E1A 12S but higher numbers of cells were required probably due to the apoptotic activity of this protein. CL3EcoR was the only clone characterised that could be rescued efficiently upon inactivation of the p16-RB pathway by HPV16 E7 and E2F-DB.
These results indicated that the predominant pathway that induced growth arrest was the p53-p21 pathway since it was most efficiently abrogated when it was inactivated. Inactivation of the p16-pRB pathway also overcame growth arrest but much less efficiently and only in some cells, indicating that pRB does not always act downstream of p53-p21 to induce senescence.
Changes in gene expression upon senescence growth arrest are reversible
To identify changes in gene expression upon senescence, RNA from growing or growth arrested CL3EcoR cells was expression profiled. To eliminate changes in expression due to the temperature shift (HS), RNA extracted from HMF3S cells grown at 34°C and after shift up to 38°C for 7 days were analysed. HMF3S cells were immortalised from the same batch of primary human mammary fibroblasts using wild type U19 SV40 LT antigen in conjunction with hTERT (Hardy et al., 2005) and continue to divide upon shift up to 38°C (Fig.1E). Genes were considered growth arrest (GA) specific if the difference in Log2Fold Change between CL3EcoR 38 versus 34 and HMF3S 38 versus 34 was <−1 or >1 (Fig. S3).
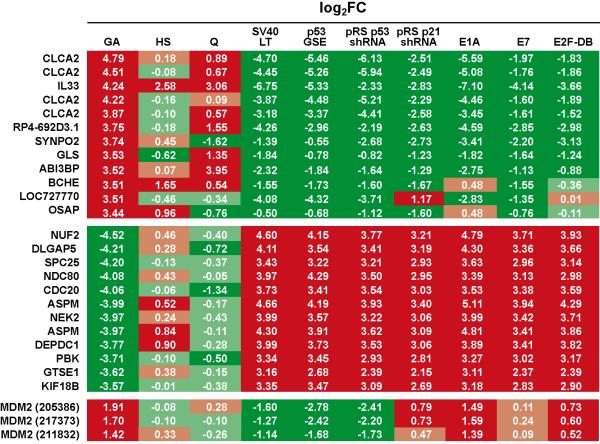
3059 up-regulated transcripts of which 816 were up-regulated >2 fold and 5005 down-regulated transcripts, 961 of which were down-regulated >2 fold were identified; indicated as GA in Tables S1 and S2. Four of the top five most highly up-regulated transcripts correspond to the same gene, CLCA family member 2, chloride channel regulator. Three of the top four most highly down-regulated transcripts NUF2, SLC25 and NDC80 are all components of the NDC80 kinetochore complex. To identify genes perturbed by serum starvation resulting in quiescence, a reversible growth arrest, CL3EcoR cells were serum starved for 7 days (Fig. S4). Many of the top senescence up-regulated genes such as IL33, ABI3P, IL1A and IL1B, were also highly up-regulated upon serum starvation (Table S1). However serum starvation did not induce a significant change in cell morphology or SA-β-galactosidase activity (Fig. S5).
If changes in expression are specific for senescence arrest, they should be reversed upon its abrogation. Triplicate independent cultures of CL3EcoR cells after bypass of senescence with SV40 LT antigen, Ad5 E1A 12S, HPV16 E7, E2F-DB, p53GSE, p53shRNA and p21shRNA were profiled, and compared to its appropriate control culture to obtain the set of differential genes which were then compared to the differential growth arrest data set (Fig. S6). Results obtained upon senescence bypass for the top 12 up- and down-regulated genes are in Fig. 2; complete data are in Tables S1 and S2. They show that when growth arrest was overcome, differential expression was reversed; down-regulated genes were up-regulated whereas up-regulated genes were suppressed. However the fold change was not always the same eg. for CLCA2, the suppression upon rescue with Ad5 E1A 12S, p53shRNA and GSE53 was about 30 fold whereas with HPV16 E7, E2F-DB and p21shRNA, the change was only 4 fold. Although these differences may be due to level of expression or the complementing gene, they most likely reflect the rescuing pathway as illustrated by the changes in expression of MDM2 (HDM2, Fig.2), an E3 ubiquitin ligase that associates with p53 and maintains it at a low level (Momand et al., 1992); up-regulation of p53 results in up-regulation of MDM2. Upon growth arrest MDM2 was up-regulated and expression of all three features (205386, 217373 and 211832) was increased. When growth arrest was abrogated using SV40 LT antigen, p53GSE or p53shRNA that directly inhibit p53 activity, MDM2 expression was reversed for all three features. However when growth arrest was abrogated with Ad5 E1A 12S, HPV16 E7 and E2F-DB or p21shRNA, none of which are known to directly act on p53, MDM2 expression levels remained up-regulated, although the fold up-regulation was reduced by HPV16 E7 and E2F-DB or upon silencing of p21CIP1.
Figure 2: Senescence specific changes in gene expression.
Log2 Fold Changes in gene expression that occur upon irreversible growth arrest are indicated as GA, upon shift up of HMF3S cells from 34°C to 38°C as HS and upon serum starvation as Q respectively. If changes in gene expression are specific for the senescence arrest, they should be reversed upon its abrogation. Up-regulated transcripts are indicated in red whereas down-regulated transcripts are in green. Results for the top 12 up- and down-regulated transcripts after complementation with the indicated constructs are shown as well as the changes in expression of the three MDM2 features.
To confirm the expression profiling data, the Human Signal Transduction Open Array™ which enabled simultaneous analysis of 600 genes by real- time qPCR, was utilised to determine the fold change in expression (FC=2ΔΔCt) due to senescence or heat shock. After filtering to eliminate growth arrest genes whose expression was very low (Ct >22), nearly 80% of the 318 genes showed concordant changes in expression by real time qPCR or expression profiling (Table 1, S3 and S4 and Fig. S7) even though the actual Log2Fold Changes obtained by the two methods were different.
Table 1. Real time qPCR analysis.
Comparison of the Log2Fold Changes in expression from expression profiling and real time qPCR for growth arrest (CL3EcoR cells at 34°C and 38°C) and heat shock (HMF3S cells at 34°C and 38°C).
| Growth arrest | Heat shock | |||
|---|---|---|---|---|
| Changes in gene expression | No. of oligos | % | No. of oligos | % |
| same direction | 254 | 79.9 | 250 | 78.6 |
| opposite direction | 64 | 20.1 | 68 | 21.4 |
Comparison of genes differentially expressed upon senescence with the meta-signature of genes over-expressed in cancer
To determine if any of the senescence arrest genes were linked to cancer, the GA list was overlapped with the meta-signatures of genes over-expressed upon neoplastic transformation and in undifferentiated cancer (Rhodes et al., 2004). The neoplastic transformation meta-signature comprises 67 over-expressed genes; 33 of these were down-regulated and 10 were up-regulated upon growth arrest (Table 2 and S5). The meta-signature of genes over-expressed in undifferentiated cancer comprises 69 genes of which 5 were up-regulated and 46 were down-regulated upon growth arrest (Table 2 and S5). This indicated that 49% and 65% of the genes over-expressed upon neoplastic transformation and in undifferentiated cancer were down-regulated upon senescence.
Table 2. Comparison with meta-signatures of neoplastic transformation and undifferentiated cancer.
Overlap of the senescence growth arrest data set with the meta-signatures of genes over-expressed upon neoplastic transformation and in undifferentiated cancer.
| Up- regulated |
Down- regulated |
Total | |
|---|---|---|---|
| Neoplastic Transformation |
10 (15%) | 33 (49%) | 67 |
| Undifferentiated Cancer |
5 (7%) | 46 (67%) | 69 |
Senescence is associated with activation of the NF-κB pathway
Our previous study suggested that the NF-κB pathway may be activated upon senescence (Hardy et al., 2005). To determine if the differential gene set comprised known NF-κB targets, they were compared to the putative NF-κB targets proposed by Gilmore (http://www.nf-kb.org). 91 NF-κB targets were found to be differentially expressed: 65 were up-regulated and 26 were down-regulated (Table 3 and S6). IL1A and IL1B were the most highly up-regulated genes. The SASP cytokine IL6 was also up-regulated; IL8 and IGFBP7 were also up-regulated but not significantly (Table S7). Interestingly IL6 expression was up-regulated to a greater extent by serum starvation. Almost all of the NF-κB targets found to be up-regulated upon senescence growth arrest were also up-regulated upon serum starvation including IL1A and B, the most highly up-regulated NF-κB targets. The modulation by serum starvation was greater than with senescence arrest; however they were not affected by heat shock. Up-regulation of these NF-κB targets was reversed when growth arrest was overcome by abrogating the p53-p21 or p16-pRB pathways (Table 3). In addition to IL1A and B and IL6, a number of other secreted protein genes were up-regulated upon senescence.
Table 3. NF-κB targets.
Top 12 up-regulated NF-κB targets upon senescence and after complementation.
| GA | Q | HS | SV40 LT |
p53 GSE |
pRS p53 |
pRS p21 |
E1A | E7 | E2F- DB |
|
|---|---|---|---|---|---|---|---|---|---|---|
| IL1A | 3.42 | 3.82 | 0.22 | −4.36 | −3.28 | −0.70 | −0.26 | −5.18 | −1.05 | −0.04 |
| IL1B | 3.33 | 3.54 | 0.06 | −4.60 | −4.13 | −1.41 | −1.48 | −5.44 | −1.98 | −0.86 |
| IL1B | 3.29 | 3.43 | 0.38 | −4.58 | −3.83 | −1.31 | −1.37 | −5.89 | −1.82 | −0.80 |
| BMP2 | 2.64 | 4.73 | 0.24 | −5.09 | −3.59 | −1.45 | −1.91 | −4.51 | −3.45 | −2.96 |
| BMP2 | 2.25 | 4.80 | 0.24 | −5.00 | −3.18 | −1.47 | −1.80 | −4.42 | −3.51 | −3.20 |
| SOD2 | 2.10 | 3.04 | 0.72 | −2.27 | −2.00 | −0.71 | −0.87 | −2.83 | −0.92 | −0.63 |
| 40118 | 2.04 | 2.14 | −0.56 | −2.97 | −2.23 | −1.74 | −1.71 | −2.96 | −1.84 | −1.41 |
| IL6 | 2.01 | 4.16 | 0.13 | −3.37 | −2.65 | −1.24 | −1.93 | −5.35 | −2.64 | −1.97 |
| AKR1C1 | 2.01 | 3.30 | −0.65 | −2.44 | −0.82 | 0.46 | −0.12 | −4.22 | −1.20 | −1.19 |
| TNFAIP3 | 1.96 | 2.42 | 0.32 | −3.27 | −3.30 | −1.68 | −1.74 | −4.35 | −1.64 | −1.60 |
| IL32 | 1.96 | 0.78 | 0.17 | −1.42 | −1.23 | −0.86 | −1.11 | −0.26 | −0.99 | −0.36 |
| 40118 | 1.93 | 2.10 | −0.38 | −2.89 | −2.36 | −1.67 | −1.76 | −2.67 | −1.79 | −1.34 |
To determine if the differentially expressed genes contained binding sites for NF-κB transcription factors, the list of GA genes was compared with a motif module map comprising a matrix of 12,254 genes and 2,394 known transcription factors (http://motifmap.googlepages.com/; Adler et al., 2006). All four NF-κB factor motifs were found to be present within the promoters of the up- and down-regulated genes (Table 4). In the up-regulated genes, the NF-κB motifs were present within the promoters of 200, 134, 124 and 114 genes, ranking them in the top 3%, 5.7%, 6.4% and 7.4% abundant motifs respectively. In the down-regulated genes, the NF-κB motifs were present within the promoters of 217, 175, 144 and 116 genes, ranking them in the top 4.2%, 5.9%, 7.5% and 10.3% abundant motifs.
Table 4. Transcription factor motifs.
Analysis of the promoter regions of the up- and down-regulated genes for NF-κB binding sites.
| No. of genes | Position out of 2394 TFs |
Rank by percent |
|
|---|---|---|---|
| UP | |||
| V_NFKB_Q6_01.wtmx | 200 | 73 | 3.0 |
| V_NFKAPPAB65_01.wtmx | 134 | 136 | 5.7 |
| V_NFKAPPAB_01.wtmx | 124 | 154 | 6.4 |
| V_NFKAPPAB50_01.wtmx | 114 | 178 | 7.4 |
|
| |||
| DOWN | |||
| V_NFKB_Q6_01.wtmx | 217 | 100 | 4.2 |
| V_NFKAPPAB65_01.wtmx | 175 | 142 | 5.9 |
| V_NFKAPPAB_01.wtmx | 144 | 180 | 7.5 |
| V_NFKAPPAB50_01.wtmx | 116 | 246 | 10.3 |
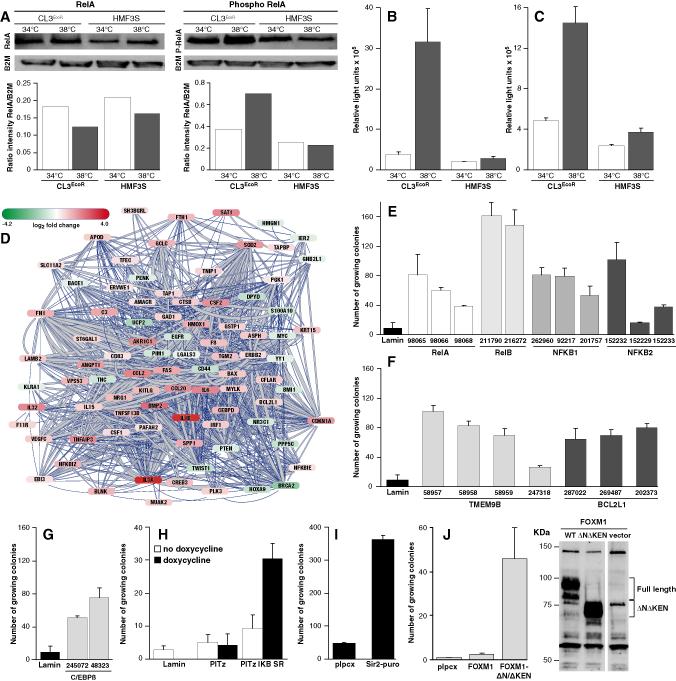
To determine if activation of NF-κB signalling was associated with increased phosphorylation of RelA, growing and senescent CL3EcoR cell lysates were analysed using an antibody specific for phosphorylated Serine 536. Growth arrest was associated with increased phosphorylation of RelA (Fig.3A). To further confirm that NF-κB signalling was activated, secretion of IL6 and IL8 was determined in culture supernatants of growth arrested CL3EcoR cells. A very large increase in the level of IL6 was observed upon growth arrest (Fig.3B); IL8 levels were also increased but not to the same extent (Fig.3C).
Figure 3: Involvement of NF-κB in senescence.
(A) Increase in phosphorylation of RelA (Ser536) in senescent cells: Nuclear proteins extracted after 12 days were analysed by western blotting using Phospho-NF-κB p65 (Ser536) (93H1) and NF-κB p65 (C22B4; Cell Signalling) antibodies.
Secretion of IL6 (B) and IL8 (C) by senescent cells: 12 hour supernatants harvested from cells grown for 21 days were analysed in triplicate by Quantiglo ELISAs (R&D Systems).
(D) Sub-network of 87 differentially expressed NF-κB target genes: The strength of interaction between genes (a combined score based on a scoring-framework, which depends on benchmarks of the different types of associations against a common reference) is indicated by the thickness of the line and the degree of up/down regulation by the colour of the node.
(E) Silencing of NF-κB transcription factor subunits: CL3EcoR cells infected in duplicate with lentiviruses expressing the indicated (V2LHS code number) pGIPZ shRNAmir silencing constructs were assayed for bypassing growth arrest. Lentiviruses were prepared and used according to Besnier et al. (2003). After puromycin selection, 0.5×105 stably transduced cells plated in T75 flasks were incubated at 38°C for 21 days and stained.
(F) Silencing of TMEM9B and BCL2L1 and (G) C/EBPβ: 0.5×105 CL3EcoR cells transduced with the indicated pGIPZ shRNAmir silencing constructs were assayed for senescence bypass at 38°C.
(H) Ectopic expression of IκB-SR: CL3EcoR cells were infected in duplicate with lentiviruses expressing Lamin A/C shRNA, pTIPz IκB-SR or an empty pTIPz vector. 0.5×105 stably transduced cells were assayed for growth at 38°C with or without doxycycline.
(I) Ectopic expression of SIRT1: CL3EcoR cells infected with retroviruses pYESir2-puro and pLPCX were assayed for growth at 38°C.
(J) Ectopic expression of FOXM1: CL3EcoR cells infected with retroviruses transducing FOXM1 and FOXM1 (ΔNΔKEN) were assayed for growth at 38°C. Ectopic expression of FOXM1 was demonstrated by western blotting, extracts prepared from cells at 38°C using a rabbit anti-FOXM1 polyclonal antibody (Abcam).
The 91 differentially expressed NF-κB targets can be linked together in a network (Fig.3D). This network was built by making use of the STRING database (Jensen et al., 2009), a database and web resource containing protein-protein interactions of both physical, functional and bibliometrical nature collected from a large number of biological resources and Cytoscape (Shannon et al., 2003), an open source software for integrating bimolecular interaction networks with high-throughput expression data. This sub-network was extracted from a ‘hairball’ of 4347 nodes (proteins) representing the 8,064 differentially expressed transcripts and 554,205 interactions.
Silencing of NF-κB subunits abrogates senescence arrest
To ascertain if activation of the NF-κB pathway had a causative role, we determined if growth arrest would be overcome upon silencing components of the NF-κB complex even though they were not significantly differentially expressed (Table S7). Human GIPZ lentiviral shRNAmir silencing constructs for RelA, RelB, NFKB1 and NFKB2 were initially tested as a pool and then individually in the senescence bypass assay. Although none of the NF-κB silencing constructs were as efficient as silencing p21CIP1, they all overcame growth arrest (Fig.3E) with some constructs yielding more colonies than others. The number of growing colonies obtained upon plating 0.5×105 stably transduced cells was very similar or slightly higher than obtained upon inactivation of the p16-RB pathway with HPV16 E7 or E2F-DB protein; 0.5×105 cells per T75 flask corresponds to plating about 5000 cells per well in 6 well plates shown in Fig.1H. In contrast plating of 0.5×105 cells in which p21CIP1 had been stably silenced produced a confluent monolayer of growing cells.
Modulation of the NF-κB pathway overcomes senescence arrest
In a parallel project, an RNAi screen has been used to identify genes whose suppression overcomes growth arrest of CL3EcoR cells. One of the shRNAs isolated, targets TMEM9B, slightly up-regulated upon growth arrest (1.3 fold) which was reversed when it was overcome (Table S7). TMEM9B is a glycosylated lysosomal transmembrane protein essential for activation of NF-κB by TNF and acts downstream of RIP1 and upstream of MAPK and IκB kinases at the level of TAK1 (Dodeller et al., 2008). It can also activate NF-κB dependent reporter constructs (Matsuda et al., 2003; Dodeller et al., 2008). Thus silencing its expression should suppress the NF-κB pathway. Four shRNAmirs targeting TMEM9B were found to bypass senescence (Fig.3F). Silencing C/EBPβ, a transcription factor linked to NF-κB activity and BCL2L1, a downstream target of NF-κB, also complemented growth (Fig.3F&G).
Suppression of NF-κB activity by ectopic expression of a non-phosphorylatable, dominant negative IκBα, the super-repressor of NF-κB (IκB-SR; Micheau et al., 2002), also overcame growth arrest (Fig.3H). This was further confirmed by ectopic expression of SIRT1, the human homologue of Sir2, a deactylase that suppresses NF-κB activity (Yeung et al., 2004). SIRT1 expression was down-regulated 2.3 fold upon growth arrest which was reversed upon complementation. Ectopic expression of SIRT1 overcame growth arrest (Fig.3I).
Constitutively active FOXM1 bypasses senescence arrest
FOXM1, a highly conserved Forkhead transcription factor, was the second most highly down-regulated transcription factor (6.32 fold), in accordance with the finding that its levels are repressed by p53, dependent upon p21CIP1 and pRB family members (Barsotti and Prives, 2009). The down-regulation was reversed upon inactivation of p53-p21 or p16-pRB pathways (Table S2).
FOXM1 expression is induced when quiescent cells re-enter the cell cycle and reaches a maximal level in S phase that is maintained throughout G2 and mitosis. During G1, the N-terminal auto-repressor domain inhibits transcriptional activity by interacting with the C-terminal transactivation domain (TAD; Park et al., 2008). This repression is relieved in G2 by phosphorylation of multiple cdk sites within the TAD by cyclinA/cdk complexes (Laoukili et al., 2008a). Phosphorylation by PLK1 has also been suggested to be important for regulating FOXM1 activity.
FOXM1 regulates expression of G2-specific genes including Cyclin B, Polo-like-kinase 1, Aurora B, CDC25B, NEK2 and many other regulators of cell cycle progression (Laoukili et al., 2007). Since many of them as well as FOXM1 were down-regulated upon growth arrest (Fig.2) and Penzo et al., (2009) have shown that acute activation of NF-κB in murine embryo fibroblasts resulted in growth arrest in association with repression of FOXM1 and cell cycle related genes that are known targets of either E2F or FOXM1, we determined whether FOXM1 would bypass senescence by expressing full length FOXM1 or a constitutively active, non-degradable, N-terminal deleted FOXM1 (ΔNΔKEN, amino acids 210-763; Laoukili et al., 2008b). Constitutively active FOXM1 (ΔNΔKEN) readily bypassed senescence whereas full length FOXM1 was inactive even though it was highly expressed (Fig. 3J).
Discussion
In addition to telomere shortening, activation of the p53 and pRB tumour suppressor pathways are key to inducing cell senescence. They have been suggested to act in a linear pathway such that p53 induces p21CIP1 which inhibits phosphorylation of pRB thereby maintaining it in a hypophosphorylated state or by parallel pathways (Ben-Porath and Weinberg, 2005). HMF3A cells undergo an irreversible growth arrest upon inactivation of the thermolabile U19tsA58 LT antigen resulting in activation of these two tumour suppressors. Inactivating p53 using SV40 LT antigen or a dominant negative suppressor peptide (p53GSE) or by silencing readily bypassed growth arrest; silencing of p21CIP1 also overcame growth arrest. In contrast inhibition of pRB activity by Adenovirus E1A or HPV16 E7 that are known to sequester the RB family of proteins or the dominant negative E2F-DB protein was more surprising; growth arrest was not readily overcome and many more transduced cells were required to isolate cells in which growth arrest was abrogated. Moreover, only one clone out of 20 analysed could be rescued efficiently upon inactivation of the p16-pRB pathway, indicating that pRB does not always act downstream of p53 and they are likely to be acting in parallel pathways to induce senescence (Ben-Porath and Weinberg, 2005).
The combination of genome-wide expression profiling and genetic complementation identified 816 up- and 961 down-regulated genes whose expression was reversed when senescence was bypassed; down-regulated genes were up-regulated whereas up-regulated genes were suppressed. Overlay of the differential data set with the meta-signatures of genes up-regulated upon neoplastic transformation or undifferentiated cancer (Rhodes et al., 2004) showed that 49% and 67% of them were down-regulated upon senescence. Hanahan and Weinberg (2000) have proposed that six independent events are required for malignant transformation and the acquisition of a limitless replicative potential is one of these events. Our results indicate that even though overcoming senescence may only be one of these events, nearly 50% of the genes are related to it, highlighting its importance as a barrier to malignant transformation. Recently it was suggested that a feedback loop between p21CIP1 and production of reactive oxygen species involving GADD45-MAPK14-GRB2-TGFBR2-TGFβ is necessary for cell senescence (Passos et al., 2010). Since many of these genes are differential in our growth arrest data set, a more systematic analysis is now underway, in collaboration with J. Passos, to determine the involvement of this pathway.
Many of the top down-regulated genes are associated with the cell cycle, comprising genes necessary for the transition from G1 to S (CDC6 and E2F1), G2 (CDC22 and CYCLIN A) and G2 to M phase (AURKB, BUB1 and KIF20A) (Whitfield et al., 2002). Association of these down-regulated genes with cell cycle control is consistent with loss of proliferative potential upon senescence mostly in G1. However, down-regulation of genes related to G2 is particularly interesting because DNA synthesis can be induced in senescent cells by fresh mitogens or by super-infection with DNA tumour viruses but the cells do not undergo mitosis suggesting there may also be a block in G2 in senescence (Gorman and Cristofalo, 1985). Using a rodent model of senescence we have previously proposed that senescence involves growth arrest in both G1 and G2 and that a block in G2 is the cause of the irreversible loss of proliferative potential (Gonos et al., 1996).
Four of the top five up-regulated transcripts correspond to CLCA2, chloride channel regulator, a direct downstream target of p53 that can induce a senescence-like arrest upon acute expression (Walia et al., 2009). The up-regulated genes also comprised a number of secreted proteins in addition to IL1A and B and IL6; IL8 and IGFBP7 were also up-regulated but not significantly. IL6, IL8 and IGFBP7 have been found to be secreted by senescent cells, act cell autonomously to induce and reinforce senescence (Kuilman and Peeper, 2009) and not be affected by quiescence (Kuilman et al., 2008). In contrast our results indicate that these secreted proteins were also strongly up-regulated by serum starvation. IL1A, one of the most highly up-regulated genes, has been suggested to be an essential cell-autonomous regulator of the senescence-associated IL6/8 cytokine network (Orjalo et al., 2009).
Our previous study predicted that senescence in these conditionally immortal cells may involve activation of NF-κB (Hardy et al., 2005). Transcription factor analysis of the promoters of the differentially expressed genes showed that NF-κB motifs were amongst the top 10% of most abundant motifs in both the up- and down-regulated genes. Increased phosphorylation of RelA-Serine 536, elevated secretion of IL6 and IL8 and bypass of senescence arrest by silencing RelA or expressing the super-repressor of NF-κB all indicate activation of canonical NF-κB signalling. However non-canonical NF-κB signalling may also be activated since silencing RelB also overcame growth arrest even more efficiently than silencing RelA. Activation of NF-κB signalling was further highlighted by the finding that 65 NF-κB targets were up- and 26 were down-regulated significantly upon growth arrest and that miR146a, a known downstream direct target of NF-κB (Taganov et al., 2006) was also highly up-regulated upon senescence. This association of NF-κB with growth arrest is in accordance with Penzo et al., (2009) who found that acute activation of NF-κB in murine embryo fibroblasts resulted in growth arrest in association with repression of FoxM1 and 20 genes essential for cell cycle progression that are known targets of either E2F or FOXM1. Comparison of these repressed genes with our differential genes indicated that all of them except CDC5A were significantly repressed. FOXM1 and E2F1, 2, 4, 6 and 8 were also down-regulated upon growth arrest which was reversed when it was overcome. Moreover constitutively active FOXM1 (ΔNΔKEN) bypassed growth arrest suggesting that loss of transcriptionally active FOXM1 may have a causative role in senescence. FOXM1 is also down-regulated upon replicative senescence in primary human fibroblasts (Hardy et al., 2005). Its expression is markedly reduced in cells from elderly patients as well as patients with progeria (Ly et al., 2000) and FoxM1 deficient mouse embryo fibroblasts undergo premature senescence (Wang et al., 2005). FOXM1 is also one of the 69 genes that comprise the meta-signature of genes over-expressed in undifferentiated cancer (Rhodes et al., 2004).
The NF-κB family of transcription factors are ubiquitously expressed and key regulators of inflammatory and immune response (Hayden and Ghosh, 2008). They also function as regulators of diverse cellular processes and the response to oxidative, physical and chemical stress. Even though previous studies to determine the role of NF-κB in senescence and ageing have yielded conflicting results, this study has delineated a central role for NF-κB activity in promoting senescence irrespective of whether it is induced via the p53-p21 or the p16-pRB pathway. It indicates that this activation results in up-regulation of stress associated genes including IL6 and IL8 that can act cell autonomously to induce and reinforce senescence. This activation could also down-regulate FOXM1 and E2F and their downstream targets that are critical for cell cycle progression particularly through the G2 phase. Although the mechanism by which activation of the p53-p21 or p16-pRB pathways results in activation of NF-κB signalling is not known, this study suggests that SIRT1 (a deactylase that can suppress NF-κB activity) and TMEM9B (a lysosomal transmembrane protein that can activate NF-κB dependent expression) are likely to be involved. SIRT1 expression was down-regulated upon senescence but reversed upon its abrogation. SIRT1 expression is known to be suppressed by miR34a (He et al., 2007), a direct downstream target of p53 (Yamakuchi et al., 2008), also up-regulated upon growth arrest. TMEM9B was slightly up-regulated upon senescence which was reversed when it was bypassed; it was also isolated in an RNAi screen to identify genes whose suppression overcomes growth arrest. Further experiments are underway to characterize the role(s) of SIRT1 and TMEM9B in the mechanism leading to activation of NF-κB signalling and to determine whether there is cross-talk between the NF-κB and p53 pathways.
Materials and Methods
Cell Culture and viral infections
HMF3A cells and derivatives thereof were maintained at 34°C ±0.5°C and temperature shift experiments performed at 38°C ±0.5°C.
RNA Expression profiling
The Affymetrix U133 plus 2 chip raw expression profiling data was averaged and normalised using the RMA algorithm (Irizarry et al., 2003). Differentially expressed genes were identified using Linear Models for Microarray Analysis. P-values were corrected for multiple-testing using the Benjamini-Hochberg correction and a corrected P-value threshold of 10−5 was used to identify significantly differentially expressed genes. Expression profiling data are available from Gene Expression Omnibus database accession number GSE24810.
Supplementary Material
Acknowledgements
We are indebted to A. M. Neville, A. Baldwin, P. Meier, M. Resnicoff and N. Perkins for advice, helpful discussions and critical reading of the manuscript. We thank J. Downward, X. Lu, P. Meier, G. Towers and C. King (the UCL shRNA library core facility) for providing reagents. We are grateful to E. Ortenberg and J. White of Biotrove Inc. for the qPCR analysis, S. Shah and A. Grigoriadis for bioinformatic analysis and R. Young for graphics. PSJ gratefully acknowledges financial support from the Wellcome Trust (078305) and an equipment grant from the Brain Research Trust.
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- Adams PD. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell. 2009;36:2–14. doi: 10.1016/j.molcel.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007;21:3244–57. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsi C, Markopoulou S, Vartholomatos G, Georgiou I, Kanavaros P, Gorgoulis VG, et al. Chronic NF-kappaB activation delays RasV12-induced premature senescence of human fibroblasts by suppressing the DNA damage checkpoint response. Mech. Ageing Dev. 2009;130:409–19. doi: 10.1016/j.mad.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–7. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–76. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Besnier C, Ylinen L, Strange B, Lister A, Takeuchi Y, Goff SP, et al. Characterization of murine leukemia virus restriction in mammals. J Virol. 2003;77:13403–6. doi: 10.1128/JVI.77.24.13403-13406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–33. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A. 1998;95:14723–8. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Campisi J. Altered profile of transcription factor-binding activities in senescent human fibroblasts. Exp Cell Res. 1994;212:132–40. doi: 10.1006/excr.1994.1127. [DOI] [PubMed] [Google Scholar]
- Dodeller F, Gottar M, Huesken D, Iourgenko V, Cenni B. The lysosomal transmembrane protein 9B regulates the activity of inflammatory signaling pathways. J Biol Chem. 2008;283:21487–94. doi: 10.1074/jbc.M801908200. [DOI] [PubMed] [Google Scholar]
- Gonos ES, Burns JS, Mazars GR, Kobrna A, Riley TE, Barnett SC, et al. Rat embryo fibroblasts immortalized with simian virus 40 large T antigen undergo senescence upon its inactivation. Mol Cell Biol. 1996;16:5127–38. doi: 10.1128/mcb.16.9.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman SD, Cristofalo VJ. Reinitiation of cellular DNA synthesis in BrdU-selected nondividing senescent WI-38 cells by simian virus 40 infection. J Cell Physiol. 1985;125:122–6. doi: 10.1002/jcp.1041250116. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hardy K, Mansfield L, Mackay A, Benvenuti S, Ismail S, Arora P, et al. Transcriptional networks and cellular senescence in human mammary fibroblasts. Mol Biol Cell. 2005;16:943–53. doi: 10.1091/mbc.E04-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius M, Hänninen M, Lehtinen SK, Salminen A. Changes associated with aging and replicative senescence in the regulation of transcription factor nuclear factor-kappa B. Biochem J. 1996;318:603–8. doi: 10.1042/bj3180603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–8. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- Kriete A, Mayo KL. Atypical pathways of NF-kappaB activation and aging. Exp Gerontol. 2009;44:250–5. doi: 10.1016/j.exger.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Kriete A, Mayo KL, Yalamanchili N, Beggs W, Bender P, Kari C, et al. Cell autonomous expression of inflammatory genes in biologically aged fibroblasts associated with elevated NF-kappaB activity. Immun Ageing. 2008;5:5. doi: 10.1186/1742-4933-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–31. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, et al. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol Cell Biol. 2008a;28:3076–87. doi: 10.1128/MCB.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008b;7:2720–6. doi: 10.4161/cc.7.17.6580. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Ly DH, Lockhart DJ, Lerner RA, Schultz PG. Mitotic misregulation and human aging. Science. 2000;287:2486–92. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- Mansfield LV. Dissecting the telomere –independent pathways underlying human cellular senescence. University of London; 1996. PhD thesis. [Google Scholar]
- Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–18. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 2002;69:1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- O’hare MJ, Bond J, Clarke C, Takeuchi Y, Atherton AJ, Berry C, et al. Conditional immortalization of freshly isolated human mammary fibroblasts and endothelial cells. Proc Natl Acad Sci USA. 2001;98:646–651. doi: 10.1073/pnas.98.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci USA. 2009;106:17031–6. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossovskaya VS, Mazo IA, Chernov MV, Chernova OB, Strezoska Z, Kondratov R, et al. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA. 1996;93:10309–14. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Wang Z, Costa RH, Tyner A, Lau LF, Raychaudhuri P. An N-terminal inhibitory domain modulates activity of FoxM1 during cell cycle. Oncogene. 2008;27:1696–704. doi: 10.1038/sj.onc.1210814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ, et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo M, Massa PE, Olivotto E, Bianchi F, Borzi RM, Hanidu A, et al. Sustained NF-kappaB activation produces a short-term cell proliferation block in conjunction with repressing effectors of cell cycle progression controlled by E2F or FoxM1. J Cell Physiol. 2009;218:215–27. doi: 10.1002/jcp.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl. Acad. Sci. U S A. 2004;101:9309–14. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, Denissov SG, Douma S, Stunnenberg HG, Bernards R, Peeper DS. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell. 2002;2:55–65. doi: 10.1016/s1535-6108(02)00085-5. [DOI] [PubMed] [Google Scholar]
- Salminen A, Huuskonen J, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Activation of innate immunity system during aging: NF-κB signalling is the molecular culprit of inflamm-aging. Ageing Res Rev. 2008;7:83–105. doi: 10.1016/j.arr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton DN, Chang E, Whittier PS, Choi D, Funk WD. Microarray analysis of replicative senescence. Curr Biol. 1999;9:939–45. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia V, Ding M, Kumar S, Nie D, Premkumar LS, Elble RC. hCLCA2 Is a p53-Inducible Inhibitor of Breast Cancer Cell Proliferation. Cancer Res. 2009;69:6624–32. doi: 10.1158/0008-5472.CAN-08-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.