Abstract
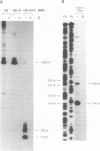
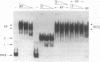
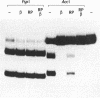
The beta protein encoded by the Streptococcus pyogenes plasmid pSM19035 is a site-specific recombinase involved in both resolution of plasmid multimers into monomers and DNA inversion. It has been proposed that the DNA region to which the beta recombinase binds to mediate recombination includes a promoter from which orf alpha and the beta gene are transcribed. We have determined the sites at which transcription of the orf alpha and the beta gene initiates in vitro and we have demonstrated that highly purified beta recombinase acts as a repressor of its own synthesis. The promoters are located within the beta recombinase binding site, which we have defined previously. The binding of the beta recombinase to its target site does not seem to exclude RNA polymerase from the promoter, despite the overlapping of their binding sites. Therefore, it is likely that the beta recombinase does not repress transcription by a mere steric hindrance on RNA polymerase binding.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behnke D., Tomich P. K., Clewell D. B. Electron microscopic mapping of deletions on a streptococcal plasmid carrying extraordinarily long inverted repeats. Plasmid. 1980 Sep;4(2):139–147. doi: 10.1016/0147-619x(80)90003-7. [DOI] [PubMed] [Google Scholar]
- Ceglowski P., Boitsov A., Karamyan N., Chai S., Alonso J. C. Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035-derived plasmids in Bacillus subtilis. Mol Gen Genet. 1993 Dec;241(5-6):579–585. doi: 10.1007/BF00279900. [DOI] [PubMed] [Google Scholar]
- Cegłowski P., Boitsov A., Chai S., Alonso J. C. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene. 1993 Dec 22;136(1-2):1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- Gourse R. L. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988 Oct 25;16(20):9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D., Prentki P., Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992 Dec;56(4):509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langert W., Meuthen M., Mueller K. Functional characteristics of the rrnD promoters of Escherichia coli. J Biol Chem. 1991 Nov 15;266(32):21608–21615. [PubMed] [Google Scholar]
- Lee J., Goldfarb A. lac repressor acts by modifying the initial transcribing complex so that it cannot leave the promoter. Cell. 1991 Aug 23;66(4):793–798. doi: 10.1016/0092-8674(91)90122-f. [DOI] [PubMed] [Google Scholar]
- Leirmo S., Gourse R. L. Factor-independent activation of Escherichia coli rRNA transcription. I. Kinetic analysis of the roles of the upstream activator region and supercoiling on transcription of the rrnB P1 promoter in vitro. J Mol Biol. 1991 Aug 5;220(3):555–568. doi: 10.1016/0022-2836(91)90100-k. [DOI] [PubMed] [Google Scholar]
- Malke H. Genetics of resistance to macrolide antibiotics and lincomycin in natural isolates of Streptococcus pyogenes. Mol Gen Genet. 1974;135(4):349–367. doi: 10.1007/BF00271149. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989 Feb 24;56(4):521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Shibuya G. I., Steitz J. A. Nucleotide sequence of gamma delta resolvase gene and demonstration that its gene product acts as a repressor of transcription. Nature. 1982 Nov 25;300(5890):381–383. doi: 10.1038/300381a0. [DOI] [PubMed] [Google Scholar]
- Rojo F., Nuez B., Mencía M., Salas M. The main early and late promoters of Bacillus subtilis phage phi 29 form unstable open complexes with sigma A-RNA polymerase that are stabilized by DNA supercoiling. Nucleic Acids Res. 1993 Feb 25;21(4):935–940. doi: 10.1093/nar/21.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F., Weise F., Alonso J. C. Purification of the beta product encoded by the Streptococcus pyogenes plasmid pSM19035. A putative DNA recombinase required to resolve plasmid oligomers. FEBS Lett. 1993 Aug 9;328(1-2):169–173. doi: 10.1016/0014-5793(93)80987-6. [DOI] [PubMed] [Google Scholar]
- Schickor P., Metzger W., Werel W., Lederer H., Heumann H. Topography of intermediates in transcription initiation of E.coli. EMBO J. 1990 Jul;9(7):2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straney S. B., Crothers D. M. Lac repressor is a transient gene-activating protein. Cell. 1987 Dec 4;51(5):699–707. doi: 10.1016/0092-8674(87)90093-6. [DOI] [PubMed] [Google Scholar]
- Wells R. G., Grindley N. D. Analysis of the gamma delta res site. Sites required for site-specific recombination and gene expression. J Mol Biol. 1984 Nov 15;179(4):667–687. doi: 10.1016/0022-2836(84)90161-x. [DOI] [PubMed] [Google Scholar]
- Whipple F. W., Sonenshein A. L. Mechanism of initiation of transcription by Bacillus subtilis RNA polymerase at several promoters. J Mol Biol. 1992 Jan 20;223(2):399–414. doi: 10.1016/0022-2836(92)90660-c. [DOI] [PubMed] [Google Scholar]
- Williams D. R., Motallebi-Veshareh M., Thomas C. M. Multifunctional repressor KorB can block transcription by preventing isomerization of RNA polymerase-promoter complexes. Nucleic Acids Res. 1993 Mar 11;21(5):1141–1148. doi: 10.1093/nar/21.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart W. L., Machida C., Ohtsubo H., Ohtsubo E. Escherichia coli RNA polymerase binding sites and transcription initiation sites in the transposon Tn3. Gene. 1983 Sep;24(1):99–113. doi: 10.1016/0378-1119(83)90135-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]