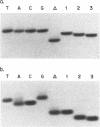
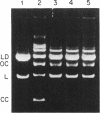
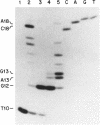
Abstract
Mutagenesis at abasic sites was investigated in E.coli and simian kidney (COS) cells using a duplex shuttle vector containing synthetic analogs of deoxyribose on the phosphodiester backbone. Lesions were positioned on opposite strands of the vector. When the tetrahydrofuranyl analog was used as the abasic site, AT or TA pairs (65-80%) were introduced at the site of the bistrand lesion. Mutagenesis occurred in the absence of SOS induction. Single base deletions (> 80%) dominated the mutational spectra for propanyl and ethanyl analogs of abasic sites lacking a ring structure. For all abasic site analogs, a small proportion of G/C and C/G pairs (6-10%) were observed. dAMP was incorporated predominantly opposite tetrahydrofuranyl sites positioned in the single strand region of a gapped duplex vector. We conclude from these studies that abasic sites positioned in a bistrand configuration are highly mutagenic in E.coli and COS cells. Repair DNA synthesis may be involved in this process.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett R. A., Swerdlow P. S., Povirk L. F. Spontaneous cleavage of bleomycin-induced abasic sites in chromatin and their mutagenicity in mammalian shuttle vectors. Biochemistry. 1993 Mar 30;32(12):3188–3195. doi: 10.1021/bi00063a034. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982 Dec 21;21(26):6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- Boosalis M. S., Mosbaugh D. W., Hamatake R., Sugino A., Kunkel T. A., Goodman M. F. Kinetic analysis of base substitution mutagenesis by transient misalignment of DNA and by miscoding. J Biol Chem. 1989 Jul 5;264(19):11360–11366. [PubMed] [Google Scholar]
- Cuniasse P., Fazakerley G. V., Guschlbauer W., Kaplan B. E., Sowers L. C. The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C and T opposite a model abasic site. J Mol Biol. 1990 May 20;213(2):303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- Cuniasse P., Sowers L. C., Eritja R., Kaplan B., Goodman M. F., Cognet J. A., Le Bret M., Guschlbauer W., Fazakerley G. V. Abasic frameshift in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Biochemistry. 1989 Mar 7;28(5):2018–2026. doi: 10.1021/bi00431a009. [DOI] [PubMed] [Google Scholar]
- Dedon P. C., Goldberg I. H. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem Res Toxicol. 1992 May-Jun;5(3):311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- Dianov G., Price A., Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992 Apr;12(4):1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentil A., Cabral-Neto J. B., Mariage-Samson R., Margot A., Imbach J. L., Rayner B., Sarasin A. Mutagenicity of a unique apurinic/apyrimidinic site in mammalian cells. J Mol Biol. 1992 Oct 20;227(4):981–984. doi: 10.1016/0022-2836(92)90513-j. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Hevroni D., Livneh Z. Bypass and termination at apurinic sites during replication of single-stranded DNA in vitro: a model for apurinic site mutagenesis. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5046–5050. doi: 10.1073/pnas.85.14.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kalnik M. W., Chang C. N., Grollman A. P., Patel D. J. NMR studies of abasic sites in DNA duplexes: deoxyadenosine stacks into the helix opposite the cyclic analogue of 2-deoxyribose. Biochemistry. 1988 Feb 9;27(3):924–931. doi: 10.1021/bi00403a013. [DOI] [PubMed] [Google Scholar]
- Kalnik M. W., Chang C. N., Johnson F., Grollman A. P., Patel D. J. NMR studies of abasic sites in DNA duplexes: deoxyadenosine stacks into the helix opposite acyclic lesions. Biochemistry. 1989 Apr 18;28(8):3373–3383. doi: 10.1021/bi00434a037. [DOI] [PubMed] [Google Scholar]
- Klinedinst D. K., Drinkwater N. R. Mutagenesis by apurinic sites in normal and ataxia telangiectasia human lymphoblastoid cells. Mol Carcinog. 1992;6(1):32–42. doi: 10.1002/mc.2940060107. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Scribner N., Kohwi Y. An ultimate chemical carcinogen, N-acetoxy-2-acetylaminofluorene, detects non-B DNA structures that are reactive with chloroacetaldehyde in supercoiled plasmid DNA. Carcinogenesis. 1988 Mar;9(3):457–461. doi: 10.1093/carcin/9.3.457. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988 Oct 15;263(29):14784–14789. [PubMed] [Google Scholar]
- Lackey D., Krauss S. W., Linn S. Isolation of an altered form of DNA polymerase I from Escherichia coli cells induced for recA/lexA functions. Proc Natl Acad Sci U S A. 1982 Jan;79(2):330–334. doi: 10.1073/pnas.79.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., Borden A., Banerjee S. K., LeClerc J. E. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 1990 Apr 25;18(8):2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Preston B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Bogenhagen D. F. Repair of a synthetic abasic site in DNA in a Xenopus laevis oocyte extract. Mol Cell Biol. 1989 Sep;9(9):3750–3757. doi: 10.1128/mcb.9.9.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya M., Ou C., Bodepudi V., Johnson F., Takeshita M., Grollman A. P. Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli. Mutat Res. 1991 May;254(3):281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- Moriya M., Takeshita M., Johnson F., Peden K., Will S., Grollman A. P. Targeted mutations induced by a single acetylaminofluorene DNA adduct in mammalian cells and bacteria. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1586–1589. doi: 10.1073/pnas.85.5.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto J. B., Gentil A., Cabral R. E., Sarasin A. Mutation spectrum of heat-induced abasic sites on a single-stranded shuttle vector replicated in mammalian cells. J Biol Chem. 1992 Sep 25;267(27):19718–19723. [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Endonuclease-resistant apyrimidinic sites formed by neocarzinostatin at cytosine residues in DNA: evidence for a possible role in mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(10):3182–3186. doi: 10.1073/pnas.82.10.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall S. K., Eritja R., Kaplan B. E., Petruska J., Goodman M. F. Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem. 1987 May 15;262(14):6864–6870. [PubMed] [Google Scholar]
- Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983 Sep 13;22(19):4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Sanderson B. J., Chang C. N., Grollman A. P., Henner W. D. Mechanism of DNA cleavage and substrate recognition by a bovine apurinic endonuclease. Biochemistry. 1989 May 2;28(9):3894–3901. doi: 10.1021/bi00435a040. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., Glickman B. W., Loeb L. A. Mutagenesis resulting from depurination is an SOS process. Mutat Res. 1982 Nov;106(1):1–9. doi: 10.1016/0027-5107(82)90186-5. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Loeb L. A. Depurination causes mutations in SOS-induced cells. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1773–1777. doi: 10.1073/pnas.78.3.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S., Grollman A. P. On the mechanism of frameshift (deletion) mutagenesis in vitro. J Biol Chem. 1993 Jun 5;268(16):11703–11710. [PubMed] [Google Scholar]
- Shibutani S. Quantitation of base substitutions and deletions induced by chemical mutagens during DNA synthesis in vitro. Chem Res Toxicol. 1993 Sep-Oct;6(5):625–629. doi: 10.1021/tx00035a006. [DOI] [PubMed] [Google Scholar]
- Steighner R. J., Povirk L. F. Bleomycin-induced DNA lesions at mutational hot spots: implications for the mechanism of double-strand cleavage. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8350–8354. doi: 10.1073/pnas.87.21.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B. S. The 'A rule' of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991 Feb;13(2):79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- Takeshita M., Chang C. N., Johnson F., Will S., Grollman A. P. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987 Jul 25;262(21):10171–10179. [PubMed] [Google Scholar]
- Vesnaver G., Chang C. N., Eisenberg M., Grollman A. P., Breslauer K. J. Influence of abasic and anucleosidic sites on the stability, conformation, and melting behavior of a DNA duplex: correlations of thermodynamic and structural data. Proc Natl Acad Sci U S A. 1989 May;86(10):3614–3618. doi: 10.1073/pnas.86.10.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]