Abstract
Polyphenol curcumin, a yellow pigment, derived from the rhizomes of a plant (Curcuma longa Linn) is a natural antioxidant exhibiting a variety of pharmacological activities and therapeutic properties. It has long been used as a traditional medicine and as a preservative and coloring agent in foods. Here, curcumin-converting microorganisms were isolated from human feces, the one exhibiting the highest activity being identified as Escherichia coli. We are thus unique in discovering that E. coli was able to act on curcumin. The curcumin-converting enzyme was purified from E. coli and characterized. The native enzyme had a molecular mass of about 82 kDa and consisted of two identical subunits. The enzyme has a narrow substrate spectrum, preferentially acting on curcumin. The microbial metabolism of curcumin by the purified enzyme was found to comprise a two-step reduction, curcumin being converted NADPH-dependently into an intermediate product, dihydrocurcumin, and then the end product, tetrahydrocurcumin. We named this enzyme “NADPH-dependent curcumin/dihydrocurcumin reductase” (CurA). The gene (curA) encoding this enzyme was also identified. A homology search with the BLAST program revealed that a unique enzyme involved in curcumin metabolism belongs to the medium-chain dehydrogenase/reductase superfamily.
Keywords: microbial screening, bioconversion
Since the dawn of civilization, natural compounds have been used as a source of medicine and foods based on their tremendous biological activities. One of the natural compounds that has been documented as having been used as a spice and a pigment since 1900 B.C. is curcumin (1). Curcumin, defined as a bis-α, β-unsaturated β-diketone (Fig. S1), has been found exclusively in the roots of Curcuma longa Linn (Zingiberaceae) (2). Curcumin has many different applications because it has a surprisingly wide variety of beneficial uses, including food coloration, cosmetics utility, and fabric dying. In addition, curcumin has a wide range of medical properties, including antitumor, anti-inflammatory, antioxidant, anticancer, and analgesic uses (3). Recently, curcumin has been used in the treatment of Alzheimer's disease (4) and malaria (5), and improvement of wound healing (6). Curcumin in the diet is partially absorbed in the intestine (7). A considerable portion of the ingested curcumin reaches the cecum and colon, where a large population of indigenous bacteria exists. Although analysis of curcumin metabolism and biosynthesis would lead to the discovery of other unknown advantages of this compound, there has neither been purification of enzymes involved in the metabolic pathway for curcumin nor identification of their genes in living organisms. In addition, no information is available regarding the effect of the intestinal bacteria on curcumin metabolism.
Studies on such curcumin-converting microorganisms and functional analyses of the curcumin-converting enzymes and genes would facilitate clarification of the metabolism of curcumin. In the present article, we report the isolation of curcumin-metabolizing microorganisms from human feces and the purification and characterization of a curcumin-converting enzyme. We are unique in demonstrating that a NADPH-dependent enzyme in an intestinal microorganism is involved in the curcumin pathway (curcumin → dihydrocurcumin → tetrahydrocurcumin).
Results
Discovery of Curcumin-Converting Microorganisms.
Microorganisms with curcumin-converting ability were isolated from feces of two healthy adults who had taken no antibiotics or other drugs for the 3 mo before sampling. We then selected the intestinal microorganism exhibiting the highest specific activity (0.00004 units/mg) as to curcumin conversion. A 16S rRNA sequence of the selected microorganism (accession number AB609595) exhibited the highest similarity (99%) to those of Escherichia coli H10407 strain [accession number FN649414 (region: 4363979..4365520)] (8), O55:H7 strain CB9615 [accession number CP001846 (region: 227062..228603)] (9), and BW2952 strain [accession number CP001396 (region: 4095850..4097391)] (10); therefore, it was identified as E. coli. The isolated microorganism was then cultured on MacConkey (11) and Levine EMB (12) agar to examine the lactose fermentation reaction and to observe the colony morphology on these media. On MacConkey agar, the isolated microorganism produced red colonies, depending upon the ability to ferment lactose: fermentation of this sugar causes the medium pH to drop, leading to darkening of the medium. Growth of the microorganism on Levine EMB agar produced black colonies with a greenish-black metallic sheen because of lactose fermentation: lactose fermentation produces acid and encourages dye absorption by colonies, which become colored black with a metallic green sheen because of the metachromatic properties of the dyes. These findings strongly supported that the isolated microorganism is E. coli.
We also detected the curcumin-converting ability of E. coli strain K-12, substrain DH10B. The specific activity (0.0005 units/mg) as to curcumin transformation in the cell-free extracts of this strain is higher than that of the isolated E. coli from human feces. Therefore, we used E. coli DH10B instead of the isolated E. coli for further experiments because of its high specific activity and whole-genome determination (13).
Addition of Various Cofactors to the Curcumin-Converting Enzyme.
Purification of the enzyme was carried out from an extract of cells cultured in the optimum medium for curcumin metabolism (SI Results). Interestingly, the curcumin-converting enzyme activity decreased during the enzyme purification. Dialysis of cell-free extracts caused complete loss of the initial enzyme activity. To determine the effect of the addition of cofactors on the enzyme activity, various kinds of cofactors at a final concentration of 0.5 mM were added to the reaction mixture, separately. The loss of activity was reversible and the activity was drastically recovered on the addition of NADPH to the assay mixture (Table S1), which allowed characterization of this enzyme in various ways after purification. Therefore, enzyme assay mixtures A and B were fortified with NADPH.
Purification of the Curcumin-Converting Enzyme.
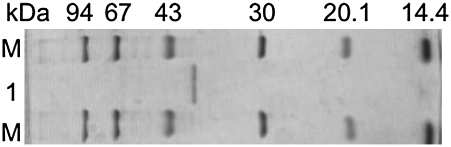
Through the purification steps described in Materials and Methods, the curcumin-converting enzyme was purified 1,100-fold, with a yield of 1.44%. The purified enzyme showed specific activity of 1.76 units/mg (Table 1). The purity of the enzyme was confirmed by migration of the protein as a single band corresponding to a molecular mass of 38 kDa on SDS/PAGE (Fig. 1).
Table 1.
Purification of the curcumin-converting enzyme
| Step | Total protein (mg) | Total activity (units) | Specific activity (units/mg) | Yield (%) |
| Cell-free extract | 3,050 | 4.88 | 0.0016 | 100 |
| DEAE Sepharose | 292 | 4.99 | 0.0171 | 102 |
| Blue Sepharose | 8.80 | 0.299 | 0.0340 | 6.13 |
| HiTrap Benzamidine | 0.52 | 0.229 | 0.440 | 4.69 |
| PIKSI Red 3 A6XL | 0.04 | 0.070 | 1.76 | 1.44 |
Fig. 1.
SDS/PAGE of the purified curcumin-converting enzyme. Protein bands were detected by staining with Coomassie brilliant blue. Lane M, marker proteins; lane 1, the purified enzyme (1 μg).
Cloning of the Gene Encoding the Curcumin-Converting Enzyme.
To identify the gene encoding the curcumin-converting enzyme, the 38-kDa band corresponding to the purified enzyme was excised from the SDS/PAGE gel and subjected to trypsin digestion, as described in SI Materials and Methods. The resulting peptides were analyzed by MALDI-TOF/MS. The peptide mass data were used to search the Mascot proteomics database (www.matrixscience.com), which revealed a significant match to YncB, which is designated in the database for E. coli strain K-12, substrain DH10B as a “predicted Zn-dependent and NAD(P)-binding oxidoreductase.” The ORF is 1,038 nucleotides long and should encode a curcumin-converting enzyme of 345 amino acids. The molecular mass of the protein encoded by curA was calculated to be 37,608 Da, which was consistent with that of the purified enzyme determined on SDS/PAGE (Fig. 1). A search with the BLAST program revealed that the deduced amino acid sequence of CurA (NADPH-dependent curcumin/dihydrocurcumin reductase) exhibits low similarity with those of alkenal double-bond reductase (AtDBR) from Arabidopsis thaliana (39%) (14) and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin13-reductase (12-HD/PGR) from guinea pig (37%) (15) (Fig. S2).
Overexpression and Purification of the Curcumin-Converting Enzyme from E. coli BL21-CodonPlus(DE3)-RIL Harboring pET-curA.
An expression plasmid (pET-curA) was constructed, as described in SI Materials and Methods and introduced into E. coli BL21-CodonPlus(DE3)-RIL for overexpression of the curcumin-converting enzyme.
Cell-free extracts prepared from the E. coli transformant carrying the yncB gene designated in the database acted on curcumin as a substrate. This finding demonstrated that this yncB gene encodes the curcumin-converting enzyme. Thus, we renamed yncB curA. To increase the amount of the soluble enzyme, various culture conditions were examined (Table S2). Under the optimum conditions given in SI Materials and Methods, the maximum level of the curcumin-converting enzyme activity in the cell-free extracts was 4.68 units/mg. We analyzed the cell-free extracts by SDS/PAGE and detected a large amount of the 38-kDa protein band consistent with the purified enzyme from E. coli DH10B; the amount of CurA was about more than 70% of the total soluble proteins. These findings indicated that overproduction of the curcumin-converting enzyme in the active form was attained.
The curcumin-converting enzyme produced in the transformed cells was purified to homogeneity through the four-step purification procedure involving three chromatography columns, as described in SI Materials and Methods (Table S3). The purified enzyme gave only one band on SDS/PAGE (Fig. S3), and apparent molecular mass of the purified enzyme was the same as that from E. coli DH10B. The molecular mass of the purified enzyme was also determined to be 37,513 Da by MALDI-TOF/MS, as described in SI Materials and Methods, this value being the same as that from E. coli DH10B (37,589 Da). These data indicated that the curcumin-converting enzyme purified from E. coli BL21-CodonPlus(DE3)-RIL carrying pET-curA is identical to that from E. coli DH10B. Because the following analyses required a large amount of the enzyme and a large amount of the purified enzyme could not be obtained from E. coli DH10B, we used purified CurA from E. coli BL21-CodonPlus(DE3)-RIL harboring pET-curA in the following experiments.
Molecular mass, UV-Absorption of the Curcumin-Converting Enzyme.
The molecular weight of the native enzyme was 82 kDa, according to the results of gel-filtration chromatography. Considering the molecular mass of 38 kDa determined on SDS/PAGE, the enzyme consists of two identical subunits.
The absorption spectrum of the colorless purified enzyme (16.3 mg/mL) in 10 mM potassium phosphate buffer (KPB) (pH 6.0) showed an absorbance maximum near 280 nm. No other absorption peak or shoulder was observed at higher wavelengths, suggesting that no cofactor was bound to the enzyme. Moreover, no metal was contained in the enzyme (SI Results).
Effects of Temperature and pH on the Activity and Stability of the Enzyme.
The effects of temperature and pH on the enzyme activity were examined. The optimal reaction temperature appeared to be 35 °C (Fig. S4A). The enzyme exhibited maximum activity at pH 5.9 (Fig. S4B).
The stability of the enzyme was investigated at various temperatures. After the enzyme had been preincubated for 30 min in 10 mM KPB (pH 6.0), an aliquot of the enzyme solution was taken and then the enzyme activity was assayed under the standard assay A condition. The enzyme was most stable from 10 to 50 °C (Fig. S4C). The stability of the enzyme was examined at various pH values. After the enzyme had been incubated at 28 °C for 30 min in the Britton-Robinson buffer (pH 2.2–12.0) (16) at a concentration of 0.1 M, an aliquot of the enzyme solution was taken and then the enzyme activity was assayed under the standard assay A condition. CurA was most stable in the pH range of 4.5 to 12.0, with 90% of the initial activity being retained even at pH 12.0 (Fig. S4D).
Identification of the Reaction Products.
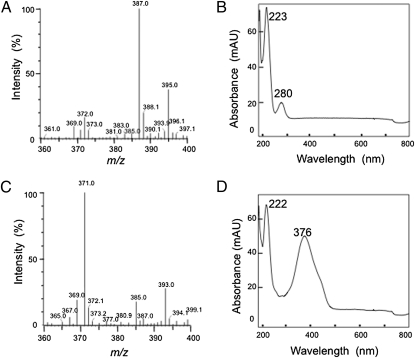
The products derived on curcumin conversion were analyzed by liquid-chromatography electrospray-ionisation mass spectrometry (LC-ESI-MS) and fast atom bombardment (FAB)-MS, as described in SI Materials and Methods. First, the reaction was carried out for 30 min at 28 °C using an excess of the purified enzyme (5 μg/mL) in standard mixture A. In this case, only one peak with the consumption of curcumin was detected at 280 nm with the HPLC method described for product purification in SI Materials and Methods. The retention time of the reaction product was found to agree with that of authentic tetrahydrocurcumin (THC) (14.04 min). Furthermore, the eluent corresponding to this peak was collected and analyzed by LC-ESI-MS, the product exhibiting a m/z value of 395, corresponding with the [M+Na]+ ion of THC (372 MW) (Fig. 2A). The UV-vis absorption spectrum of the reaction product agreed with that of authentic THC, and was the same as that of THC reported previously (17) (Fig. 2B). The FAB mass spectrum also gave ion at m/z 373.1656, which is in very good agreement with the molecular formula of THC, [C21H25O6]+. These findings demonstrated that the reaction product was THC. Next, the reaction was carried out for 10 min at 28 °C using a low concentration of the purified enzyme (0.5 μg/mL) in standard mixture A. Surprisingly, another peak with a retention time of 14.43 min was detected with a HPLC method. The new peak was not detected when the reaction was carried out completely, suggesting that the new peak is that of an intermediate product. The eluent corresponding to the new peak was collected and analyzed by LC-ESI-MS. The new product exhibited a m/z value of 393, consistent with the [M+Na]+ ion of dihydrocurcumin (DHC) (370 MW) (Fig. 2C). Moreover, the new product exhibited strong UV-absorption at 376 nm, and the UV-vis absorption spectrum of the new product was the same as that of DHC reported previously (17) (Fig. 2D). The FAB mass spectrum also gave ion at m/z 371.1511, which is in a very good agreement with the molecular formula of DHC, [C21H23O6]+. These findings suggested that the new intermediate product was DHC. Moreover, no other compounds, including hexahydrocurcumin and octahydrocurcumin, were detected on HPLC, despite an increased reaction time and the use of a large amount of the enzyme in the reaction mixture. Thus, the reaction products on curcumin metabolism were identified as DHC and THC. These findings corroborated that the curcumin-converting enzyme converted curcumin to DHC as an intermediate product, followed by further reduction to THC as an end product.
Fig. 2.
The reaction products, dihydrocurcumin (DHC), and tetrahydrocurcumin (THC). (A) LC-ESI-MS spectrum of THC in the positive ion mode. (B) UV-vis spectrum of THC. (C) LC-ESI-MS spectrum of DHC in the positive ion mode. (D) UV-vis spectrum of DHC.
Substrate Specificity.
The ability of the enzyme to catalyze the reduction of various compounds was examined using standard assay B. Among the tested substances, described in SI Materials and Methods, curcumin was the most suitable substrate for the enzyme. The curcumin-converting enzyme acted on only a few substances [i.e., 3-octene-2-one (59.7% of the activity toward curcumin), 3-hepten-2-one (28%), resveratrol (14.8%), and trans-2-octenal (8.13%)]. The reduction of curcumin followed Michaelis-Menten-type kinetics, the Km and Vmax values being 0.029 mM and 9.35 units/mg, respectively.
Discussion
Despite the impressive applications (e.g., as foods, dyes, cosmetics, and medicines) of natural compounds and their metabolites, biochemical and genetic information on their biosynthesis, biotransformation, and biodegradation is limited. Although among natural products curcumin is well known as one of the very useful substances having an important variety of functions, particularly from the medicine perspective, there have not been many efforts regarding its biodegradation and metabolism. So far, two phases of curcumin metabolism in the livers and intestines of rats and humans, and plasma of mice, have been reported (17–20). Phase I metabolism comprises the reduction of the four double bonds of the heptadiene-3, 5-dione structure: namely, curcumin → DHC → THC → hexahydrocurcumin → octahydrocurcumin (7, 17–20). During phase II, curcumin and its reduced metabolites are conjugated with a monoglucuronide, a monosulfate and a mixed sulfate/glucuronide: namely, conjugated curcumin → conjugated DHC → conjugated THC → conjugated hexahydrocurcumin → conjugated octahydrocurcumin (7, 17–20). To the best of our knowledge, however, no studies on purification and characterization of these enzymes or on cloning of the genes encoding these enzymes have been reported. Thus, the detailed mechanism of curcumin metabolism remains unclear. Moreover, no study on the effect of curcumin on the human gut flora has been reported, although curcumin has been taken orally as a food agent or medicine since ancient times. Therefore, we addressed the question of whether intestinal microorganisms that can metabolize curcumin exist or not. Our success in isolating the microorganisms exhibiting curcumin-converting activity from human feces is unique. Among the microorganisms isolated, the selected one exhibiting the highest curcumin-converting ability was identified as E. coli. In addition, we found that E. coli strain K-12, substrain DH10B, whose whole-genome sequence had already been determined, possessed higher curcumin-converting activity than the isolated E. coli. Therefore, E. coli DH10B was used for the further experiments.
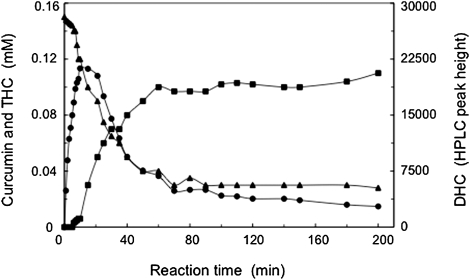
We purified the enzyme CurA, which is responsible for curcumin transformation. During the enzyme purification, the curcumin-converting enzyme activity was lost on dialysis. However, we found that the addition of NADPH increased the activity, but the addition of NADH did not. These findings suggest that the enzyme catalyzes NADPH-dependent conversion of curcumin. Based on mass determination by means of LC-ESI-MS, FAB-MS, and UV-vis absorption spectra (17), we also identified two reaction products (DHC and THC), depending on the consumption of curcumin. Moreover, the time course of curcumin conversion revealed the order of product formation (i.e., curcumin is first reduced to DHC, followed by the latter's sequential reduction to THC) (Fig. 3). Thus, in the two reaction steps catalyzed by CurA, DHC is first a product and then a substrate. In this investigation, we discovered the two-step curcumin metabolic pathway (curcumin → DHC → THC) catalyzed by CurA (Fig. 4). The products were generated from curcumin through reductive destruction of the chromophoric diarylheptatrienone chain. The lack of production of further reduced reaction products than THC by CurA suggested that CurA catalyzes the reduction of only compounds with C = C double bonds (i.e., not C = O double bonds). Although a large number of different compounds were tested as potential substrates for CurA, CurA appears to have a narrow substrate spectrum and to preferentially act on curcumin. Considering the substrate specificity, reaction, and NADPH-dependency, we named the purified enzyme (CurA) NADPH-dependent curcumin/dihydrocurcumin reductase.
Fig. 3.
Time course of curcumin conversion and generation of products. Curcumin (▲), DHC (●), and THC (■).
Fig. 4.
Curcumin metabolic pathway.
CurA exhibited sequence similarity with some enzymes belonging to the medium-chain dehydrogenase/reductase (MDR) superfamily, which contains many different families including the alcohol dehydrogenases family and leukotriene B4 dehydrogenase family (21). All enzymes belonging to the MDR superfamily use NAD(H) or NADP(H) as a cofactor (22), and catalyze oxidation or reduction reactions biologically (23). These characteristics support the requirement of NADPH as a cofactor for the reduction of curcumin/dihydrocurcumin by CurA. On the other hand, several (i.e., not all) of the MDR superfamily members have one zinc ion with a catalytic function in the active site (22), which is necessary for their activities. Based on the following findings, CurA belongs to Zn-independent families of the MDR superfamily: (i) CurA showed no absorption peak or shoulder except near 280 nm; (ii) CurA contained no metal (SI Results); (iii) chelating reagents did not inhibit CurA at all (SI Results); and (iv) the addition of different metals including zinc to the reaction mixture did not increase the enzyme activity significantly (SI Results), suggesting that CurA is metal-independent. It is expected that CurA contains an NADPH-binding site in its structure. Based on comparison of CurA with 3D structural information of AtDBR (14) and 12-HD/PGR (15), we here propose Asp-51, Val-162, Gly-182, Lys-186, Asn-225, Cys-247, Phe-282, and Asn-333 comprise the NADPH-binding site of CurA. Interestingly, CurA was found to be inhibited by thiol-specific reagents (e.g., p-chloromercuribenzoate and 5,5′-dithio-bis-2-nitrobenzoate) as a result of inhibition studies (Table S4). To confirm the effects of such reagents on Cys-247, which is located in the proposed NADPH-binding residues of CurA, we constructed a point mutant (C247A) by substituting alanine for Cys-247 in CurA, and purified the C247A mutant enzyme to homogeneity, as described in SI Materials and Methods (Fig. S3). The C247A mutant enzyme exhibited a reduction of about 50% in activity (2.73 units/mg), but was not inhibited by 5,5′-dithio-bis-2-nitrobenzoate at all (2.64 units/mg), whereas wild-type enzyme was completely inhibited (SI Results). These results indicated that Cys-247 is sole target of 5,5′-dithio-bis-2-nitrobenzoate. Considering that Cys-247 was proposed as one of the NADPH-binding sites of CurA based on comparison of the corresponding residues constituting the NADPH-binding sites of AtDBR (14) and 12-HD/PGR (15), CurA would lose the activity because of the binding of 5,5′-dithio-bis-2-nitrobenzoate to Cys-247 followed by the inhibition of NADPH-binding to Cys-247. Like other typical MDR superfamily members, CurA has a glycine-rich motif, 157AATGPVG163 (Fig. S2), shared by several oxidoreductases (AXXGXXG), which is known to participate in binding of the pyrophosphate group of NAD(P)+ or NADPH (24). So far, known MDR superfamily members play important roles in biological oxidation/reduction and exist in all organisms (25). However, the majority of the enzymes indicated to belong to the MDR group are depicted as putative gene products identified by genome sequencing and are poorly characterized (26). The present discovery of CurA in E. coli and its functional identification will allow further elucidation of the evolution and distribution of unknown enzymes in the MDR superfamily, which will pave the way for their use for the biotransformation of natural compounds and new chemical inputs into the environment.
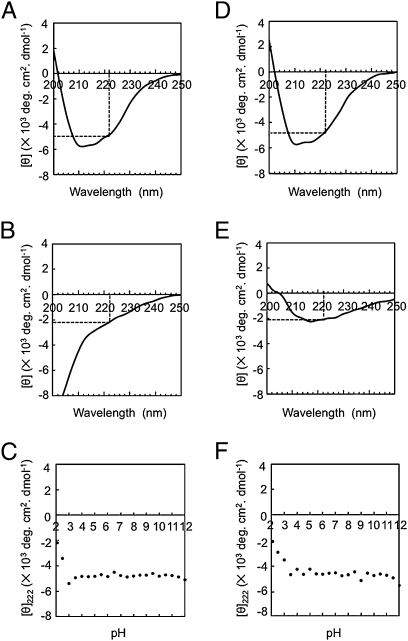
During the characterization of CurA, we surprisingly found that it shows a wide pH stability range (pH 4.5–12.0), particularly including alkaline pH values. To obtain information on the secondary structure (27) of CurA, far-UV circular dichroism (CD) spectra were measured (SI Materials and Methods). The CD spectra of the enzyme in a broad pH value range (3.5–12.0) were similar to that under usual pH conditions (pH 6.0) (Fig. 5A), but those at pHs 2.1 to 3.0 were different (Fig. 5B). These findings suggested that the ellipticity at 222 nm (secondary structure) of CurA did not change at pHs 3.5 to 12.0 (Fig. 5C). On CD measurement after the pH had been shifted from various pH values (pH 2.1–12.0) to 6.0, the CD spectra of CurA (Fig. 5D) were similar to one another for the shift from pH 3.5 to 12.0, but not from pH 2.1 to 3.0 (Fig. 5E), and the ellipticities at 222 nm for pH 3.5 to 12.0 were also similar to one another (Fig. 5F). The data for the pH stability of the CurA activity and the pH stability of the CurA secondary structure demonstrated that CurA is structurally stable from pH 4.5 to 12.0 (Table S5).
Fig. 5.
CD spectra of the curcumin-converting enzyme and the ellipticity in different pH values. The CD spectra of CurA treated in 10-mM Britton-Robinson buffer pH 6.0 (A) and pH 2.1 (B); 0.1 mg/mL of the purified enzyme was treated at different pH values in 10-mM Britton-Robinson buffer and CD spectra were measured. The mollar ellipticity (θ) was then calculated for the different pHs. θ222 was denoted by dotted line. (C) θ222 at different pH values. The CD spectra of CurA after the pH was shifted from 6.0 to 6.0 (D) and from 2.1 to 6.0 (E); 0.1 mg/mL of the purified enzyme was treated at different pH values in 10-mM Britton-Robinson buffer and then shifted to the usual pH 6.0 using 40-mM Britton-Robinson buffer (pH 6.0), and then the CD spectrum was measured and θ222 was denoted by a dotted line. (F), θ222 after the pH had been shifted from various values to 6.0.
In humans and animals, four reduction products (i.e., THC and hexahydrocurcumin as major products, and DHC and octahydrocurcumin as minor ones) (7, 17–20) were detected, although the nature of the enzymes involved in the formation of such products is unknown. In microorgamisms, we first found that both DHC and THC are the products, using the purified enzyme (Fig. 2). It was previously found that THC is more potent than curcumin as to anti-inflammatory (28), antidiabetic, and antihyperlipidemic (29) activity, and equally potent as to antioxidant activity (30, 31). THC exhibits its biological activities at a very low concentration both in vivo and in vitro. For example, 50 μM THC can inhibit the metastasis of HT1080 cancer cells in vitro (32). THC also exhibits antioxidant activity at a concentration of 18.7 μM in vitro, using the radical scavenging activity of 2,2-diphenyl-1-picrylhydrazyl (33). In the mouse brain, THC at a concentration of 1.286 μM has an anti-inflammatory effect and can prevent Alzheimer's disease by inhibiting IL-1β (34). These findings would uncover the mystery of a reduced age-adjusted prevalence of Alzheimer's disease (35) and Parkinson disease (36) in India, where the people consume curcumin in their diet. It is speculated that curcumin in the human intestine is converted into THC by E. coli CurA and that the produced THC is responsible for many various activities in the human body. Further studies are required to understand the participation of the common human commensal E. coli in curcumin conversion. Our research here shed new light on the role of human intestinal microorganisms in the mechanism of curcumin metabolism in vivo. CurA is a thus-far unknown enzyme found to produce THC from curcumin, and therefore it would be a potent catalyst for the production of a large amount of the value-added product in vitro.
Materials and Methods
Isolation of Curcumin-Converting Microorganisms from Human Feces.
We isolated microorganisms with curcumin-degrading ability from human feces. Fresh human fecal samples were inoculated into test tubes containing 10 mL of a synthetic medium comprising 0.05% (wt/vol) KH2PO4, 0.05% K2HPO4, 0.1% NaCl, 0.05% MgSO4·7H2O, 0.2% yeast extract, 0.05% curcumin and 5% Tween20, and then incubated at 37 °C for 24 h. Tween20 was added to enhance curcumin solubility in the culture medium. Drops of the incubated liquid cultures were spread on agar plates (of the above-mentioned medium), and the microorganisms that grew were isolated. Each of the isolated strains was then inoculated into a test tube including 10 mL of the same medium. The cells were harvested by centrifugation, washed twice with 0.1 M KPB (pH 6.0), and then suspended in the same buffer. The suspended cells were disrupted by sonication at 10 W for 10 min with an ultrasonic disrupter (Microson Ultrasonic Cell Disrupter XL; Misonix, Inc.). The cell debris was removed by centrifugation at 0 to 4 °C. The curcumin-converting abilities of the isolated strains were measured by means of the cell-free extract reaction. The reaction was carried out as described in the SI Materials and Methods subsection, Enzyme assays, using standard assay A in the absence of NADPH.
Purification of the Curcumin-Converting Enzyme from E. coli DH10B.
All purification steps were carried out at 0 to 4 °C. KPB (pH 6.0) was used throughout the purification. Centrifugation was performed for 20 min at 16,200 × g.
Step 1: Preparation of cell-free extracts.
Washed cells from 24 L of broth culture were suspended in 500 mL of 0.1 M KPB (pH 6.0) and then disrupted by sonication at 200 W for 30 min with an Insonator model 201 M (Kubota). The cell debris was removed by centrifugation.
Step 2: DEAE-Sepharose Fast Flow column chromatography.
Ammonium sulfate was added to the resulting supernatant solution to give 80% saturation. After centrifugation of the suspension, the precipitate was dissolved in 0.1 M KPB (pH 6.0) and then dialyzed against 10 mM KPB (pH 6.0). The resultant dialyzed solution was applied to a DEAE-Sepharose Fast Flow column (5 × 40 cm) equilibrated with 10 mM KPB (pH 6.0). Protein was eluted from the column with 0.5 L of 10 mM KPB (pH 6.0). The enzyme was collected as the flow-through fraction, followed by the addition of ammonium sulfate to give 80% saturation. The precipitate was collected by centrifugation, and dissolved in 0.1 M KPB (pH 6.0), followed by dialysis against 10 mM KPB (pH 6.0).
Step 3: Blue Sepharose 6 Fast Flow column chromatography.
The dialyzed solution was applied to a Blue Sepharose 6 Fast Flow column (3 × 15 cm) equilibrated with 10 mM KPB (pH 6.0). Protein was eluted by increasing KCl linearly from 0 to 2 M in the same buffer. The active fractions were collected, and then ammonium sulfate was added to give 80% saturation. After centrifugation of the suspension, the precipitate was dissolved in 0.1 M KPB (pH 6.0), followed by dialysis against 10 mM KPB (pH 6.0).
Step 4: Hitrap Benzamidine FF (high sub) column chromatography.
Because the structure of nicotinamide and/or adenine in the parts of NADPH resembled that of the benzamidine, the enzyme solution from step 3 was applied to a Hitrap Benzamidine FF (high sub) column (1.6 × 2.5 cm) equilibrated with 10 mM KPB (pH 6.0). Protein was eluted from the column by increasing KCl linearly from 0 to 1 M in the same buffer. The active fractions were combined and then precipitated with ammonium sulfate at 80% saturation. After centrifugation of the suspension, the precipitate was collected by centrifugation and then dissolved in 0.1 M KPB (pH 6.0). The resultant solution was dialyzed against 10 mM KPB (pH 6.0).
Step 5: Mimetic Red 3 A6XL column chromatography.
The enzyme solution from step 4 was applied to a PIKSI (R) M Test Kit 0013 Mimetic Red 3 A6XL column (1 × 1 cm) (Affinity Chromatography Ltd.) equilibrated with 10 mM KPB (pH 6.0). Protein was eluted from the column by increasing KCl linearly from 0 to 2 M in the same buffer. The homogeneity of the purified protein was confirmed by SDS/PAGE.
Nucleotide Sequence Accession Number.
The nucleotide sequence data reported in this paper appear in the DNA Data Base in Japan/GenBank database under accession numbers AB609595 and AB583756.
Description of SI Materials and Methods.
The materials used in this study, identification of a curcumin-converting microorganism, culture conditions for E. coli DH10B, and the two enzyme assay systems used to measure CurA activity are described in SI Materials and Methods. Additionally, electrophoresis, peptide mass fingerprinting, cloning and overexpression of the gene encoding CurA, purification of the curcumin-converting enzyme from E. coli BL21-CodonPlus(DE3)-RIL harboring pET-curA, molecular mass determination, metal analysis, substrate specificity, CD analysis, identification of the reaction products, time course of curcumin conversion and product formation, and site-directed mutagenesis were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Kentaro Shiraki for help with the circular dichroism spectra analysis and for the valuable advice; Dr. Yoichi Takase for fast atom bombardment mass spectrometry analysis; and Mrs. Mitsue Arimoto for the help with MALDI-TOF/MS analysis.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the DNA Databank of Japan (DDBJ) database (accession nos. AB609595 and AB583756).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016217108/-/DCSupplemental.
References
- 1.Aggarwal BB, Sundaram C, Malani N, Ichikawa H. Curcumin: The Indian solid gold. Adv Exp Med Biol. 2007;595:1–75. doi: 10.1007/978-0-387-46401-5_1. [DOI] [PubMed] [Google Scholar]
- 2.Sharma RA, Gescher AJ, Steward WP. Curcumin: The story so far. Eur J Cancer. 2005;41:1955–1968. doi: 10.1016/j.ejca.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: From ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Alloza M, Borrelli LA, Rozkalne A, Hyman BT, Bacskai BJ. Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. J Neurochem. 2007;102:1095–1104. doi: 10.1111/j.1471-4159.2007.04613.x. [DOI] [PubMed] [Google Scholar]
- 5.Nandakumar DN, Nagaraj VA, Vathsala PG, Rangarajan P, Padmanaban G. Curcumin-artemisinin combination therapy for malaria. Antimicrob Agents Chemother. 2006;50:1859–1860. doi: 10.1128/AAC.50.5.1859-1860.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panchatcharam M, Miriyala S, Gayathri VS, Suguna L. Curcumin improves wound healing by modulating collagen and decreasing reactive oxygen species. Mol Cell Biochem. 2006;290:87–96. doi: 10.1007/s11010-006-9170-2. [DOI] [PubMed] [Google Scholar]
- 7.Ravindranath V, Chandrasekhara N. In vitro studies on the intestinal absorption of curcumin in rats. Toxicology. 1981;20:251–257. doi: 10.1016/0300-483x(81)90056-1. [DOI] [PubMed] [Google Scholar]
- 8.Crossman LC, et al. A commensal gone bad: Complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol. 2010;192:5822–5831. doi: 10.1128/JB.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, et al. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS ONE. 2010;5:e8700. doi: 10.1371/journal.pone.0008700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferenci T, et al. Genomic sequencing reveals regulatory mutations and recombinational events in the widely used MC4100 lineage of Escherichia coli K-12. J Bacteriol. 2009;191:4025–4029. doi: 10.1128/JB.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macconkey A. Lactose-fermenting bacteria in feces. J Hyg (Lond) 1905;5:333–379. doi: 10.1017/s002217240000259x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin M. Differentiation of E. coli and A. aerogenes on a simplified eosin-methylene blue agar. J Infect Dis. 1918;23:43–47. [Google Scholar]
- 13.Durfee T, et al. The complete genome sequence of Escherichia coli DH10B: Insights into the biology of a laboratory workhorse. J Bacteriol. 2008;190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youn B, et al. Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970. J Biol Chem. 2006;281:40076–40088. doi: 10.1074/jbc.M605900200. [DOI] [PubMed] [Google Scholar]
- 15.Hori T, et al. Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop. J Biol Chem. 2004;279:22615–22623. doi: 10.1074/jbc.M312655200. [DOI] [PubMed] [Google Scholar]
- 16.Britton HTS, Robinson RA. Universal buffer solutions and the dissociation constant of veronal. J Chem Soc. 1931;458:1456–1462. [Google Scholar]
- 17.Hoehle SI, Pfeiffer E, Sólyom AM, Metzler M. Metabolism of curcuminoids in tissue slices and subcellular fractions from rat liver. J Agric Food Chem. 2006;54:756–764. doi: 10.1021/jf058146a. [DOI] [PubMed] [Google Scholar]
- 18.Pan MH, Huang TM, Lin JK. Biotransformation of curcumin through reduction and glucuronidation in mice. Drug Metab Dispos. 1999;27:486–494. [PubMed] [Google Scholar]
- 19.Ireson C, et al. Characterization of metabolites of the chemopreventive agent curcumin in human and rat hepatocytes and in the rat in vivo, and evaluation of their ability to inhibit phorbol ester-induced prostaglandin E2 production. Cancer Res. 2001;61:1058–1064. [PubMed] [Google Scholar]
- 20.Ireson CR, et al. Metabolism of the cancer chemopreventive agent curcumin in human and rat intestine. Cancer Epidemiol Biomarkers Prev. 2002;11:105–111. [PubMed] [Google Scholar]
- 21.Persson B, Hedlund J, Jörnvall H. Medium- and short-chain dehydrogenase/reductase gene and protein families: The MDR superfamily. Cell Mol Life Sci. 2008;65:3879–3894. doi: 10.1007/s00018-008-8587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nordling E, Jörnvall H, Persson B. Medium-chain dehydrogenases/reductases (MDR). Family characterizations including genome comparisons and active site modeling. Eur J Biochem. 2002;269:4267–4276. doi: 10.1046/j.1432-1033.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 23.Vitale A, Rosso F, Barbarisi A, Labella T, D'Auria S. Properties and evolution of an alcohol dehydrogenase from the Crenarchaeota Pyrobaculum aerophilum. Gene. 2010;461:26–31. doi: 10.1016/j.gene.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Pereira PJB, et al. Structure of human biliverdin IXbeta reductase, an early fetal bilirubin IXbeta producing enzyme. Nat Struct Biol. 2001;8:215–220. doi: 10.1038/84948. [DOI] [PubMed] [Google Scholar]
- 25.Jörnvall H, Höög JO, Persson B. SDR and MDR: Completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 1999;445:261–264. doi: 10.1016/s0014-5793(99)00130-1. [DOI] [PubMed] [Google Scholar]
- 26.Dick RA, Kensler TW. The catalytic and kinetic mechanisms of NADPH-dependent alkenal/one oxidoreductase. J Biol Chem. 2004;279:17269–17277. doi: 10.1074/jbc.M400427200. [DOI] [PubMed] [Google Scholar]
- 27.Strickland EH. Circular dichroism of horseradish peroxidase and its enzyme-substrate compounds. Biochim Biophys Acta. 1968;151:70–75. doi: 10.1016/0005-2744(68)90162-9. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay A, Basu N, Ghatak N, Gujral PK. Anti-inflammatory and irritant activities of curcumin analogues in rats. Agents Actions. 1982;12:508–515. doi: 10.1007/BF01965935. [DOI] [PubMed] [Google Scholar]
- 29.Pari L, Murugan P. Antihyperlipidemic effect of curcumin and tetrahydrocurcumin in experimental type 2 diabetic rats. Ren Fail. 2007;29:881–889. doi: 10.1080/08860220701540326. [DOI] [PubMed] [Google Scholar]
- 30.Sugiyama Y, Kawakishi S, Osawa T. Involvement of the β-diketone moiety in the antioxidative mechanism of tetrahydrocurcumin. Biochem Pharmacol. 1996;52:519–525. doi: 10.1016/0006-2952(96)00302-4. [DOI] [PubMed] [Google Scholar]
- 31.Osawa T, Sugiyama Y, Inayoshi M, Kawakishi S. Antioxidative activity of tetrahydrocurcuminoids. Biosci Biotechnol Biochem. 1995;59:1609–1612. doi: 10.1271/bbb.59.1609. [DOI] [PubMed] [Google Scholar]
- 32.Yodkeeree S, Garbisa S, Limtrakul P. Tetrahydrocurcumin inhibits HT1080 cell migration and invasion via downregulation of MMPs and uPA. Acta Pharmacol Sin. 2008;29:853–860. doi: 10.1111/j.1745-7254.2008.00792.x. [DOI] [PubMed] [Google Scholar]
- 33.Somparn P, Phisalaphong C, Nakornchai S, Unchern S, Morales NP. Comparative antioxidant activities of curcumin and its demethoxy and hydrogenated derivatives. Biol Pharm Bull. 2007;30:74–78. doi: 10.1248/bpb.30.74. [DOI] [PubMed] [Google Scholar]
- 34.Begum AN, et al. Curcumin structure-function, bioavailability, and efficacy in models of neuroinflammation and Alzheimer's disease. J Pharmacol Exp Ther. 2008;326:196–208. doi: 10.1124/jpet.108.137455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganguli M, et al. Apolipoprotein E polymorphism and Alzheimer disease. Arch Neurol. 2000;57:824–830. doi: 10.1001/archneur.57.6.824. [DOI] [PubMed] [Google Scholar]
- 36.Muthane U, Yasha TC, Shankar SK. Low numbers and no loss of melanized nigral neurons with increasing age in normal human brains from India. Ann Neurol. 1998;43:283–287. doi: 10.1002/ana.410430304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.