Abstract
Background. Parvovirus 4 (PARV4) is a recently identified human virus that has been found in livers of patients infected with hepatitis C virus (HCV) and in bone marrow of individuals infected with human immunodeficiency virus (HIV). T cells are important in controlling viruses but may also contribute to disease pathogenesis. The interaction of PARV4 with the cellular immune system has not been described. Consequently, we investigated whether T cell responses to PARV4 could be detected in individuals exposed to blood-borne viruses.
Methods. Interferon γ (IFN-γ) enzyme-linked immunospot assay, intracellular cytokine staining, and a tetrameric HLA-A*0201–peptide complex were used to define the lymphocyte populations responding to PARV4 NS peptides in 88 HCV-positive and 13 HIV-positive individuals. Antibody responses were tested using a recently developed PARV4 enzyme-linked immunosorbent assay.
Results. High-frequency T cell responses against multiple PARV4 NS peptides and antibodies were observed in 26% of individuals. Typical responses to the NS pools were >1000 spot-forming units per million peripheral blood mononuclear cells.
Conclusions. PARV4 infection is common in individuals exposed to blood-borne viruses and elicits strong T cell responses, a feature typically associated with persistent, contained infections such as cytomegalovirus. Persistence of PARV4 viral antigen in tissue in HCV-positive and HIV-positive individuals and/or the associated activated antiviral T cell response may contribute to disease pathogenesis.
There are 4 parvoviruses known to infect humans: adeno-associated virus, human bocavirus, B19, and parvovirus 4 (PARV4) [1]. PARV4 was first identified in 2005 in an injecting drug user (IDU) with acute onset of symptoms including fatigue, vomiting, arthralgias, and confusion [2]. PARV4 has been found in the livers of 7/18 advanced hepatitis C virus (HCV)–positive patients [3] and in the bone marrow of 31/58 human immunodeficiency virus (HIV)–positive individuals [4, 5].
PARV4 has a 5 kb linear genome of single-stranded DNA that contains 2 open reading frames (ORFs): ORF1 and ORF2 are genetically and likely functionally homologous to NS and VP proteins, respectively, in other parvoviruses. They share greatest nucleotide identity (85%) with a chimpanzee variant recently found in Cameroon [6] but are also similar to parvoviruses in wild boars, pigs, and cows (porcine and bovine hokovirus) [7, 8]. These sequence relationships likely justify the classification of these viruses into a new genus. PARV4 has 3 known genotypes: genotypes 1 and 2 are found mainly in Western countries, whereas genotype 3 has only been detected in sub-Saharan Africa [9–12].
Most studies to date have looked for the presence of viral DNA in various populations. PARV4 DNA has been detected in 0–5% of healthy blood donors in 8 studies (median: 1%) [13–20] and in up to 85% of IDU HIV-positive individuals with AIDS [15]. The frequency of coinfection with HCV has varied from 8% to 30% [15, 20, 21]. A recently developed enzyme-linked immunosorbent assay (ELISA) has allowed the comparison between DNA detection, which is rarely positive in peripheral blood, and the presence of antibodies to PARV4 [18]. Thirty-three percent of HCV-positive individuals and 67% of HIV/HCV-coinfected individuals were seropositive. It has been suggested that frequent coinfection of PARV4 in HIV- and HCV-positive individuals is due to a shared transmission route [15]. Parenteral transmission is supported by the high prevalence of PARV4 DNA and antibodies in hemophiliacs [15, 18] and the relatively low prevalence in non-IDU men who have sex with men (MSM) [18]. Nonetheless, low frequencies of seropositivity are found among these HCV-negative MSM. Additionally, high incidence of PARV4 infection has been described in healthy infants from Ghana [9], in the general population in Central Africa, and in HIV-1–positive individuals without parenteral risk factors [10, 22], suggesting alternative transmission routes.
The T cell immune response plays a major role in viral clearance, for example, in HCV [23]. Previous studies on parvovirus B19, a more common human infection, have shown that this family of viruses can trigger surprisingly strong and long-lived cellular responses (∼900 spot-forming units per million peripheral blood mononuclear cells [SFU/106 PBMCs] per pool of 10 peptides, detected up to 32 months after onset of symptoms) especially to the nonstructural protein NS1 of B19 [24, 25].
In view of the importance of cellular immunity in controlling infection, and since the T cell immune response to PARV4 has not been studied previously, we investigated whether HCV- and HIV-positive individuals made such a response. Stimulated T cells produce interferon γ (IFN-γ) as part of the antiviral response, and this cytokine can be detected using enzyme-linked immunospot (ELISpot) assays to measure magnitude and specificity. We present here the first data on the breadth, magnitude, specificity, and phenotype of the T cell responses to PARV4.
METHODS
Study Subjects and Sample Collection
Informed patient consent was obtained from 4 groups of individuals: 88 HCV-positive (HIV-negative) individuals (Supplementary Table), 13 HIV-positive (HCV-negative) individuals, 7 acute B19-seropositive (immunoglobulin M [IgM]) individuals, and 9 healthy individuals for control tests (local ethical approvals: COREC O4.OXA.010, 06/Q1604/2, and CO2.113). Blood samples were obtained while patients were not undergoing HCV treatment (except for one patient, with no PARV4-specific response); of those previously treated (43/88), samples were a minimum of 5 months post-treatment (range: 5–77 months, median: 25). PBMCs were obtained from heparinized blood by density gradient centrifugation and were either tested directly or frozen until needed.
Synthetic Peptides
One hundred thirty-one peptides of 15 amino acids in length, overlapping by 10 amino acids spanning the entire PARV4 NS protein of the prototype genotype 1 sequence (AY622943), were synthesized (>70% pure, fluorenylmethyloxycarbonyl chemistry [Weatherall Institute of Molecular Medicine, Oxford]), weighed, and dissolved in dimethyl sulfoxide (DMSO [Sigma]). These were combined into 13 pools of 10 peptides each.
IFN-γ ELISpot Assays
ELISpot assays were carried out ex vivo unless mentioned, with PARV4 NS pools and individual peptides as described elsewhere [26]. Then 2 × 105 PBMCs were plated per well in duplicate and incubated overnight at 37°C, 5% CO2, with peptides at 1 μmol/L. Wells containing PBMCs with DMSO or concavidin A (20 μg/mL [Sigma]) were used as negative or positive controls, respectively. The plates were read on an automated ELISpot reader (AID), and results were presented as SFU/106 PBMCs. A well was considered positive if above 55 SFU/million PBMC per pool of peptides after subtraction against the average of duplicate background wells. This cutoff has been extensively evaluated using negative controls in ongoing vaccine trials in our group [27] and in HIV ELISpots [28]. This value is also above both the mean value +3 standard deviations (= 14) and the highest count seen in the controls of this study (healthy and B19-positive individuals).
Serological Screening
Serum samples from 84 of 88 individuals were tested for anti-PARV4 immunoglobulin G (IgG; and additionally IgM in a subset) as described elsewhere [18].
PBMC Culture
PBMCs were pulsed with 25 μmol/L peptide for 30 minutes before incubating at 37°C, 5% CO2, for 10–14 days, supplementing every 3 days with natural human T cell growth factor (10% vol/vol [Helvetica Healthcare]). Cells were harvested and assayed by intracellular cytokine staining (ICS). After 14 days of culture, PBMCs could be maintained with HLA-matched B cell lines (BCL) pulsed with PARV4 peptide (12.5 μmol/L for 1 h at 37°C) and 3 healthy donor PBMCs to use as feeders. BCLs and healthy PBMCs were irradiated with 30 Gy. Cells were resuspended with the individual PBMCs to culture at a 1:1 ratio.
Intracellular Cytokine Staining, Surface Staining, and Flow Cytometry Analysis
Cells were counted and aliquoted at ∼106 per condition. Negative and positive controls were set up with DMSO and with phorbol myristate acetate (.05 μg/mL [Sigma]) and ionomycin (.5 μg/mL [Sigma]), respectively. Samples were stimulated with 3 μmol/L peptide. Then 2 μg/mL brefeldin A (Sigma) was added to each tube, and they were incubated at 37°C, 5% CO2, for 5–6 h. Cells were surface stained with fluorochrome-conjugated antibodies and processed for analysis or, for ICS, were permeabilized (BD Cytofix/Cytoperm) and then stained for intracellular markers for Fluorescence-Activated Cell Sorting (FACS) analysis. Fluorochrome-conjugated antibodies used were live/dead marker L34955, CD4-QDot 605 (Invitrogen), CD8-APC, CD8-PE-Cy7, IFNγ–Alexa Fluor 700, CCR7-PE-Cy7 (BDPharmingen), CD8-PerCP-Cy5.5, IFNγ–FITC (BD Biosciences), CD45 RA-APC (AbCam), and HLA-A*0201-RMT-PE tetramer (synthesized as described elsewhere [29]). All samples were processed on a BD LSR II and analyzed using FlowJo software.
Lymphocytes were gated on forward and side scatter, and then live CD3+ cells were gated. The FACS plots presented in this study are gated on CD3+ cells uness indicated.
DNA Extraction and HLA Typing
DNA was extracted using the Gentra Puregene kit (Qiagen) from PBMCs or from whole blood, as described by the manufacturer. HLA typing was performed using the Sequence-Specific Primer method (Weatherall Institute of Molecular Medicine, Oxford) [30].
Epitope Prediction
Bioinformatics programs Syfpeithi (http://www.syfpeithi.de) and Bimas (http://www-bimas.cit.nih.gov/molbio/hla_bind/) were used to predict potential epitopes. These were recorded if their score was above 20 for Syfpeithi and above 50 for Bimas. The peptide RMTENIVEV identified through experiments was a 9 amino acid epitope that scored 27 in Syfpeithi and 474 in Bimas.
Statistics
Statistics were calculated using the Mann-Whitney test and Fisher exact test using GraphPad Prism software.
RESULTS
HCV-Positive Individuals Have Strong T Cell Responses to PARV4 NS Peptides That Are Maintained Over Time
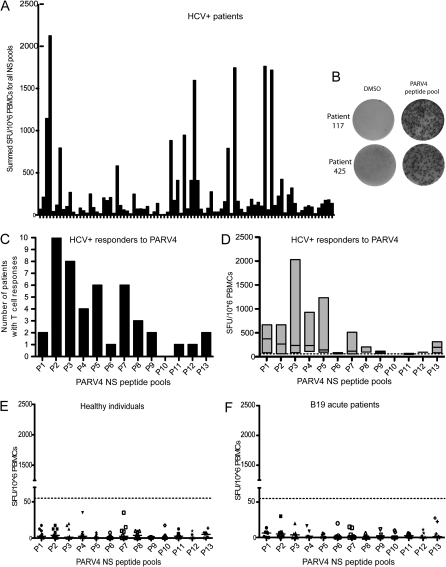
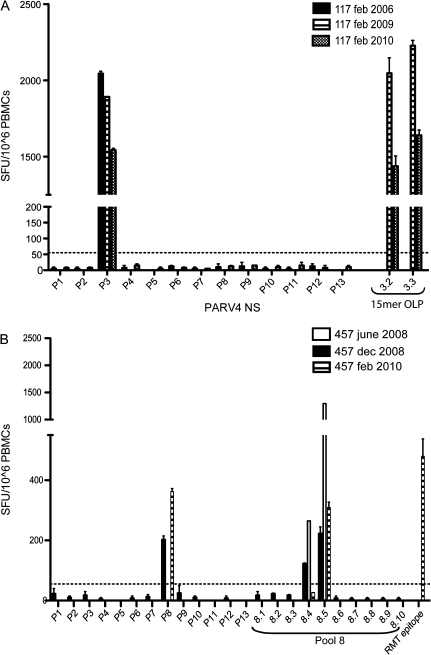
PBMCs from 88 randomly selected HCV-positive individuals attending a hospital as outpatients, 9 healthy individuals, and 7 individuals with acute parvovirus B19 infection were assayed by ELISpot for the production of IFN-γ in response to 13 pools of 10 PARV4 NS peptides. Summed responses of all HCV-positive individuals to the entire NS protein are shown in Figure 1A. Twenty-three HCV-positive individuals (26%) had T cell responses to individual pools higher than the predefined cutoff 55 SFU/106 PBMC (see Methods), and 18 had T cell responses higher than 250 SFU/106 PBMC, with a maximum of 2045 SFU/106 PBMC per peptide pool (Figure 1C–1D). Examples of the highest responses seen in this assay are presented in Figure 1B. PBMC samples from one time point were also confirmed in other assays such as ICS (see below). Additionally, a selection of responders in the cohort were tested longitudinally: 6/7 individuals with positive responses remained positive across periods of 3 months to 3 years (Table 1 and Figure 2). The T cell response of the 7th individual was recovered upon culture of the PBMCs with peptide (data not shown). Likewise, 3/3 individuals with no response remained negative over time. Responses to the N-terminal portion of the NS protein were both higher in magnitude and more frequent (pools 1–7: 37 responses, median: 6; pools 8–13: 9 responses, median: 1.5; Mann-Whitney test, P = .036; Figures 1C–1D). Some individuals with T cell responses to pools were tested against individual peptides; in total, 13 individual peptides were identified (data not shown and see below).
Figure 1.
Analysis of PARV4 T cell responses in HCV-infected patients and controls. T cell responses were assessed by IFN-γ ELISpot assays with PARV4 NS peptides. (A) T cells from 88 HCV-infected patients to each peptide pool are summed so that each column represents the response of one patient to the entire NS protein. (B) These photos of ELISpot wells are examples of the highest levels of responses and their negative control wells (DMSO). Peptide pool responses for patients 117 and 425 are 410 and 248 SFU/well, respectively. (C–D) These graphs show the distribution and the magnitude of the T cell responses across the NS protein. (C) Each column represents the number of patients responding to each pool. These are concentrated at the N-terminus of the protein. (D) Each column represents the range of responses of all responders and the median to that peptide pool. PARV4 NS responders indicate preferential recognition of N-terminal pools of peptides. (E–F) T cell responses of 9 healthy individuals (E) and 7 B19-positive patients (F) to PARV4 NS peptide pools are displayed. Each point represents the T cell response of one individual to the peptide pool on the x-axis. The highest response was 35 SFU/106 PBMC. The dashed line marks the cutoff for positivity.
Table 1.
T Cell Responses to PARV4 Are Maintained Over Time
| Patient No. | First Time Point |
Second Time Point |
||
| Sample Date | Result | Sample Date | Result | |
| 117 | Feb-06 | + | Feb-09 | + |
| 123 | Oct-06 | − | Nov-09 | − |
| 146 | Jul-08 | + | Oct-09 | + |
| 162 | Feb-06 | − | May-09 | − |
| 289 | Mar-07 | − | May-09 | − |
| 324 | Oct-06 | + | Nov-09 | − |
| 382 | Apr-07 | + | Nov-09 | + |
| 422 | Apr-08 | + | Oct-08 | + |
| 433 | Feb-08 | + | May-08 | + |
| 457 | Dec-08 | + | Feb-10 | + |
NOTE. A positive response (+) is defined by at least 1 pool eliciting a T cell response >55 SFU/106 PBMC. Only the response of patient 324 was not detectable ex vivo at the second time point (data not shown). –, the absence of response.
Figure 2.
Longitudinal analysis of T cell responses to PARV4 NS. T cell responses of patients 117 (A) and 457 (B) are illustrated here as examples of sustained responses over time. The dates next to the patient numbers represent the sample date. (”457 june 2008” sample: these cells were cultured with peptide for 22 days before the assay.) OLP, overlapping peptides.
As controls, 9 healthy “low-risk” individuals were tested; the maximum median of all individuals for one single pool was 3.75 SFU/106 PBMC, with responses reaching a maximum of 35 SFU/106 PBMC per peptide pool (ie, under the predefined cutoff [Figure 1E]). Seven individuals with acute parvovirus B19 infection also had low responses, with a maximum median of all individuals for one single pool of 6.25 SFU/106 PBMC and an individual maximum of 30 SFU/106 PBMC (Figure 1F). The T cell responses of healthy controls and individuals with acute B19 infection did not differ significantly (Mann-Whitney test, P = .61).
Analysis of Antibody Responses in Relation to T Cell Responses
Serum samples from 84 of 88 HCV-positive individuals and 12 of 16 control individuals (5 B19 positive and 7 healthy) were available for serological tests. Anti-PARV4 antibodies were found in 22 HCV-positive individuals (26% seroprevalence) and in no control individuals. Fourteen individuals had both antibodies and T cell responses, 7 showed T cell responses but were seronegative, 8 were seropositive for PARV4 antibodies, and 55 had neither (data not shown). To investigate the frequent discrepancies in T cell and serological reactivity, we repeated and retested antibody levels and T cell responses at several time points. For example, individual 117 was tested for antibodies on serum samples from 2 time points, and both were negative. On the other hand, PBMC samples from these same time points were both positive and at very high levels (2045 and 1890 SFU/million PBMC per peptide pool [Figure 2, antibody data not shown]). These 7 individuals lacking anti-PARV4 IgG were also negative in an additional ELISA for anti-PARV4 IgM. Overall, the correlation between T cell responses and IgG detection was significant (Fisher exact test, P < .0001), although clearly individuals may possess T cell responses without detectable antibody and vice versa.
HIV-Positive Individuals Elicit a Similar Rate of Response as the HCV-Positive Individuals
We next investigated whether T cell responses to PARV4 could be detected in individuals with another persistent infection. We studied 13 HIV-positive HCV-negative individuals for IFN-γ production in response to PARV4 NS. Figure 3 shows that T cell responses were detected at a comparable frequency and level to the HCV cohort; 3 individuals had responses over 55 SFU/106 PBMC per peptide pool. This figure shows that, as in HCV-positive individuals, the responses targeted the N-terminal half of the NS protein. Anti-PARV4 antibodies were detected in 2/13 HIV-positive individuals, only one of whom also demonstrated a T cell response to PARV4 peptides. Thus, an immune response to PARV4 was detectable in HIV-positive individuals and comparable to those in the HCV-positive cohort.
Figure 3.
T cell responses to PARV4 NS protein in HIV-infected individuals. Results are displayed as in Figures 1E–1F. Three of 13 HIV-positive patients had a positive response to PARV4 NS peptide pools (N067, N090, and R010, with CD4+ T cell counts of 430, 350, and 460 cells/μL, respectively), and the distribution of responses across the NS protein is shown. Individuals with no responses had CD4+ T cell counts between 130 and 1300 cells/μL (median: 430 cells/μL).
Identification of PARV4-Specific CD8+ T Cells and an HLA-A*0201–Restricted PARV4 Epitope RMTENIVEV
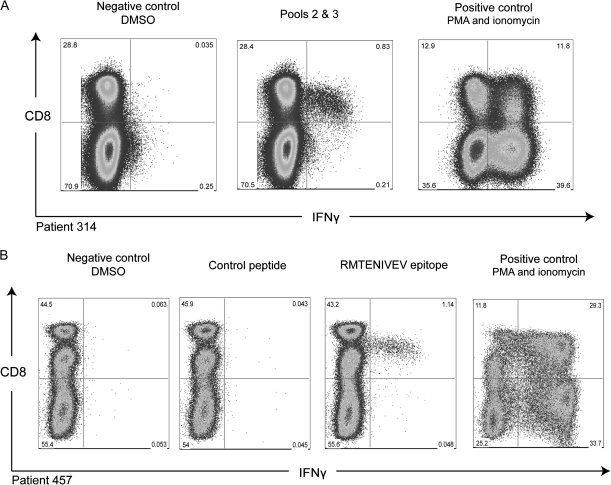
The nature of the T cell immune response to PARV4 peptides was further characterized through ICS assays. Due to restricted PBMC samples, not all individuals could be tested. Figure 4 shows that CD8+ T cells produced IFN-γ in response to PARV4 NS peptides in 2 individuals. Individual 314 made a strong CD8+ T cell response to NS pools 2 and 3, constituting 2.8% of total CD8+ T cells. Individual 457, who was HLA-A*0201 positive, made a similar CD8+ T cell response (2.6%) to peptides 8.4 (WWEEGRMTENIVEVA) and 8.5 (RMTENIVEVAKAVLG). The overlapping portion of these 2 peptides contained the HLA-A*0201 anchor residues methionine at P2 and valine at P9 [32], and the epitope RMTENIVEV scored highly using binding algorithms (see Methods). This peptide was synthesized and tested in functional assays. Figure 4 (bottom panel) shows that this peptide triggers strong production of IFN-γ. Thus this epitope was synthesized on 2 separate occasions: first as a 15mer and second as a 9mer, both triggering T cell responses. No other individual was found to respond to this peptide.
Figure 4.
Demonstration of PARV4 NS peptides triggering the production of IFN-γ by CD8+ T cells. Short-term PBMC lines for patients 314 (upper panel) and 457 (lower panel) elicit a CD8+ T cell response to 2 pools of peptides and one 9mer epitope, respectively. The control peptide is a tetanus toxin peptide [31].
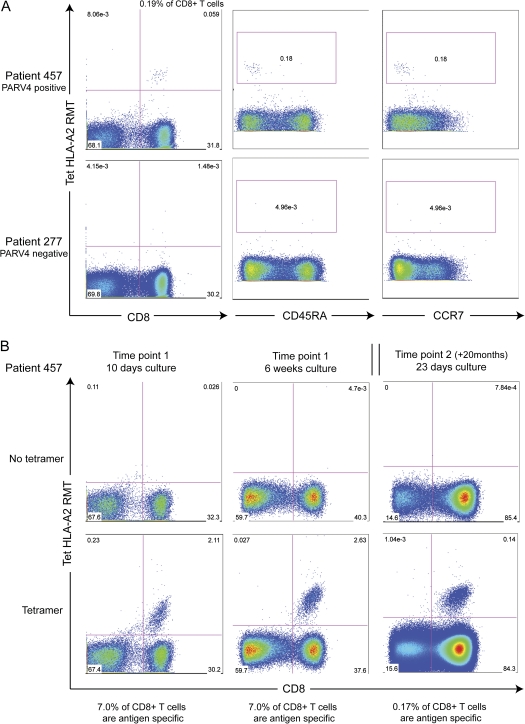
To further quantify and phenotype the T cell response to the RMTENIVEV epitope, a fluorescent-labeled tetrameric HLA-A*0201–peptide complex (HLA-A*0201-RMT tetramer) was synthesized and used to stain the PBMC of individual 457. Staining revealed that .19% of peripheral CD8+ T cells (or .06% of CD3+ cells) specifically recognized this epitope ex vivo (Figure 5A, left panels). Ex vivo cells were stained for CCR7 and CD45RA to identify their memory phenotype. As shown in Figure 5A (middle and right panels), cells stained with HLA-A*0201-RMT tetramer in this individual were CCR7 and CD45RA negative, suggesting that they are effector memory T cells. After in vitro culture with peptide for 10 days and 6 weeks, 7.0% of CD8+ T cells were antigen specific (Figure 5B, left and middle panels). A second sample taken 20 months after the first was cultured for 23 days, and antigen-specific cells were still detected (.17% of CD8+ T cells [Figure 5B, right panels]). This provides evidence that the cells were maintained over time in vivo and were able to proliferate in culture.
Figure 5.
HLA-A*0201–restricted PARV4 RMTENIVEV-specific T cells have an effector memory phenotype and proliferate in culture in vitro. (A) FACS plots for patient 457 (top panel), from whom the epitope was determined (HLA-A*0201 and positive response for peptides 8.4–8.5), and for patient 277 (lower panel, ”negative” = no response on ELISpot assays to PARV4 NS). Left-hand plots are gated on CD3+ cells. The middle and right-hand panels assess the phenotype of the epitope-specific cells and are gated on CD3+ CD8+ T cells. CD45RA and CCR7 are used as memory markers. Antigen-specific cells are visible in patient 457 at a frequency of .19% and do not present CD45RA or CCR7 on their surface. (B) After short-term culture, tetramer HLA-A*02-RMT–positive cells have expanded to 7% of CD8+ T cells. This appears to plateau as the level is unchanged after 6 weeks (middle panels). Tetramer-positive cells are still visible at a second time point (right panels) in the same patient, although the percentage decreases. Samples from time points 1 and 2 were taken 20 months apart.
DISCUSSION
We detected strong T cell responses to PARV4 NS in 23/88 HCV-positive individuals (26%) and in 3/13 HIV-positive individuals, 2 cohorts of individuals exposed to blood-borne viruses. These data suggest that PARV4 infections are relatively common in these clinical groups and that viral antigen persists to trigger and maintain these responses. We also identify the first (HLA-A*0201–restricted) PARV4 peptide epitope, RMTENIVEV, and show that CD8+ T cells specific for this epitope in one individual have an effector memory phenotype. The T cell responses are PARV4 specific, as there were no T cell responses to PARV4 NS in healthy individuals or in acute B19 seroconverters. Up to 60% of the adult population has been exposed to B19 [33], and our data therefore exclude the possibility of antigenic cross-reactivity with B19 parvovirus. Cross-reactivity with HCV is unlikely for several reasons. Responses were detected in an HCV-negative HIV-positive group; the key epitope, RMTENIVEV, does not have a clear homologue in HCV; T cell responses to HCV-derived peptides in this patient group with persistent HCV infection are of extremely low frequency [34]. Additionally, there is a strong association between PARV4 serostatus and PARV4 T cell response (see Results).
The strong T cell responses reached 2000 SFU/million PBMCs to a single 15mer peptide; thus PARV4 appears to be highly immunogenic, at least in certain individuals. These T cell levels are comparable to other low-load persistent viruses such as cytomegalovirus (CMV) [35] and Epstein-Barr virus (EBV) [36].
Most of the responses were concentrated at the N-terminus of the NS protein in both HCV-positive and HIV-positive cohorts. This protein is poorly characterized functionally, although it contains 2 known motifs: a rolling circle replication motif, found at the N-terminus, and an adenosine triphosphatase domain [11, 37, 38]. The latter was represented in pool 7 of the NS peptides and was shown to frequently elicit T cell responses. Within the N-terminus of the NS protein there was no immunodominant epitope.
T cell responses were found at a similar frequency to that previously found for PARV4 antibodies (33%) [18] and for PARV4 DNA (30%) [21] in other HCV-positive individuals. There was no relationship found between immune reactivity to PARV4 and age (Mann-Whitney, P = .71; Supplementary Table). Parvovirus B19 has been shown to persist at very low levels in bone marrow, skin, and synovia [39–41]. PARV4 may behave in a similar manner, having been detected in autopsy tissue or at very low viral load in tissue without viremia [4]. The T cell responses observed against PARV4 were sustained for periods up to 3 years, similar to that observed for T cell responses to parvovirus B19 [42]. This continuity supports the hypothesis that PARV4 is indeed a low-level persistent virus, providing a continuous supply of antigen to stimulate CD8+ T cells. PARV4-specific CD8+ T cells have an effector memory phenotype (CCR7–CD45RA– [Figure 5]), suggesting recurrent antigen exposure as seen in CMV [43, 44]. The study of HIV-positive individuals shows that T cell responses to PARV4 NS are detectable even in individuals with a degree of immune suppression (average CD4+ T cell count of the 3 responders was 410 cells/μL).
Of the 84 HCV-positive individuals tested in both assays, and 21 with T cell responses to PARV4, only 14 also had antibodies to PARV4. Moreover, 8 more individuals had antibodies to PARV4, with no T cell responses. There could be several reasons for this. First, the detection thresholds of the 2 assays are not directly comparable, as one detects circulating antibody and the other, individual cytokine-secreting cells, associated with different sensitivities. The lack of IgM in individuals lacking IgG suggests that they were not acutely infected. There may be an issue of differential specificity, since the ELISpot assay identifies immune responses to the PARV4 NS protein, whereas the ELISA detects antibodies to PARV4 VP protein. As VP is a capsid protein, it is expected that it would be a primary target for antibodies, whereas CD8+ T cells may preferentially recognize the NS protein, once expressed intracellularly. So, potentially, where there is antibody but no T cells detected, this may indicate induction of VP-specific T cells rather than NS-specific T cells. A similar phenomenon has been reported for parvovirus B19 [45, 46]. Further T cell studies using VP peptides are in progress to address this issue.
Both assays look at the peripheral blood T cells in individuals. While antibodies may be freely circulating, it is possible that T cells activated by PARV4 are compartmentalized to tissues, as seen in CMV, EBV, and HCV infections where CD8+ T cells are enriched in the liver [47]. Notably, PARV4 DNA has also been found in the liver, among other tissues [3–5]. The lack of VP-specific antibodies is surprising but was consistent as both assays were repeated for several individuals at multiple time points. There may have been a loss of circulating antibody over time, as occurs with HCV after spontaneous control, where T cells may be detected over longer periods [48]—sensitive analysis of antigen-specific memory B cells might reveal low-frequency populations, as we have previously described after hepatitis B virus vaccination [49]. Maintenance of T cell responses in such seronegative persons might then reflect low-level persistence of antigen in tissue or potentially continued limited transcription of NS—analogous to the setting in CMV infection [50]. Antigenic variation within parvoviruses resulting from distinct circulating genotypes may also impact on the sensitivity of both assays, although this should be limited as there is minimal amino acid sequence variability among the 3 genotypes [10].
Planned experiments on longitudinal samples will allow us to look at any changes in antibody levels and T cell responses. In this study, no polymerase chain reaction tests for DNA were carried out, as previous studies in comparable cohorts revealed that DNA was only detected at very low frequency in peripheral blood [3, 4, 8].
Parenteral infection appears to be the principal route of transmission without being exclusive, as viral DNA has been found in a small percentage of healthy adults [13–20], in healthy infants in Ghana [9], and in HIV-positive individuals who had no history of parenteral exposure [10]. A recent study has also reported that up to 37% of the general population in some countries in Central Africa are seropositive for PARV4, suggesting additional transmission routes [22]. The safety of blood products administered to vulnerable populations warrants further investigation into this blood-borne agent. So far, there is no clear link between PARV4 infection and disease, largely complicated by the burden of disease in HCV- and HIV-infected individuals. Exploring the role of T cells in unconventional persistent viral infections such as B19 or PARV4 will contribute to the understanding of the antiviral immune response.
This study is the first to identify T cell responses and to define a CD8+ T cell epitope for the novel parvovirus PARV4. Until recently this virus was only detected by looking for viral DNA. Through the availability of serological assays [18] and now T cell assays, the prevalence of PARV4 in different populations and its effect on the immune system can be more readily assessed.
Supplementary Data
Supplementary data are available at http://jid.oxfordjournals.org/ online.
Funding
This work was supported by the Wellcome Trust (including WT091663MA); the National Institutes of Health (NIAID IU19AI082630/01); and the Medical Research Council, United Kingdom. The development and use of the serological assay for anti-PARV4 antibodies was supported by an unrestricted investigator-initiated grant from Baxter Healthcare and by the National Institutes of Health, National Institute of Child Health and Human Development (R01 HD41224). Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Supplementary Material
Acknowledgments
We would like to thank the Medical Research Council, United Kingdom; the Wellcome Trust; the National Institute for Health Research Biomedical Research Centre (Oxford); the James Martin 21st Century School (Oxford); Baxter Healthcare; and the National Institutes of Health, National Institute of Child Health and Human Development, for funding this research. The HLA typing was carried out by Timothy Rostron, and the peptides were synthesized by Katalin Di Gleria (Weatherall Institute of Molecular Medicine, Oxford).
References
- 1.Brown KE. The expanding range of parvoviruses which infect humans. Rev Med Virol. 2010;20:231–44. doi: 10.1002/rmv.648. [DOI] [PubMed] [Google Scholar]
- 2.Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J Virol. 2005;79:8230–6. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider B, Fryer JF, Reber U, et al. Persistence of novel human parvovirus PARV4 in liver tissue of adults. J Med Virol. 2008;80:345–51. doi: 10.1002/jmv.21069. [DOI] [PubMed] [Google Scholar]
- 4.Manning A, Willey SJ, Bell JE, Simmonds P. Comparison of tissue distribution, persistence, and molecular epidemiology of parvovirus B19 and novel human parvoviruses PARV4 and human bocavirus. J Infect Dis. 2007;195:1345–52. doi: 10.1086/513280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longhi E, Bestetti G, Acquaviva V, et al. Human parvovirus 4 in the bone marrow of Italian patients with AIDS. AIDS. 2007;21:1481–3. doi: 10.1097/QAD.0b013e3281e38558. [DOI] [PubMed] [Google Scholar]
- 6.Sharp CP, Lebreton M, Kantola K, et al. Widespread infection of chimpanzees and gorillas with homologues of human parvoviruses B19, PARV4 and human bocavirus in the wild. J Virol. 2010;84:10289–96. doi: 10.1128/JVI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adlhoch C, Kaiser M, Ellerbrok H, Pauli G. High prevalence of porcine hokovirus in German wild boar populations. Virol J. 2010;7:171. doi: 10.1186/1743-422X-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SK, Woo PC, Tse H, et al. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J Gen Virol. 2008;89:1840–8. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- 9.Panning M, Kobbe R, Vollbach S, et al. Novel human parvovirus 4 genotype 3 in infants, Ghana. Emerg Infect Dis. 2010;16:1143–6. doi: 10.3201/eid1607.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmonds P, Douglas J, Bestetti G, et al. A third genotype of the human parvovirus PARV4 in sub-Saharan Africa. J Gen Virol. 2008;89:2299–302. doi: 10.1099/vir.0.2008/001180-0. [DOI] [PubMed] [Google Scholar]
- 11.Fryer JF, Delwart E, Bernardin F, Tuke PW, Lukashov VV, Baylis SA. Analysis of two human parvovirus PARV4 genotypes identified in human plasma for fractionation. J Gen Virol. 2007;88:2162–7. doi: 10.1099/vir.0.82620-0. [DOI] [PubMed] [Google Scholar]
- 12.Botto S, Bergallo M, Sidoti F, et al. Detection of PARV4, genotypes 1 and 2, in healthy and pathological clinical specimens. New Microbiol. 2009;32:189–92. [PubMed] [Google Scholar]
- 13.Fryer JF, Delwart E, Hecht FM, et al. Frequent detection of the parvoviruses, PARV4 and PARV5, in plasma from blood donors and symptomatic individuals. Transfusion. 2007;47:1054–61. doi: 10.1111/j.1537-2995.2007.01235.x. [DOI] [PubMed] [Google Scholar]
- 14.Fryer JF, Kapoor A, Minor PD, Delwart E, Baylis SA. Novel parvovirus and related variant in human plasma. Emerg Infect Dis. 2006;12:151–4. doi: 10.3201/eid1201.050916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmonds P, Manning A, Kenneil R, Carnie FW, Bell JE. Parenteral transmission of the novel human parvovirus PARV4. Emerg Infect Dis. 2007;13:1386–8. doi: 10.3201/eid1309.070428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallerini D, Barozzi P, Quadrelli C, et al. Parvoviruses in blood donors and transplant patients, Italy. Emerg Infect Dis. 2008;14:185–6. doi: 10.3201/eid1401.070610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biagini P, Dussol B, Touinssi M, et al. Human parvovirus 4 in kidney transplant patients, France. Emerg Infect Dis. 2008;14:1811–2. doi: 10.3201/eid1411.080862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp CP, Lail A, Donfield S, et al. High frequencies of exposure to the novel human parvovirus PARV4 in hemophiliacs and injection drug users, as detected by a serological assay for PARV4 antibodies. J Infect Dis. 2009;200:1119–25. doi: 10.1086/605646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuke PW, Parry RP, Appleton H. Parvovirus PARV4 visualisation and detection. J Gen Virol. 2009;91:541–4. doi: 10.1099/vir.0.014852-0. [DOI] [PubMed] [Google Scholar]
- 20.Lurcharchaiwong W, Chieochansin T, Payungporn S, Theamboonlers A, Poovorawan Y. Parvovirus 4 (PARV4) in serum of intravenous drug users and blood donors. Infection. 2008;36:488–91. doi: 10.1007/s15010-008-7336-4. [DOI] [PubMed] [Google Scholar]
- 21.Fryer JF, Lucas SB, Padley D, Baylis SA. Parvoviruses PARV4/5 in hepatitis C virus–infected patient. Emerg Infect Dis. 2007;13:175–6. doi: 10.3201/eid1301.060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp C, Vermeulen M, Nébié Y, et al. Changing epidemiology of human parvovirus 4 infection in sub-Saharan Africa. Emerg Infect Dis. 2010;16:1605–7. doi: 10.3201/eid1610.101001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertolino PMG, Bowen DG. Immunological parameters influencing adaptive immune responses to the hepatitis C virus. epatitis C disease immunobiology and clinical applications. New York: Springer; 2007. pp. 39–70. [Google Scholar]
- 24.Tolfvenstam T, Oxenius A, Price DA, et al. Direct ex vivo measurement of CD8(+) T-lymphocyte responses to human parvovirus B19. J Virol. 2001;75:540–3. doi: 10.1128/JVI.75.1.540-543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isa A, Norbeck O, Hirbod T, et al. Aberrant cellular immune responses in humans infected persistently with parvovirus B19. J Med Virol. 2006;78:129–33. doi: 10.1002/jmv.20514. [DOI] [PubMed] [Google Scholar]
- 26.Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AV, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–65. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folgori ABE, Aston S, Smith K, et al. 1051 phase I trial of a highly immunogenic T cell vaccine for hepatitis C virus based on novel adenoviral vectors from rare serotypes. J Hepatol. 2009;50:S382. [Google Scholar]
- 28.Streeck H, Frahm N, Walker BD. The role of IFN-gamma Elispot assay in HIV vaccine research. Nat Protoc. 2009;4:461–9. doi: 10.1038/nprot.2009.7. [DOI] [PubMed] [Google Scholar]
- 29.Altman JD, Moss PA, Goulder PJ, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–6. [PubMed] [Google Scholar]
- 30.Bunce M. PCR-sequence-specific primer typing of HLA class I and class II alleles. Methods Mol Biol. 2003;210:143–71. doi: 10.1385/1-59259-291-0:143. [DOI] [PubMed] [Google Scholar]
- 31.Falugi F, Petracca R, Mariani M, et al. Rationally designed strings of promiscuous CD4(+) T cell epitopes provide help to Haemophilus influenzae type b oligosaccharide: a model for new conjugate vaccines. Eur J Immunol. 2001;31:3816–24. doi: 10.1002/1521-4141(200112)31:12<3816::AID-IMMU3816>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–6. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 33.Heegaard ED, Brown KE. Human parvovirus B19. Clin Microbiol Rev. 2002;15:485–505. doi: 10.1128/CMR.15.3.485-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauer GM, Barnes E, Lucas M, et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology. 2004;127:924–36. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Gillespie GM, Wills MR, Appay V, et al. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8(+) T lymphocytes in healthy seropositive donors. J Virol. 2000;74:8140–50. doi: 10.1128/jvi.74.17.8140-8150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callan MF. The evolution of antigen-specific CD8+ T cell responses after natural primary infection of humans with Epstein-Barr virus. Viral Immunol. 2003;16:3–16. doi: 10.1089/088282403763635401. [DOI] [PubMed] [Google Scholar]
- 37.Astell CR, Mol CD, Anderson WF. Structural and functional homology of parvovirus and papovavirus polypeptides. J Gen Virol. 1987;68:885–93. doi: 10.1099/0022-1317-68-3-885. [DOI] [PubMed] [Google Scholar]
- 38.Ding C, Urabe M, Bergoin M, Kotin RM. Biochemical characterization of Junonia coenia densovirus nonstructural protein NS-1. J Virol. 2002;76:338–45. doi: 10.1128/JVI.76.1.338-345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heegaard ED, Petersen BL, Heilmann CJ, Hornsleth A. Prevalence of parvovirus B19 and parvovirus V9 DNA and antibodies in paired bone marrow and serum samples from healthy individuals. J Clin Microbiol. 2002;40:933–6. doi: 10.1128/JCM.40.3.933-936.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norja P, Hokynar K, Aaltonen LM, et al. Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc Natl Acad Sci U S A. 2006;103:7450–3. doi: 10.1073/pnas.0602259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassinotti P, Burtonboy G, Fopp M, Siegl G. Evidence for persistence of human parvovirus B19 DNA in bone marrow. J Med Virol. 1997;53:229–32. [PubMed] [Google Scholar]
- 42.Isa A, Kasprowicz V, Norbeck O, et al. Prolonged activation of virus-specific CD8+ T cells after acute B19 infection. PLoS Med. 2005;2:e343. doi: 10.1371/journal.pmed.0020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kern F, Surel IP, Faulhaber N, et al. Target structures of the CD8(+)–T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J Virol. 1999;73:8179–84. doi: 10.1128/jvi.73.10.8179-8184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stone SF, Price P, French MA. Cytomegalovirus (CMV)-specific CD8+ T cells in individuals with HIV infection: correlation with protection from CMV disease. J Antimicrob Chemother. 2006;57:585–8. doi: 10.1093/jac/dkl049. [DOI] [PubMed] [Google Scholar]
- 45.Modrow S, Dorsch S. Antibody responses in parvovirus B19 infected patients. Pathol Biol (Paris) 2002;50:326–31. doi: 10.1016/s0369-8114(02)00302-4. [DOI] [PubMed] [Google Scholar]
- 46.Norbeck O, Isa A, Pohlmann C, et al. Sustained CD8+ T-cell responses induced after acute parvovirus B19 infection in humans. J Virol. 2005;79:12117–21. doi: 10.1128/JVI.79.18.12117-12121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward SM, Jonsson JR, Sierro S, et al. Virus-specific CD8+ T lymphocytes within the normal human liver. Eur J Immunol. 2004;34:1526–31. doi: 10.1002/eji.200324275. [DOI] [PubMed] [Google Scholar]
- 48.Takaki A, Wiese M, Maertens G, et al. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med. 2000;6:578–82. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 49.Ward SM, Phalora P, Bradshaw D, Leyendeckers H, Klenerman P. Direct ex vivo evaluation of long-lived protective antiviral memory B cell responses against hepatitis B virus. J Infect Dis. 2008;198:813–7. doi: 10.1086/591094. [DOI] [PubMed] [Google Scholar]
- 50.Simon CO, Holtappels R, Tervo HM, et al. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J Virol. 2006;80:10436–56. doi: 10.1128/JVI.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.