Abstract
The accurate and complete selection of candidate genomic regions from a DNA sample before sequencing is critical in molecular diagnostics. Several recently developed technologies await substantial improvements in performance, cost, and multiplex sample processing. Here we present the utility of long padlock probes (LPPs) for targeted exon capture followed by array-based sequencing. We found that on average 92% of 5,471 exons from 524 nuclear-encoded mitochondrial genes were successfully amplified from genomic DNA from 63 individuals. Only 144 exons did not amplify in any sample due to high GC content. One LPP was sufficient to capture sequences from <100–500 bp in length and only a single-tube capture reaction and one microarray was required per sample. Our approach was highly reproducible and quick (<8 h) and detected DNA variants at high accuracy (false discovery rate 1%, false negative rate 3%) on the basis of known sample SNPs and Sanger sequence verification. In a patient with clinical and biochemical presentation of ornithine transcarbamylase (OTC) deficiency, we identified copy-number differences in the OTC gene at exon-level resolution. This shows the ability of LPPs to accurately preserve a sample's genome information and provides a cost-effective strategy to identify both single nucleotide changes and structural variants in targeted resequencing.
Keywords: DNA sequencing, mitochondrial disease, statistical analysis, target capture, copy number detection
DNA variant discovery is performed in an increasing number of individuals with specific disorders. In addition, thousands of disease candidate regions have been prioritized through linkage mapping and functional approaches. Studying hundreds of candidate genes in hundreds of samples is nontrivial given the high standards of accuracy and completeness in medical resequencing. To select genomic regions at a fraction of the cost of traditional sample preparations, novel DNA sequence capture methods are under development that include hybridization-based target enrichment (e.g., microarrays, beads) and in-solution methods (1–11). Hybrid capture is quickly scalable, shows high levels of uniformity, and has been applied in exome sequencing (5, 12). In-solution methods, and in particular molecular inversion probes (MIPs) (6–8, 13), provide the highest target specificity (>98%) at comparably lower costs and DNA requirement, which has advantages when studying many samples. However, MIPs’ target size limitation of 191 bp (8) could lead to amplification failures or overlooked sample variants if an exonic SNP is located in the probes’ annealing regions, which become part of the final amplification sequence. To capture most human exons (≤500 bp) using only a single capture probe per exon, we have developed an in-solution method using long padlock probes (LPPs) (14). Here we describe an optimized protocol for LPPs and the application to the capture of 524 candidate genes (5,471 exons) in a simple and robust single-tube assay. The nuclear-encoded mitochondrial genes selected are associated with many different disorders including diabetes, cancer, and neurodegeneration (15). We studied 63 medical cases and controls and performed both sample quality control and resequencing using data from a single hybridization to a DNA microarray.
Results
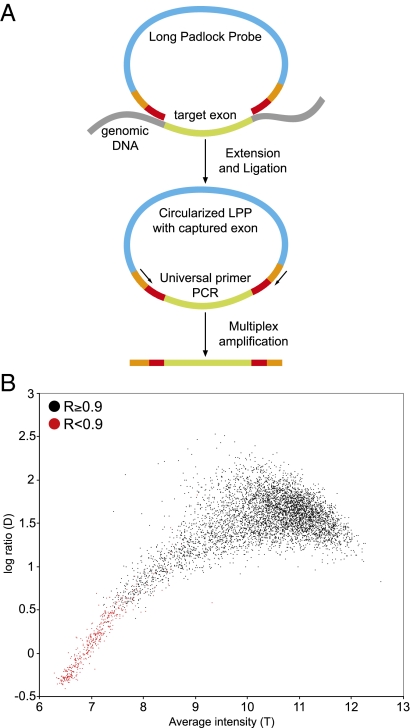
We designed LPPs to target all 5,471 exons of the 524 candidate genes (Dataset S1). Each probe is 320 bases in length and contains two sequences at both ends that hybridize to their complementary sites on genomic DNA (Fig. 1A). The gap between the probes’ 5′ and 3′ ends ranged from 92 to 546 bases and was filled by polymerization from the 3′ end using the target sequence as the template. After gap fill and formation of a circular DNA molecule by ligation, the target regions were multiplex amplified using a primer pair common to all probes. The 5,471 exons were targeted with 5,619 LPPs (Dataset S2) that included only a small number of additional probes (1.7%) to capture larger exon sequences. By comparison, a MIP-based capture would require the design of overlapping probes for nearly half of the target exons (8). We established the LPP-based capture assay from 0.5-μg genomic DNA template and all 5,619 probes in a single-tube assay. This, combined with a shortened assay time (<8 h), which was achieved through optimization of our experimental protocol (Materials and Methods), aided the parallel sample processing.
Fig. 1.
DNA sequence capture and sample quality assessment. (A) A genomic DNA sample is incubated with thousands of single-stranded long padlock probes (LPPs) each of which target a specific genomic region (e.g., intronic sequence flanking an exon). Following annealing, gap filling by a DNA polymerase and probe circularization by ligation, the captured targets are amplified in multiplex using a primer pair common to all probes. The entire capture pool of one sample is hybridized to a resequencing microarray containing the complementary sequences. (B) A statistical analysis of array quality measures (R, D, and T) is used in combination to monitor the capturing yield and identify failed targets (red dots) in each sample preparation.
We designed a unique high-density resequencing microarray (5 μm feature size) covering the complete coding sequences of the 524 genes (>0.8 Mb) to sequence the captured exons at single-base resolution. The array hybridization data were also used to assess the success of the LPP-based technology to capture the targeted exons in each sample. The capture pools were directly hybridized to this microarray (1 array per sample) and we performed data analysis with a statistical software developed for resequencing arrays (16). First, to identify rare instances of hybridization failure, we verified the overall signal quality on each array by measuring the average differences in signal intensities across all reference match (RM) and alternative match (AM) probes. Second, because all sample sequences are amplified together, we evaluated the quality and quantity of each target capture using three measures. These include the reference call rate (R) to measure the overall sequence similarity between an amplified exon (amplicon) and its reference sequence; the median log ratio (D) to measure the average signal-to-noise ratio of all probes of each amplicon; and the median average intensity (T) to measure the quantity of each amplicon in each array hybridization (Fig. 1B). Poorly amplified sequences showed lower values for R, D, and T, which are correlated. In well-amplified targets, R was close to 1, whereas D and T had nonlinear associations, which could represent effects of DNA hybridization kinetics (e.g., DNA quantity, sequence composition). Together, these measures provided immediate feedback on each amplicon and the success of sample preparations.
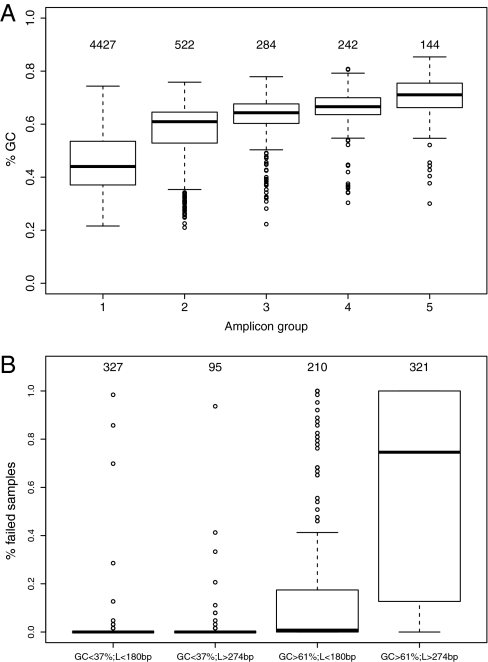
The performance of the LPP-based capture was measured in 63 samples with 353,997 sample amplicons (Dataset S3). On average, 91.6% (SD 2.3%) of the 5,619 targets were successfully amplified on the basis of the stringent threshold of R ≥ 0.9. Only 144 targets had R < 0.9 in all samples, and that could have been related to probe design. However, we identified a positive correlation between amplification failures and a target's GC content (P < 0.0001), which was highest (average 71%) for the 144 missed targets compared with the average of 44% for the 4,427 targets that amplified consistently in all samples (Fig. 2A). For targets with a GC content of up to 65% (5,014 amplicons), the capture yield in multiplex amplifications was 96.6% (SD 1.5%) and was reduced to about half (51.2%; SD 9.9%) for the higher GC targets (>65–86%, 605 amplicons). In addition to amplicons that completely failed (144) or successfully amplified (4,427) in all samples, we found 1,048 amplicons (18% of 5,619) below our detection threshold (R ≥ 0.9) in only a subset of samples. These sample-specific failures could be explained by variants that interfere with the target capture in only some samples. We did not find support for this hypothesis in an analysis of common SNPs mapping to the probes’ annealing regions, but rare single nucleotide variants and rearrangements as a cause of these sample-specific failures cannot be ruled out. We also examined the capture performance related to amplicon length (Fig. S1) and in comparison of length and GC content, which revealed a tendency for amplification failures in longer amplicons with higher GC content (P < 0.0001) (Fig. 2B). Our method was highly reproducible (R2 = 0.999) as measured by replicate sample preparations (Fig. S2). The capturing performance was reproducible using either genomic or whole-genome–amplified (WGA) DNA (Fig. S3 and Dataset S3), which is useful if only very small amounts of genomic DNA are available from clinical samples.
Fig. 2.
LPPs capture performance related to GC content and length. (A) The box plots show the distribution in GC content (y axis) of five groups of exons, defined on the basis of amplification success (R ≥ 0.9) in 63 DNA samples. These groups contain exons that amplified in all samples (group 1: 4,427 exons), exons that failed in only a subset of samples including in 1–10 samples (group 2: 522 exons), 11–40 samples (group 3: 284 exons), 41–62 samples (group 4: 242 exons), and exons that failed in all samples (group 5: 144 exons). Group 1 with the highest amplification success had the lowest mean GC content compared with groups 2–5 (P < 0.0001). (B) The box plots show the fraction of failed samples in four amplicon groups that we defined on the basis of amplicon length and GC content. Amplicons with lower GC content (<37%; <20th percentile) amplified successfully irrespective of their length, whereas amplicons with higher GC content (>61%; >80th percentile) had a tendency for amplification failures and in particular in longer amplicons (>274 bp; >80th percentile).
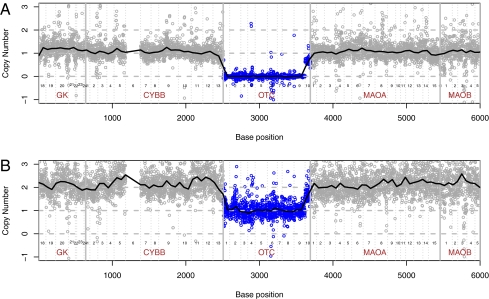
An analysis of all target exons grouped by genes revealed a complete amplification failure of 9 out of 10 exons of the ornithine transcarbamylase (OTC) gene (Xp11.4) in a male child (P44, Dataset S3) with clinical and biochemical features of OTC deficiency. OTC encodes a mitochondrial protein and point mutations and deletions (complete or partial) have been identified in patients with this inborn error of metabolism (17). The analysis of the child's unaffected mother identified a single-copy deletion of the same nine exons confirming an inherited deletion (Fig. 3). On the basis of the presence of a deletion breakpoint in OTC intron 9 and the capture of CYBB exons located upstream of OTC, we estimated a maximal deletion region of 610 kb. This result was confirmed through array-based comparative genomic hybridization (aCGH), revealing a 172- to 212-kb deletion spanning OTC and the retinitis pigmentosa GTPase regulator (RPGR) gene (Fig. S4). Our results indicate that LPP-based capturing accurately preserves a sample's genome information. This enables the targeted discovery of small genomic rearrangements at high resolution (e.g., intragenic deletion, copy-number variants) using a downstream sequencing technology.
Fig. 3.
Detecting copy number differences with resequencing arrays. A male child with OTC deficiency (A) and his healthy mother (B) had a single-copy deletion of 9 of 10 OTC exons (Xp21.1). Each circle represents the normalized array intensity value for 1 base of that sample. The 6,000 sequenced bases are concatenated from a 12.9 Mb genomic interval and represent exons of OTC and flanking candidate genes. The vertical lines indicate exon and gene boundaries. CYBB exon 6 failed in all samples. The dashed horizontal lines represent the normalized intensity values for two, one, and zero copies. The solid horizontal line shows a smoothed estimate of the normalized base intensities (100-bp window size) that we calculated as a ratio of background adjusted sample intensities to pooled background adjusted female reference intensities. The normalized intensities are scaled so that within regions of 1 copy number in the child (A) and 2 copies in the mother (B), the corresponding median value across positions of the normalized intensities equals 1 and 2, respectively.
We were then interested in the ability of resequencing arrays to detect single nucleotide variants. For positions in amplicons with R ≥ 0.9, we performed an automated clustering analysis of the 63 sample arrays and assigned a position-specific quality score to measure the confidence of each base call (16). Similar to Bainbridge et al. (11), the variants discovered were compared with known HapMap SNPs and to dbSNP. On the basis of five HapMap samples (Table 1), we estimated a false negative rate (FNR) of 2.9% and a false discovery rate (FDR) of 1.3%. A slightly lower performance was found in the analysis of variant positions in 38 patient samples that we had verified previously using Sanger capillary sequencing (16). Manual inspection revealed that most discordant base calls were related to limitations in the microarray analysis, which could be improved as more samples are studied (Fig. S5). In addition, dbSNP reported rs4534 in NA18507 as homozygous variant T, whereas our analysis and a recent study by Ng et al. (5) identified this sample position as homozygous reference C. It has been hypothesized that a substantial portion of apparent single nucleotide differences are due to the presence of similar sequences (i.e., paralogs, pseudogenes) in the genome (18). Highly specific target capturing of annotated candidate genes using LPPs could be used to identify such positions.
Table 1.
DNA variant discovery using resequencing arrays
| NA11840 | NA12156 | NA12878 | NA18507 | NA19625 | 38 cases | |
| (i) Number of sample-specific variants | 214 | 224 | 228 | 234 | 180 | 465 |
| (ii) Variant calls in this study | 212 | 220 | 225 | 224 | 180 | 453 |
| Overlap of variant calls in i and ii | 210 | 218 | 221 | 223 | 176 | 445 |
| False negative rate (FNR), % | 1.9 | 2.7 | 3.1 | 4.7 | 2.2 | 4.3 |
| False discovery rate (FDR), % | 1.0 | 0.9 | 1.8 | 0.4 | 2.2 | 1.8 |
| Concordance to known variants, % | 97.9 | 98.4 | 97.7 | 97.7 | 96.4 | 94.7 |
We used multiarray clustering for base calling of each position with the R package mclust (26) and ranked the confidence of each base call using a position-specific quality score (cutoff 0.67) (16). This analysis focused on verified DNA variants with a minor allele frequency larger than 0.05 in five HapMap individuals (columns 2–5) and in a pool of 38 medical cases (column 6) that we confirmed through Sanger sequencing (16). The variant calls from this study (ii) were compared with all previously reported sample variants in each sample (i) to estimate this method's performance. Concordance is the percentage of all variant and reference base calls in each sample that agreed with previously known base calls.
Discussion
DNA variant discovery relies on complementary methods such as whole-genome approaches in some and targeted resequencing in many individuals. Here we combined LPP-based target capture in single tubes with direct sequencing using microarrays (Fig. 1) to identify DNA variants in 524 genes in 63 samples. This approach captured on average 92% of all candidate gene exons (completeness), identified DNA variants at high accuracy (FDR 1%, FNR 3%, Table 1), and detected copy-number changes at exon-level resolution (Fig. 3). Our assay was quick (<8 h) and reproducible and we required only a single capture reaction from 0.5 μg of DNA and one array per sample, which are all increasingly important parameters in genetic research. In addition we established statistical algorithms for resequencing array data analysis (16) that enable quality control checks for each sample and amplicon in the parallel sample preparations.
Similar to the multiplexing capabilities of MIPs (8), LPP-based capturing is scalable to include larger sets of candidate genes such as the ∼1,200 nuclear-encoded mitochondrial genes (15) and perhaps all human exons in a single tube reaction as only minute amounts of probe are required per target (14). An important feature of LPPs is the ability to capture larger sequences of currently up to 546 bp using a single probe resulting in a probe/target ratio for human exonic targets of 1 for LPP and 1.4 for MIP. In addition to relatively fewer probes in capture reactions, the larger length of LPPs provide more flexibility in the primer design because the most optimal annealing sites cannot always be found in sequences immediately flanking a target exon. For probe construction, we presently rely on standard synthesis of 60-mer oligonucleotides ($10/probe) but millions of capture reactions can be performed with such a probe set at very low cost. Technological advances in oligonucleotide synthesis will make the production of LPPs more economical for much larger numbers of genomic targets.
The pools of captured targets can be sequenced directly with microarrays as shown here or with sequencing-by-synthesis technology (e.g., Illumina/Solexa, ABI/SOLiD, Roche/454) (19). An important parameter that remains to be addressed for LPPs in conjunction with these platforms is capture uniformity, which is the variability in sequence coverage across target regions. Capture uniformity will depend on both the capture technology and the platform used for sequencing. On the basis of the signal intensity range of amplicons in the array hybridizations (measure T) in the 5 HapMap samples, we estimated that on average 85% (SD 4%) of the successfully captured exons (R ≥ 0.9) were distributed within a 5-fold range and 94% (SD 2%) within a 10-fold range, which is significantly improved over MIPs (58% within 10-fold) (12). In comparison, hybrid capture methods can achieve higher target uniformity (12) but typically between 20 and 35% of the sequence reads are off target (specificity), which compromises the coverage of the desired targets. The lower specificity of hybrid capture also leads to the capture of duplicated or highly similar sequences (e.g., paralogs and pseudogenes), which causes overcoverage of some targets at the expense of others but can also lead to errors in base calling and genotyping. For LPPs, the probability for off-target captures is reduced because the two probe ends are tethered and amplification occurs only after both ends hybridized with optimum efficiency. In this study, LPPs targeted each exon predicted for the 524 genes (20) compared with hybrid capture where an estimated <5–15% of the desired regions are omitted due to repetitive elements that are homologous to these sequences (12). To further improve the capture uniformity and increase coverage of the low-copy targets, LPPs (and MIPs) could be grouped into probe sets with similar capture performance (8, 14) such as for targets with similar GC content (Fig. 2). The performance of LPPs is constrained by the efficiency of the gap-closing reaction (polymerization and ligation), which is sequence dependent and varies for different gaps, and which can lead to amplification failures for GC-rich, longer targets. We found that adding 0.75 M betaine (Sigma) to the gap-fill reaction improved the overall efficiency of the assay (Fig. S6). Betaine reduces the formation of GC-rich secondary structures and is a known PCR enhancer (21, 22). We further suggest extending the gap-fill time while reducing the annealing time, which increased capture yields and reliability and leads to an overall reduction in the assays’ running time.
A common feature of mitochondrial diseases is heterogeneity (defects in different genes cause a similar phenotype) and pleiotropy (one gene is associated with different phenotypes) (15) and the discovery of reliable gene-disease associations will depend on high-quality sequencing in increasing numbers of patients. As more complex mitochondrial diseases are studied, maximal efforts should be placed on sequence accuracy and completeness as well as recording this wealth of information in public databases. One early strategy relies on whole-exome capture in well-phenotyped individuals (23), while coding regions that are hard to target by hybrid capture (e.g., homologous sequence) and exons with insufficient read coverage can be studied with LPPs. These two DNA sequence capture technologies are complementary (one method cannot fully measure what the other measures and they support each other) and applied together allow to maximize sequence coverage and to control the false discovery rate for rare (minor allele frequency, MAF < 1%) variants. Similarly, because of different biases and error rates of the current sequencing technology (11, 16, 24, 25), high-quality variant discovery in medical samples may be best performed through a combination of technologies. A comparison of different sequencing technologies (e.g., array based vs. sequencing by synthesis) is our next goal to identify the cause of sequencing errors inherent to each method and to improve the protocols for targeted medical resequencing.
Materials and Methods
Selection of Candidate Genes and Study Population.
We selected 524 nuclear genes on the basis of evidence of the localization of their gene products to human mitochondria (Dataset S1). A subset of these genes cause hereditary mitochondrial disorders and we prioritized additional disease candidate genes through an integrative network analysis of mitochondrial diseases and genes (15). We studied 63 samples (Dataset S3) that included 47 medical cases with a mitochondrial disease such as OTC deficiency and mtDNA maintenance disorders. Written consent was obtained from the patients’ families and approved by the institutional review boards. We also studied 16 healthy controls including five HapMap samples (NA11840, NA19625, NA12156, NA12878, and NA18507) obtained from Coriell Cell Repositories and one genomic reference DNA (G147A; Promega). These samples were used to establish our LPP-based capturing assay. To obtain starting material from very small amounts of genomic DNA (∼10 ng), we performed whole-genome amplification (WGA) using the REPLI-g mini kit (Qiagen) following Qiagen’s protocol. DNA was quantified using PicoGreen reagent (Invitrogen). A subset of amplicons in each sample was inspected by 1.2% agarose gel electrophoresis to monitor the success of target amplifications and to develop methods for sample quality control and resequencing (16).
Design and Construction of LPPs.
The increased length of LPP, which we engineered through modifications of the probes’ backbone, allows the capture of DNA sequences of up to 546 bp in length. In this study, we targeted all 5,471 exons from 524 nuclear-encoded mitochondrial candidate genes and designed LPPs for each exon according to our previous protocol (14). The 5,471 exons included all exons from the 524 genes as predicted in the Ensembl database that we downloaded through Ensembl's Biomart (NCBI Build 36) (20). We designed the two oligonucleotide primers required for each probe using an in-house program based on Primer3. Primers were placed in intronic regions flanking an exon (50-bp window) to capture the entire exon and intronic splice sites. Shorter exons separated by short introns were paired and amplified together as one amplicon. Larger exons amplified using multiple capture probes required that the ligation arm of one probe be located on the same sequence but on the opposite strand as the extension arm of the next probe. This design maximized the overlap region of consecutive probes (∼30 bp in size) to minimize the potential loss of exonic sample sequence. In addition, targeted sequences were chosen to avoid exonic SNPs and to have similar length (range 17–28 bp, average 24 bp) and GC content (range 8–92%, average 46%). We designed in total 5,619 LPPs to target all 5,471 candidate gene exons (Dataset S2). On the basis of our modified Primer3 design criteria, ∼42% of our target amplicons are larger than 231 bp. Capturing these targets using the alternative MIP technology would require multiple overlapping probes, due to MIPs target size limitation of 191 bp plus 40 bp targeting arms (8). We prepared the LPPs in 14 sets of 384 probes and 1 set of 243 probes pooled by the amplicons’ GC content. In the final capture pool containing all 5,619 probes the number of probes with GC > 0.6 was increased proportionally with GC content. The subpools can be easily combined at variable amounts in a single tube reaction volume to test the target uniformity requirements of different sequencing platforms.
Multiplex DNA Sequence Capture with LPPs.
We mixed 500 ng of genomic DNA or WGA DNA with 20 fmole of LPPs (2–5 attomole each) in 1× ampligase buffer (Epicentre) in a 10-μL volume. The mixture was heated to 98 °C for 3 min, followed by 85 °C for 30 min, 60 °C for 1 h, and 56 °C for 2 h. For probe extension and ligation, a 10-μL mixture of 0.3 mM dNTP, 2 mM NAD, 0.75 M betaine, 1× ampligase buffer, 5 units ampligase (Epicentre), and 0.8 units Phusion polymerase (NEB) was added to the reaction and the reaction was incubated at 56 °C for 60 min followed by 72 °C for 20 min. To completely eliminate linear DNA molecules, we added 2 μL of a mixture of the total of six exonucleases including 3.5 units exo I (USB), 18 units exo III (USB), 4 units exo T7 (USB), 0.4 units exo T (NEB), 3 units RecJf (NEB) and 0.2 units lambda exo (Epicentre). The reaction was placed at 37 °C for 20 min, 80 °C for 10 min, and 95 °C for 5 min. The circled molecules with captured sequences were PCR amplified in three 50-μL reactions containing 1× Phusion GC buffer (NEB), 0.2 mM dNTP, 0.01 units/μL Phusion polymerase, and 0.5 μM of each of the two common amplification primers. PCR was performed at 98 °C for 3 min followed by 30 cycles of 98 °C for 10 s, 64 °C for 20 s, and 72 °C for 30 s and a final extension for 5 min at 72 °C. We also found that increasing polymerization and ligation time from 15 min (14) to 1 h increased capture yield. Under this condition, a 4-h annealing time is sufficient and comparable to overnight annealing. Together these improvements are significant because the assay can now be performed reliably in less than 8 h for rapid and reliable sample processing.
DNA Sequencing with Resequencing Arrays.
In collaboration with Affymetrix we designed a unique high-density DNA resequencing microarray (5 μm feature size). This resequencing array format allows the interrogation of the entire coding region and exon-splice sites of the 524 candidate genes (5,471 exons) with total 816,817 sequence bases. Each array contains ∼6.5 million 25-mer probes, with 8 probes targeting each nucleotide position including 4 probes for sense and antisense strand. The 4 probes for each strand differ only at the center positions in A, T, G, and C, respectively, and match the reference sequence in the flanking 24 bases. We describe the probes completely complementary to the reference sequence as reference match probes (RMs) and the probes for the other three alternative alleles as alternative match probes (AMs). We expect highest intensities in the probes that are completely complementary to the target sequence.
To evaluate the quality and quantity of the amplified targets, we calculated three measures for each amplicon (16): (i) the median of the average (log2) probe intensity across all bases on the amplicon, denoted by T; (ii) the median of the log ratios across all bases on the amplicon, denoted by D; and (iii) the percentage of bases on the amplicon presenting highest intensities in the RM probes (reference call rate), denoted as R. A joint analysis of these three measures among all amplicons is informative about the target amplification and hybridization experiments. Specifically, to identify failed amplicons across all 353,997 amplicons (5,619 amplicons in 63 samples) that were not suitable for base calling, we used a simple criterion for the success of target amplifications of R ≥ 0.9. This measure is based on estimated variant frequencies in protein-coding regions of diploid genomes, with more than 99% of all positions being reference bases. We evaluated this cutoff by manual inspection of gel-electrophoresis data on 5,694 singleplex amplicons in a previous study (16). Following our quality screening, on average 91.6% of all sample amplicons with R ≥ 0.9 (324,261) proceeded to sequence data analysis and base calling. R values between 0.5 and 0.9 indicate some amplification, but the overall lower signal intensity of these amplicons on the arrays could prevent accurate base calling. The target amplification rates based on R > 0.7 and R > 0.8 are shown in Dataset S3.
As with the amplicon quality assessment, base calling and DNA variant detection was performed using a subset of our toolkit sequence robust multiarray analysis (SRMA) (16). In brief, for each SNP position and each array, we calculated log ratios for RM and AM probes: δ = θRM − θAM, θ denoting log2 of intensity, given the known reference and alternative alleles. We calculate the log ratios for sense and antisense strands separately. We then used a single-position multiarray bivariate Gaussian mixture model to make base calls for all samples. There can be three classes of interest: homozygous reference, heterozygous variant, and homozygous variant. We used the R package mclust (26) to perform multiarray classification at common SNP positions (MAF > 0.05). The multiarray classification results are susceptible to batch effects presenting multiple clusters for the reference samples and to weak probe performance, making the heterozygous samples more similar to the reference samples. We inferred genotypes from the classification results using constraints on the location of the genotype groups to merge reference clusters and by leveraging information from high confidence homozygous variant calls to identify additional heterozygous variants based on Hardy–Weinberg equilibrium.
The 63 individuals studied included 5 HapMap samples with a total of 1,318 known SNP positions (dbSNP131). A quality score q was assigned to each base for each individual in the clustering analysis and a high-quality score of larger than 0.67 was used for the comparison. We further included in this analysis 38 previously Sanger-verified DNA variants in 39 candidate genes and 38 samples (16). These positions are a subset of variants queried by the new 5-μm array and had a minor allele count (MAC) greater than 4 among the 38 samples. In addition to score q evaluating the quality of base calls for each sample position, a second quality score Q was used to measure the probe performance at each position (four probes per strand). Measuring the ability of probe quartets to discriminate between reference and alternative base signals is important, because the design of resequencing arrays requires the tiling of all probe sequences to complement a reference. Our position-specific Q score identified 12% of the 1,318 nucleotide positions with at least one probe quartet with suboptimal performance. At these positions, SRMA base calling is still possible when all samples are reference. Only 2% of the 1,318 positions had both probe quartets affected, limiting the identification of variants.
Supplementary Material
Acknowledgments
We thank Mamta Maheshwary and Keyi Liu for technical assistance and Rita Horvath, Kate Rauen, and Iris Schrijver for providing DNA samples. This work was supported by Grant R01 EY016240 from the National Eye Institute (to R.W.D. and C.S.) and Grants P30 CA016672 and 3U24CA143883 (to W.W.).
Footnotes
Conflict of interest statement: S.K., M.N.M., and R.W.D. are named on a patent application for work described in this paper.
This article is a PNAS Direct Submission.
Data deposition: Raw data are available via the European Bioinformatics Institute's repository ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession no. E-MTAB-499.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018981108/-/DCSupplemental.
References
- 1.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 2.Dahl F, et al. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci USA. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredriksson S, et al. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 2007;35:e47. doi: 10.1093/nar/gkm078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng J, et al. High-throughput, high-accuracy array-based resequencing. Proc Natl Acad Sci USA. 2009;106:6712–6717. doi: 10.1073/pnas.0901902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tewhey R, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bainbridge MN, et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11:R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mamanova L, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 13.Hardenbol P, et al. Highly multiplexed molecular inversion probe genotyping: Over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnakumar S, et al. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci USA. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scharfe C, et al. Mapping gene associations in human mitochondria using clinical disease phenotypes. PLoS Comput Biol. 2009;5:e1000374. doi: 10.1371/journal.pcbi.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, et al. Identification of rare DNA variants in mitochondrial disorders with improved array-based sequencing. Nucleic Acids Res. 2011;39:44–58. doi: 10.1093/nar/gkq750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shchelochkov OA, et al. High-frequency detection of deletions and variable rearrangements at the ornithine transcarbamylase (OTC) locus by oligonucleotide array CGH. Mol Genet Metab. 2009;96:97–105. doi: 10.1016/j.ymgme.2008.11.167. [DOI] [PubMed] [Google Scholar]
- 18.Musumeci L, et al. Single nucleotide differences (SNDs) in the dbSNP database may lead to errors in genotyping and haplotyping studies. Hum Mutat. 2010;31:67–73. doi: 10.1002/humu.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller CW, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27:1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 20.Flicek P, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38(Database issue):D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rees WA, Yager TD, Korte J, von Hippel PH. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry. 1993;32:137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- 22.Henke W, Herdel K, Jung K, Schnorr D, Loening SA. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng SB, et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harismendy O, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder M, Du J, Gerstein M. Personal genome sequencing: Current approaches and challenges. Genes Dev. 2010;24:423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. JASA. 2002;97:611–631. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.