Coleman and Chisholm (1) analyzed shotgun metagenomic data and concluded that populations of marine Prochlorococcus were indistinguishable between the North Atlantic and the North Pacific Oceans except in their phosphorus (P) utilization genes. They attributed these findings to recent horizontal gene transfer (HGT) of P genes in the genome of the Atlantic Prochlorococcus, favored by the stronger P-limitation in the Atlantic vs. the Pacific Ocean. We analyzed the datasets in their article (1) and found that the most abundant surface Prochlorococcus ecotype is indistinguishable between the two sampling sites [Hawaii Ocean Time Series (HOT) for Pacific and Bermuda Atlantic Time Series (BATS) for Atlantic] in most, if not all, of the P-utilization genes identified previously to be more abundant at BATS (1). In particular, the two corresponding populations encode the phoBR genes (phosphate two-component response regulators), whereas they both lack the phn operon (phosphonate utilization) (Table 1). phn genes become relatively more abundant with higher depth at BATS, which accounts for the results reported previously (1), but this is likely associated with non-Prochlorococcus populations. For instance, the reads of the BATS-50m small-insert library encoding Prochlorococcus-like phn genes have sister reads matching more frequently non-Prochlorococcus than Prochlorococcus genomes. Similarly, the higher abundance of phoBR in deeper (but not surface) waters at BATS vs. HOT is associated with non-Prochlorococcus taxa (Table 1) and the presence of a mixture of Prochlorococcus ecotypes, which are not all shared between BATS and HOT (Fig. 1). Only phoA [putative alkaline phosphatase (2)] clearly shows higher abundance at BATS-20m vs. HOT-25m; however, this is inconsistent with the results for the remaining pho genes, all cells encode an alternative dedA-type alkaline phosphatase, and many phoA-encoding reads likely originate from non-Prochlorococcus organisms, including viruses (e.g., note that phoA is twice as abundant at BATS-50m vs. BATS-100m, although total Prochlorococcus signal is similar).
Table 1.
Analysis of pyrosequencing reads encoding P-utilization genes in each sample
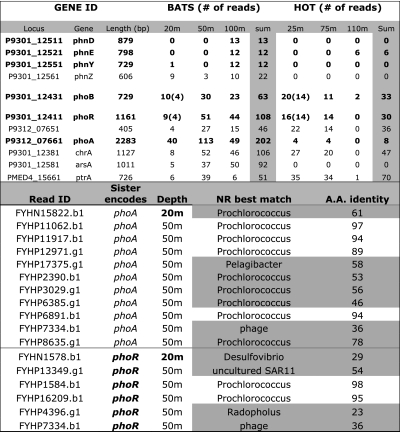 |
(Upper) The number of reads encoding P-related genes for each 454 shotgun dataset, identified essentially as described previously (1). High-light adapted Prochlorococcus abundance is about six times higher at HOT-25m vs. BATS-20m and three times higher at BATS-50m or BATS-100m vs. BATS-20m; thus, the numbers shown must be divided by 6 and 3 for HOT-25m and BATS-50m/BATS-100m, respectively, to normalize for population abundance. Only genes identified previously (1) to be more abundant at BATS and not hypothetical are shown. (Lower) Phylogenetic affiliation of the pho-encoding clones from BATS. The table shows the genome that provided the best Blastx match in nr database (fourth column) for each Sanger read (first column) that does not encode a pho gene [otherwise Prochlorococcus was the best match because the Prochlorococcus pho operon was used as reference sequence to recruit reads (1)] and whose sister read encodes a pho gene (second column). Note that many reads (<50% of the total, highlighted in gray) had non-Prochlorococcus genomes as best match or matched Prochlorococcus with low amino acid identity, indicating that they probably originate from an non-Prochlorococcus genetic background. These results contrast with a genome average of less than “9.3% of the putative Prochlorococcus clones at BATS matched Prochlorococcus on one end and a different taxon on the other end (1).” Appropriate clone data for HOT are not available for comparison (1). Also note that numbers in parentheses in the upper panel denote the number of reads that map on the phoBR-encoding contig from HOT-25m at the 95% nucleotide identity cutoff level [which selects for reads originating from the abundant population represented by Fig. 1 relatively to the approach taken previously that did not discriminate between Prochlorococcus ecotypes (1)]. These data, especially when also considering the fraction of BATS gene sequences originating from non-Prochlorococcus organisms (lower panel results), show that the BATS-20m Prochlorococcus population encodes similar phoBRA gene content compared with its HOT-25m counterpart.
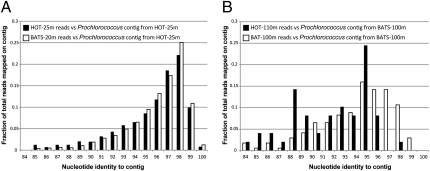
Fig. 1.
Comparing Prochlorococcus populations from HOT and BATS. Graphs show the coverage plots, performed essentially as described previously (4), of a high-light adapted Prochlorococcus contig assembled from HOT-25m (A) and BATS-100m (B) 454 shotgun datasets by selected 454 datasets (figure key). The contigs represent the population that makes up more than 80% of the total high-light adapted Prochlorococcus population within each sample. Note that the two surface populations (i.e., HOT-25m and BATS-20m) are indistinguishable from each other when compared against the same reference contig from HOT-25m (A). However, the populations from deeper waters are clearly differentiated/divergent from each other when compared against the reference contig from BATS-100m (B). These results indicate that the deep population within each site is probably heterogeneous [i.e., composed of different subpopulations (ecotypes)] and not identical (and directly comparable) between BATS and HOT. Contigs are available by the authors upon request.
The reason(s) for the differential presence of pho/phn genes and Prochlorococcus ecotypes in deeper waters may be related to factors other than P-limitation, such as seasonal deep-water mixing and depth-stratified dissolved organic matter content, as hypothesized previously (3). Such seasonal fluctuations are also more consistent with recent HGT than long-lived P-limitation. The fact that surface populations (20–25 m) do not show significant differences in P-gene abundance and that BATS was sampled at the beginning of the winter deep-water mixing whereas HOT was stably stratified during sampling (1) strongly support these interpretations. Thus, the basis for the higher abundance (if any) of P-genes in the Atlantic Ocean is likely more complicated than previously proposed (1). Surface Prochlorococcus P-genes might be phylogenetically (in contrast to presence/absence) distinct between BATS and HOT, due, for instance, to fine-tuning to in situ phosphorus species and/or concentrations, as suggested previously (1). However, additional data are necessary to establish the true phylogenetic identity of every P-gene sequence analyzed and rule out alternative explanations, such as sample-specific variations, before robust conclusions can emerge. Our results also highlight the need to better understand the genomic variability and taxonomic relationships of different Prochlorococcus ecotypes to more fully resolve the ecological underpinnings of population and gene distributions.
Footnotes
The authors declare no conflict of interest.
References
- 1.Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kathuria S, Martiny AC. Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ Microbiol. 2011;13:74–83. doi: 10.1111/j.1462-2920.2010.02310.x. [DOI] [PubMed] [Google Scholar]
- 3.Martinez A, Tyson GW, Delong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12:222–238. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- 4.Konstantinidis KT, DeLong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]