Abstract
The association between embryos of the spotted salamander (Ambystoma maculatum) and green algae (“Oophila amblystomatis” Lamber ex Printz) has been considered an ectosymbiotic mutualism. We show here, however, that this symbiosis is more intimate than previously reported. A combination of imaging and algal 18S rDNA amplification reveals algal invasion of embryonic salamander tissues and cells during development. Algal cells are detectable from embryonic and larval Stages 26–44 through chlorophyll autofluorescence and algal 18S rDNA amplification. Algal cell ultrastructure indicates both degradation and putative encystment during the process of tissue and cellular invasion. Fewer algal cells were detected in later-stage larvae through FISH, suggesting that the decline in autofluorescent cells is primarily due to algal cell death within the host. However, early embryonic egg capsules also contained encysted algal cells on the inner capsule wall, and algal 18S rDNA was amplified from adult reproductive tracts, consistent with oviductal transmission of algae from one salamander generation to the next. The invasion of algae into salamander host tissues and cells represents a unique association between a vertebrate and a eukaryotic alga, with implications for research into cell–cell recognition, possible exchange of metabolites or DNA, and potential congruence between host and symbiont population structures.
Keywords: photosymbiont, endosymbiosis, amphibian, chlorophyte
Mutualistic endosymbiosis occurs in many protist and even metazoan groups. One of the most prominent forms of endosymbiosis is between photosynthetic microbes and eukaryotic hosts. Photosynthetic endosymbionts and plastids are hosted by many eukaryote lineages, including several invertebrate animals (1). However, despite the ability of pathogenic organisms to enter vertebrate cells (2), there is an apparent absence of mutualist endosymbionts in vertebrates (3). The inability of symbionts to enter vertebrate host cells may be attributable to the adaptive immune system (3), a gnathostome synapomorphy that recognizes and destroys foreign cells (4). Associations between microbes and vertebrate embryos are possible candidates for intracellular symbioses because such early associations may precede an adaptive immune response.
The symbiosis between algae and embryos of Ambystoma maculatum (spotted salamander) was first reported more than 120 y ago (5). These salamanders spend most of their adult lives underground but emerge for semiannual spring breeding congregations in vernal pools. Eggs are deposited in these pools, where embryos develop through metamorphosis. Once transformed, the young salamanders leave these pools to live underground. Algae live in direct association with embryos inside salamander egg capsules, which are contained in large jelly masses (6). Individual egg capsules appear green due to dense accumulations of algae surrounding the embryo (Fig. 1A). The mutual benefit of this facultative association has been clearly established through exclusion experiments. Clutches raised in the dark do not accrue detectable algae (6–9). The presence of algae in these experiments correlates with earlier hatching (6, 9), decreased embryonic mortality (7, 8), more synchronous hatching (8, 9), and reaching a larger size (8) and later developmental stage (9) at hatching. Additionally, algal growth is minimal in egg capsules after embryos are removed (8), indicating that the embryos, and not the egg capsules, aid algal growth. Algae are thought to benefit from nitrogenous wastes released by the embryos [ref. 6; supported by research in the closely related Ambystoma gracilis (10)], whereas salamander embryos benefit from increased oxygen concentrations associated with the algae [refs. 11–13; however, others (14) found no oxygen benefit using older techniques]. Presence of the algae also correlates with decreased cilium-mediated rotation of early-stage embryos and increased muscular contractions during later embryonic stages (9), both of which may be secondary effects of modulated oxygen levels.
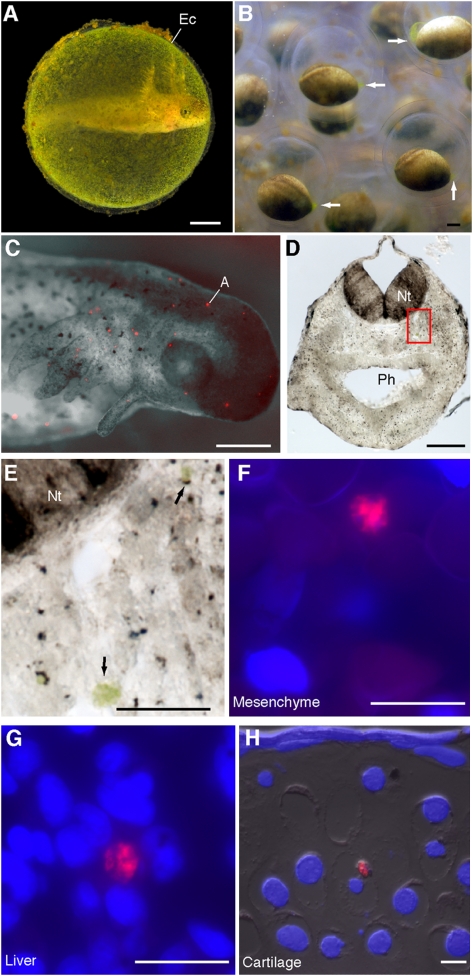
Fig. 1.
Algal cells from the egg capsule invade salamander tissues. (A) A Stage-44 embryo inside its egg capsule (Ec). (B) Still frame taken from time-lapse recordings of Stage-15 embryos. Concentrations of algae occur synchronously, adjacent to the blastopore (arrows). (C) Black-and-white and fluorescent overlay of a Stage-39 embryo removed from its egg capsule; lateral view of the head. Red spots are autofluorescing cells (A) described in text. (D and E) Scattered algal cells embedded in Stage-37 cranial mesenchyme appear green under light microscopy. Boxed region in D shown magnified in E. (F–H) FISH-stained algal cells embedded in (F) Stage-37 cranial mesenchyme, (G) Stage-46 liver, and (H) Stage-46 ceratohyal chondrocyte. Nt, neural tube; Ph, pharynx. (Scale bars: 1 mm in A–D; 50 μm in E; 20 μm in F–H.)
Despite these physiological studies, structural and developmental aspects of this symbiosis remain unclear. Early attempts to identify or culture the algae from oviducts of gravid female salamanders failed, discouraging further investigations into the anatomical associations between host and symbiont (8, 15). All previous research describes the relationship between the algae and embryo as an ectosymbiotic mutualism, with no intratissue or intracellular stages.
In this study we show that algae invade embryonic salamander tissues (Figs. 1 and 2) and cells (Figs. 3 and 4) during embryonic development. This intracellular invasion resembles the photosynthetic endosymbiosis that has occurred in many protists (16) and metazoan invertebrates (1, 17, 18) but has not been reported in a vertebrate host.
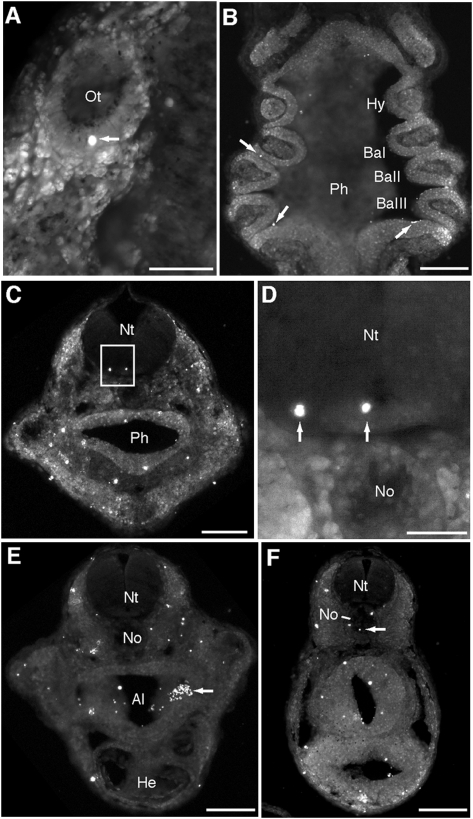
Fig. 2.
Distribution of algal cells in salamander embryos. (A and B) Coronal and (C–F) transverse vibratome sections imaged for chlorophyll autofluorescence. Boxed region in C shown in higher magnification in D. Arrows indicate autofluorescent algal cells. Individual algae were embedded in (A) the otic capsule (Ot); (B) the pharynx (Ph) and pharyngeal clefts; (C and D) several regions of the head, including the neural tube (Nt); and several regions of the anterior (E) and posterior trunk (F), including the alimentary canal (Al) and notochord (No). Ba, branchial arch; He, heart; Hy, hyoid arch. (Scale bars: 100 μm in A and D; 1 mm in B, C, E, and F.)
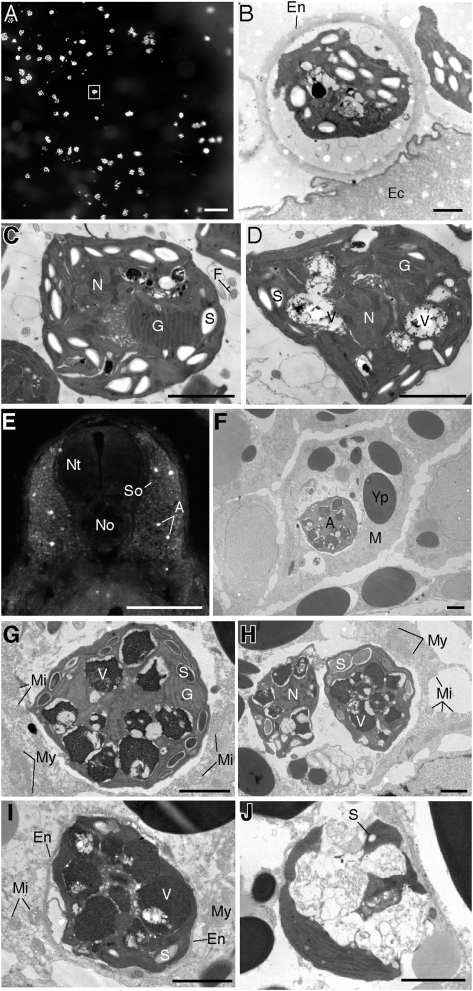
Fig. 3.
Intracapsular and intracellular algae. (A) Whole-mount image of egg capsule (Ec) showing clusters of algal cells. (B) TEM image of algal cells encased by an outer envelope (En; boxed region in 3A) on the inner border of the innermost egg capsule. (C and D) TEM images of intracapsular algae revealing starch granules (S), chloroplast grana (G), nuclei (N), and vacuoles (V). (E) Coronal vibratome section through the trunk region of a Stage-37 embryo. White spots are autofluorescing cells embedded in the somites (So). (F–J) Intracellular algae found within somites. (F) Algal cell embedded inside a salamander myocyte (M). (G) Higher magnification of intracellular alga in F. Salamander mitochondria (Mi) and myofibrils (My) occur in the surrounding host cytoplasm. (H) Paired algal cells within a host cell. (I) Algal cell surrounded by a thickened outer envelope. (J) Degraded algal cell without an outer envelope. F, flagella; Nt, neural tube; No, notochord; Yp, yolk platelet. (Scale bars: 100 μm in A; 2 μm in B–D; 1 mm in E; 2 μm in F–J.)
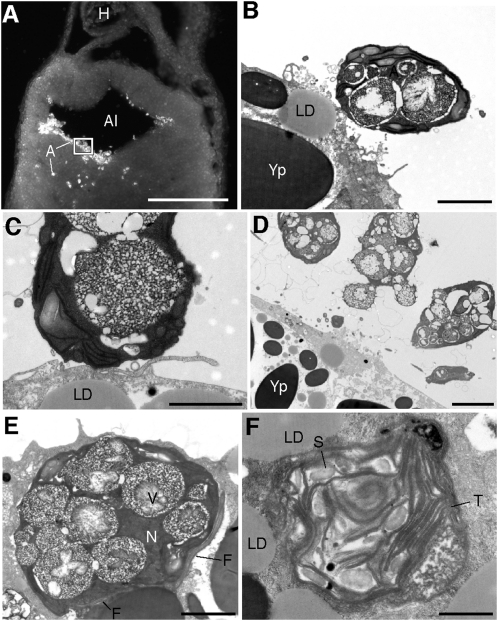
Fig. 4.
Algal cells within the alimentary canal during the process of cellular invasion. (A) Fluorescent image of a coronal vibratome section through a Stage-35 embryo showing an aggregation of algae (A) inside the alimentary canal (Al) and invading the surrounding endoderm. (B–F) TEM images from boxed region in A. (B–D) Algal cells in the alimentary canal, adjacent to salamander endoderm. (E) Algal cell inside endodermal cell of the alimentary canal containing many large vacuoles (V) and paired flagella (F). (F) Free thylakoid membranes (T) and starch granules (S) inside cytoplasm of host cell shown in E. H, heart; LD, lipid droplet; N, nucleus; Yp, yolk platelet. (Scale bars: 1 mm in A; 2 μm in B and C; 5 μm in D; 2 μm in E; 1 μm in F.)
Results
Phylogenetic reconstruction, using chloroplast 16S (1,362 bp; GenBank accession no. HM590633) and nuclear 18S (1,714 bp; GenBank accession no. HM590634) rDNA sequences from intracapsular algae, places the algae in the Chlorophyceae, specifically the Chlamydomonadales (Fig. S1). Two algal clones of each PCR amplification had >99% sequence identity to each other, and the 16S and 18S rDNA phylogenies were congruent. These observations suggest that intracapsular algal cells in our samples comprise a single species. The published literature on this association (6, 8, 9) refers to the algae as “Oophila amblystomatis” (Lambert ex Printz) (19) on the basis of an informal species designation by Lambert (6), although—to date—no formal taxonomic description exists. Previous studies reported additional bacteria and protists within the egg capsules (6, 14). Our 18S rDNA amplification clones included sequences from ciliate and cercozoan protists, and our 16S rDNA amplification clones included sequences for the prokaryotes Pedobacter sp., Acidovorax sp., Duganella zoogloeoides, Janthinobacterium sp., Verrucomicrobiales sp., Chitinophaga sp., and Flavobacterium sp. However, no other green algae were identified from the egg capsules aside from the conserved Oophila sequences.
Intratissue algae were first detected through their chlorophyll autofluorescence (480 nm excitation) in a Stage-39 embryo (Fig. 1C). Autofluorescent cells were scattered throughout the embryo, with a high concentration in the anterior end of the ventral abdomen. Algal cell entry into host tissues was verified through PCR amplification of symbiont 18S rDNA (20), time-lapse photography, and fluorescence microscopy.
The timing of algal cell entry was determined through algal 18S rDNA PCR. Embryos were extracted from their egg capsules and rinsed thoroughly before DNA extraction. Oophila-specific 18S rDNA was amplified and sequenced from Harrison (21) Stage-26 (pharyngula), and older, salamander embryos. It was not detected in Stage-17 (neurula), or earlier, embryos (Fig. S2). However, algae were detected microscopically on the innermost egg capsule during these early stages (Fig. 3 A and B).
Gilbert (6) described a concentration of algae around the salamander blastopore (“proctodeum”). Time-lapse photography of A. maculatum embryos revealed this concentration forming in Stage-15 embryos (Fig. 1B and Movie S1). This concentration of algal cells precedes their detection inside salamander tissues through PCR, indicating that the algal bloom outside the blastopore precedes subsequent algal cell invasion into the salamander embryo host.
By imaging chlorophyll autofluorescence, we identified algal cells in embryonic epidermis, optic cup, neural tube, cranial mesenchyme, presumptive lens, somitic myotome, and otic capsule between Stages 35 and 44 (Figs. 2, 3E, and 4A). Autofluorescing cells were more abundant in the yolk and within the alimentary canal (Fig. 4A). Intratissue autofluorescent cells and algae from the intracapsular fluid both fluoresced red under the same excitation wavelength (480 nm). Many of the intratissue autofluorescing cells appeared green under light microscopy in vibratome sections (Fig. 1 D and E). Autofluorescing cells were no longer detectable in feeding larvae (Stage 46, 3 to 4 wk of development). Although many algal cells invaded the alimentary canal and surrounding tissues, most algae persist in the intracapsular fluid and are released into the surrounding water upon hatching.
The genetic identity of intratissue algae was verified through fluorescence in situ hybridization (FISH) using an oligonucleotide probe that targets Oophila 18S rRNA. FISH-positive algal cells were detected in several Stage-37 tissues, including the cranial mesenchyme (Fig. 1F) and endoderm. By Stage 46, FISH-positive cells were found in the larval liver (n = 2; Fig. 1G) and ceratohyal cartilage (n = 1; Fig. 1H) after the cessation of detectable algal autofluorescence. We detected few algal cells in our Stage-46 sample through FISH, in comparison with the widespread distribution of algae during embryonic Stages 35–44.
Transmission electron microscopy (TEM) was used to determine the ultrastructure of algae attached to the egg capsules (Fig. 3B), motile algae within the egg capsule fluid (Fig. 3 C and D), algae within the alimentary canal (Fig. 4 B–D), and algae embedded in salamander tissues (Figs. 3 F–J and 4E). Algal cells were initially present in small clusters on individual egg capsules of early embryos (Fig. 3A). Algae on the egg capsules were encased in an outer envelope (Fig. 3B) and lacked flagella. These cells were recessed from the outer envelope walls.
Vibratome sections containing autofluorescent cells were dissected and prepared for TEM, allowing ultrastructural analysis of 26 algal cells inside salamander tissues during Stages 35, 37, and 42 (e.g., Fig. 3 F–I). Each intratissue alga contained starch granules inside the chloroplast, chloroplast grana, and numerous thylakoid membranes (e.g., Fig. 3G), which were also found in intracapsular algae (Fig. 3 C and D). These conserved ultrastructural features also verify the green algal identity of intratissue autofluorescent cells (22).
Surprisingly, many autofluorescent algal cells were embedded within the cytoplasm of differentiated salamander host cells. Intracellular algae were found in autofluorescent somitic cells (n = 11; Fig. 3 F–I) and embedded within endodermal cells surrounding the alimentary canal (n = 13; Fig. 4E). Intracellular algae often occurred in close proximity to host cell mitochondria (Fig. 3 G–I) and occasionally bordered host cell nuclei. Host cells showed no signs of cellular necrosis or apoptosis and were structurally similar to adjacent, alga-free, cells. Although most intracellular algal cells appeared intact (Fig. 3 G–I), several were degraded (Fig. 3J). All host somite cells were undergoing myocyte differentiation, apparent through myofibril formation within the cytoplasm (Fig. 3 G–I). Most intracellular algae occurred as single cells, although three pairs of intracellular algae were also observed (Fig. 3H). The absence of detectable mitosis or cytokinesis in any of the singular or paired intracellular algae suggests that these paired cells invaded the salamander host cell together.
Several changes to algal cells were associated with host tissue and cellular invasion. Relative area measurements of starch granules and vacuoles were compared between intracapsular (n = 26), intracellular (n = 24), and extracellular algae found within the embryonic alimentary canal (n = 18). There were no differences in starch granule sizes [one-way ANOVA, F(2,56) = 2.89, P > 0.05]; however, starch granules constituted more cross-sectional area of intracapsular algae (10.5% SD 9.2%) than intracellular (5.9% SD 4.2%) or alimentary canal (3.4% SD 3.2%) algae [one-way ANOVA, F(2,56) = 4.70, P < 0.05]. Additionally, vacuoles constituted increasing cross-sectional area of intracapsular (11.0% SD 8.5%), intracellular (48.3% SD 13%), and alimentary canal (56.0% SD 15%) algae [F(2,65) = 135.18, P < 0.05]. Both intracapsular and invaded algae had paired flagella (Fig. 4E), although these were apparent in only a few intracellular algae.
An electron-translucent “perisymbiont zone” (20) surrounded several intracellular algae (Fig. 3 G–J). These perisymbiont zones were not associated with vesicle membranes, suggesting no distinct host symbiosome surrounding each alga (20). Five intracellular algae were encased in an outer envelope (Fig. 3I) similar to the envelope of encysted algae on the innermost egg capsule wall of early-stage embryos (Fig. 3B). Many intracellular algae that did not have an outer envelope appeared degraded (e.g., Fig. 3J), with vacuolar contents recessed and occasionally extruded from the cell.
Salamander endoderm adjacent to algal cells in the alimentary canal had indistinct plasma membranes and dissociated cytoplasm in regions of host–symbiont cellular contact, consistent with an active process of cellular invasion (Fig. 4 B–D). Algal cells within the alimentary canal occurred in loose aggregates (Fig. 4D), often adjacent to the boundary between two endodermal cells. Algae were not found in the alimentary canal of Stage-46 larvae either through autofluorescence, FISH, or TEM imaging.
Nine salamander cells containing algae also contained small collections of acellular thylakoid membranes and starch granules within their cytoplasm (Fig. 4F). These free thylakoids were more common in endodermal cells (n = 7) than in somitic cells (n = 2). They superficially resemble the intracellular kleptoplasts acquired by some sacoglossan mollusks (e.g., Elysia chlorotica) from their algal food (e.g., Vaucheria litorea) (23). However kleptoplasts are often enclosed in distinct vesicular membranes (17, 23), whereas the free thylakoids in salamander cells are not.
We extracted DNA from reproductive tissues of three adult female and three adult male A. maculatum. In female salamanders, Oophila-specific 18S rDNA was amplified from the posterior oviduct (two of three), anterior oviduct (one of three), and posterior ovary (one of three) but was not found in the medial oviduct, anterior or medial ovary. In adult male salamanders, Oophila-specific 18S rDNA was amplified from the Wolffian (one of three) and Müllerian (one of three) ducts but not from the anterior or posterior testes (Fig. S2).
Discussion
The invasion of green algae into salamander cells reveals that intracellular symbiosis of a phototroph can occur in a vertebrate host. This resembles endosymbioses between cyanobacterial Prochloron sp. and didemnid ascidian hosts (24), dinoflagellates of the genus Symbiodinium and cnidarian, poriferan, mollusk, platyhelminth, and foraminiferan hosts (1, 25), and between the green alga Chlorella and ciliate, poriferan, and cnidarian hosts (26). However, the Ambystoma–alga symbiosis is unique in several features. In the wild, nearly all intratissue algal–animal symbioses are obligatory and consist of photosynthate transfer from symbiont to host (17, 27). However, previous research has shown that the Ambystoma–alga association is facultative (6, 8, 15) and may not result in photosynthate transfer (28). Algal–animal associations are typically limited by the availability of carbon dioxide and photons. The latter are undoubtedly in short supply within the opaque tissues of a primarily fossorial salamander species.
Previous research on invertebrate–alga associations provides ready-made experimental hypotheses for the Ambystoma–alga symbiosis. Two reoccurring features of intracellular photosymbionts include photosynthate transfer and lateral gene transfer. Unlike the case in many invertebrate photosymbionts, photosynthate transfer from algae to A. maculatum has not been detected (27). However, the negative result of this earlier research is not conclusive (10). The methods used to detect carbon fixed by photosynthesis (27) should have revealed algal cells invading host tissues, but did not. Our data on this symbiosis justifies experimentally readdressing the possibility of photosynthate transfer by tracing labeled carbon isotopes. Such experiments will require careful attention to the occurrence of photosynthate transfer vs. algal cell digestion within the host.
Recent research has also revealed lateral gene transfer of algal nuclear genes into the sea slug E. chlorotica (29), which may be associated with maintaining kleptoplast function (however, see ref. 30). Because amphibian nuclei readily acquire foreign DNA from the cytoplasm (31), some scattered intracellular algae found in A. maculatum embryos may have transferred heritable algal DNA. However, unlike E. chlorotica kleptoplasts, there is no obvious role for transferred algal genes in the A. maculatum genome.
Structural Changes to Invading Algae.
Algal endosymbionts, such as Chlorella sp., often have a wide range of ultrastructural variation (32). This variation also was apparent in our ultrastructural analyses of Oophila sp. Several structural changes correlate with alimentary canal and cellular invasion. Algae found both within the alimentary canal and inside host cells had a proportional increase in vacuoles and decrease in starch granules compared with intracapsular algae. Vacuoles reached a peak size in algae found within the alimentary canal and often appeared extruded from individual algal cells (Fig. 4D). The vacuoles of intracapsular green algae in Ambystoma gracile hosts have been shown to be sites of ammonia waste storage in the form of proteins (10). This may account for larger vacuole sizes of invading algae, if these algae are following a nitrogenous waste gradient. However, these vacuoles also resemble the accumulation bodies of encysting dinoflagellates, which are a form of autophagic vesicle (22). Therefore, the increase of vacuole size may be associated with alimentary canal and cellular invasion, programmed cell death (33), or even algal cell encystment (22, 26).
Most intracellular algae were in direct contact with the host cytoplasm and not enclosed in a distinct vesicular membrane. This is similar to the association between Platymonas convolutae (a prasinophycean green alga) and acoel flatworms of the genus Convoluta sp., but unlike the association of Chlorella sp. (a trebouxiophycean green alga) with cnidarian hydras (26), which are contained in a symbiosomal membrane.
Several intracellular algae were enclosed in a thickened envelope. A structurally similar envelope surrounds algae found on the innermost egg capsule of early-stage embryos (before tissue invasion; Fig. 3 A and B). Despite their structural similarity, we do not know whether these envelopes are independently derived or whether they represent a lineage of encysted algal cells. These envelopes are similar to those formed by encysting diatoms (22), indicating that they may be involved in a process of algal cell encystment after invasion.
The process of algal cell integration in the Ambystoma–alga symbiosis differs from that described in other animal–alga symbioses. Invertebrate hosts typically ingest their algal symbionts, and cellular integration is often a result of partial digestive assimilation (17, 18, 27). However, in A. maculatum, algae primarily enter salamander embryos through the blastopore before the formation of a patent stomodeum and therefore before active feeding is possible. Additionally, some algal cells are embedded within epithelial or mesenchymal tissues that are far from the alimentary canal, consistent with algae having entered salamander embryos directly by penetrating their embryonic integument. Algal cells leave direct sunlight by entering opaque salamander tissues. Behavioral stimuli, such as gradients of nitrogenous waste, which may instigate algal proliferation (8, 10), could serve as the behavioral cue for tissue and cellular invasion against this light gradient.
Possibility of Vertical Transmission.
Previous research using light microscopy did not find algae in the oviducts of A. maculatum (15) and failed to culture algae from oviduct (6, 15) or oocyte (15) rinses. Additionally, clutches grown in alga-free tap water failed to grow algae in comparison with those raised in pond water (6, 15). This evidence against vertical transmission has led to a general acceptance that the algae are derived from the environment (34). However, neither the process of environmental acquisition (15) nor free-living Oophila sp. from vernal pools (35) has been described. Additionally, previous studies have had difficulty in obtaining alga-free clutches from either the laboratory or the wild (6, 8, 11).
Although our data are consistent with a process of vertical transmission, this process is unlikely to be the dominant mode of algal acquisition. The amplification of algal 18S rDNA from the oviducts, the encysted algae on the innermost egg capsule (Fig. 3B), and the similar encystment of some intracellular algae (Fig. 3I) indicate possible oviductal transmission. However, we did not amplify 18S rDNA consistently from adult reproductive tracts (Fig. S2), nor did we find algae concentrated in the reproductive tracts of embryos or early-stage larvae. Mixed modes of vertical and horizontal acquisition of symbionts are common. These are often revealed through incongruent phylogenetic topologies between host and symbiont populations (20). Similar phylogenetic comparisons between Ambystoma-Oophila populations could reveal the extent of vertical or horizontal acquisition in different salamander populations and test the controversial (6) species designation “Oophila amblystomatis” (Lambert ex. Printz) (19).
Endosymbiosis.
The reciprocal benefits of hatchling survival and growth to the embryo, and population growth to the algae, reveal that this symbiosis is a true mutualism (6, 8, 9). However, the material benefit of algal symbionts has been controversial. Hutchison and Hammen (14) found no net gain of oxygen from the algae and instead attributed the algal contribution to unknown “growth factors” supplied to the embryo. More refined microelectrode studies have since shown that intracapsular algae do produce a net increase of oxygen (11, 12), which confers a meaningful physiological benefit (13). The unexpected intracellular association shown in our study may reveal a benefit to the embryo other than oxygen production. Most intracellular algae, and algae of the alimentary canal, disappear by early larval stages. Their possible absorption may confer a metabolic benefit to their host.
Intracellular symbionts have been reported for many metazoan taxa, but we know of no other observation for vertebrates (3). As suggested in the Introduction, the embryonic association of the Ambystoma–alga symbiosis may preclude an adaptive immune response that would otherwise remove invading algal cells. V(D)J recombination in B and T cells, as indicated by RAG-1 protein in the thymus, does not occur until 6–8 wk after fertilization in the axolotl Ambystoma mexicanum (4), well after the initial invasion of algae in A. maculatum. The number of potential antibodies used in adult salamander antigen response is restricted (36), which may account for their remarkable regenerative abilities and acceptance of allo- and xenografts (37). The combination of an embryonic symbiosis and an inefficient immune system may, in part, account for the acceptance of an intracellular symbiont in A. maculatum. However, the lack of other intracellular symbiont examples in vertebrates may simply be due to a lack of investigation. The intracellular symbiosis described here reveals unanticipated complexity in the Ambystoma–alga system with implications for the ecology, evolution, and development of both host and symbiont.
Materials and Methods
Animal Care.
Animal procedures followed Dalhousie University Committee on Laboratory Animals protocol (09-029). Embryo clutches were collected from the Halifax Regional Municipality, Nova Scotia, Canada under permits from The Wildlife Division of the Nova Scotia Department of Natural Resources.
Fluorescence and TEM.
Individual embryos were anesthetized in tricaine methanesulfonate (pH 7.4), fixed overnight at 4 °C in Karnovsky's solution (2% paraformaldehyde, 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4), cut in 100-μm sections in 1% agarose (wt/vol) on a Vibratome 1000 (Automatic Tissue Sectioning Systems), and imaged for fluorescence on a Zeiss Axio Observer Z1 compound microscope. Color images were acquired on a Nikon AZ0 stereo dissecting microscope. Tissue regions containing bright autofluorescing cells were manually dissected from vibratome sections, postfixed in 2% osmium tetroxide in 0.1 M cacodylate buffer (30 min), dehydrated in an ethanol series, and embedded in Spurr's resin (Structure Probe). Polymerized resin blocks were trimmed and sectioned to a thickness of 70 nm, stained with saturated (2%) uranyl acetate in 50% (vol/vol) ethanol, counterstained with Reynold's lead citrate, and imaged on an FEI Tecnai-12 transmission electron microscope. Additional algae from the intracapsular fluid (Fig. 1A) were aspirated with a 32-gauge needle and processed in parallel to the salamander tissues. All image area measurements were made with Image J (http://rsbweb.nih.gov/ij/). One-way ANOVAs of area measurements with Games-Howell post hoc tests were run on SPSS (version 18).
Fluorescent in Situ Hybridization.
We designed an oligonucleotide probe specific for Oophila 18S rRNA (5′-TCTCTCAAGGTGCTGGCGA-3′) based on the regions of low RNA folding complexity in eukaryotes (38). The horse radish peroxidase-conjugated Oophila-specific probe, along with a positive control bacterial 16S rRNA-targeted probe (EUB338) (39) and negative control bacterial sense probe (NON338) (40), were purchased from biomers.net.
Salamander tissues were fixed in 4% paraformaldehyde (PFA) overnight, rinsed in sterile phosphate buffered saline (PBS), and stored in 100% methanol (−20 °C). Prehybridization steps followed Zelzer et al. (41) for mRNA in situ hybridizations and included rehydration into PTw (PBS with 0.1% Tween-20), 15 min in 0.02 M HCl, 10 min in 3% H2O2, 10 min in 5 μg/mL proteinase-K in PBS (37 °C), 2 × 5 min in 0.1 M triethanolamine (pH 7.0–8.0; Sigma), then 2 × 5 min in 0.1 M triethanolamine solution containing 2.5 μL/mL acetic anhydride. Slides were washed 2 × 5 min in PBS in between each step up to the triethanolamine treatment. Slides were then refixed in 4% PFA for 5 min, rinsed 5 × 1 min in PBS, dehydrated in a graded ethanol series, and air-dried for 30 min.
Each slide was covered with 150 μL hybridization buffer [900 mM NaCl, 20 mM Tris (pH 7.5), 0.02% SDS, 25% (vol/vol) formamide, 1% blocking reagent, and 10% (wt/vol) dextran sulfate] and included an unlabeled helper oligonucleotide (5′-GTCATCAAAAGAACGCTCGCC-3′) (42) and HRP-conjugated probe (0.15 ng/mL final concentration each). Slides were covered with parafilm strips during the hybridization and tyramide signal amplification (TSA) steps. Slides were hybridized for 3 h in a humidity chamber (46 °C), followed by a brief rinse in prewarmed wash buffer [20 mM Tris (pH 7.5), 159 mM NaCl, 0.01% SDS, and 5 mM EDTA; 46 °C] and an additional 15 min in wash buffer (46 °C). Slides were soaked in TNT buffer [0.1 M Tris (pH 7.5), 150 mM NaCl, and 0.074% Tween 20] for 15 min (25 °C). A TSA plus fluorescence kit (PerkinElmer) was used for signal amplification, following the manufacturer's instructions for tetramethylrhodamine or cyanine-3 tyramide in 40% (wt/vol) dextran sulfate. Slides were covered with 150 μL of amplification reagent and incubated for 45 min in the dark (25 °C) before sequential washes (20 min and 15 min) with TNT buffer (46 °C) and 1.5 mg/mL DAPI counterstaining (5 min; 25 °C). Slides were cover-slipped with 150 μL of AF1 (Citifluor) and sealed with nail polish.
Supporting Methods.
SI Materials and Methods provides additional methods pertaining to the online supporting material. These methods include amplification and sequencing of nuclear 18S and chloroplast 16S rDNA of Oophila, Oophila-specific 18S rDNA amplification in salamander tissues, phylogenetic reconstructions, and in vivo time-lapse microscopy.
Supplementary Material
Acknowledgments
We thank Joe Martinez, John Gilhen, David Hewitt, and Robin Kodner. This work was partially completed in the laboratories of James Hanken, Alastair Simpson, and John Archibald. Michelle Leger and Yana Eglit provided useful comments on an earlier draft. R.K. is an American Association of Anatomists Scholar. E.K. is supported by the Tula Foundation. A.A.H is supported in part by Natural Sciences and Engineering Research Council (NSERC) Grant 298366-2009 to Alastair Simpson. This research was funded by the American Association of Anatomists (R.K.), an NSERC grant (to B.K.H.), and National Science Foundation Grant MCB-0848083 (to R.P.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. HM590633 (Oophila sp. 16S) and HM590634 (Oophila sp. 18S)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018259108/-/DCSupplemental.
References
- 1.Venn AA, Loram JE, Douglas AE. Photosynthetic symbioses in animals. J Exp Bot. 2008;59:1069–1080. doi: 10.1093/jxb/erm328. [DOI] [PubMed] [Google Scholar]
- 2.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 3.Douglas A. The Symbiotic Habit. Princeton: Princeton Univ Press; 2010. [Google Scholar]
- 4.Flajnik M, Du Pasquier L. Evolution of the immune system. In: Paul W, editor. Fundamental Immunology. 6th Ed. Philadelphia: Wolters Kluwer, Lippincott Williams & Wilkins; 2009. pp. 56–124. [Google Scholar]
- 5.Orr H. Note on the development of amphibians, chiefly concerning the central nervous system; with additional observations on the hypophysis, mouth, and the appendages and skeleton of the head. Q J Micro Sci N S. 1888;115:483–489. [Google Scholar]
- 6.Gilbert PW. Observations on the eggs of Ambystoma maculatum with especial reference to the green algae found within the egg envelopes. Ecology. 1942;23:215–227. [Google Scholar]
- 7.Breder R. The courtship of the spotted salamander. Bull N Y Zool Soc. 1927;30:51–56. [Google Scholar]
- 8.Gilbert PW. The alga-egg relationship in Ambystoma maculatum. A case of symbiosis. Ecology. 1944;25:366–369. [Google Scholar]
- 9.Tattersall G, Spiegelaar N. Embryonic motility and hatching success of Ambystoma maculatum are influenced by a symbiotic alga. Can J Zool. 2008;86:1289–1298. [Google Scholar]
- 10.Goff LJ, Stein JR. Ammonia: Basis for algal symbiosis in salamander egg masses. Life Sci. 1978;22:1463–1468. doi: 10.1016/0024-3205(78)90641-0. [DOI] [PubMed] [Google Scholar]
- 11.Bachmann M, Carlton R, Burkholder J, Wetzel R. Symbiosis between salamander eggs and green algae: Microelectrode measurements inside eggs demonstrate effect of photosynthesis on oxygen concentration. Can J Zool. 1985;64:1586–1588. [Google Scholar]
- 12.Pinder A, Friet S. Oxygen transport in egg masses of the amphibians Rana sylvatica and Ambystoma maculatum: Convection, diffusion and oxygen production by algae. J Exp Biol. 1994;197:17–30. doi: 10.1242/jeb.197.1.17. [DOI] [PubMed] [Google Scholar]
- 13.Valls JH, Mills NE. Intermittent hypoxia in eggs of Ambystoma maculatum: Embryonic development and egg capsule conductance. J Exp Biol. 2007;210:2430–2435. doi: 10.1242/jeb.003541. [DOI] [PubMed] [Google Scholar]
- 14.Hutchison V, Hammen C. Oxygen utilization in the symbiosis of embryos of the salamander, Ambystoma maculatum and the alga, Oophila amblystomatis. Biol Bull Mar Biol Lab Woods Hole. 1958;115:483–489. [Google Scholar]
- 15.Gatz J. Algal entry into the eggs of Ambystoma maculatum. J Herpetol. 1973;7:137–138. [Google Scholar]
- 16.Lane CE, Archibald JM. The eukaryotic tree of life: Endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Trench R. The cell biology of plant-animal symbiosis. Annu Rev Plant Physiol. 1979;30:485–531. [Google Scholar]
- 18.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York: John Wiley; 1965. [Google Scholar]
- 19.Printz H. Chlorophyceae. In: Engler A, Prantl K, editors. Die natürlichen Pflanzenfamilien [The Natural Plant Families] Vol. 3. Leipzig, Germany: W. Engelmann; 1928. pp. 1–463. [Google Scholar]
- 20.Bright M, Bulgheresi S. A complex journey: Transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison R. Harrison stages and description of normal development of the spotted salamander, Ambystoma punctatum (Linn) In: Wilens S, editor. Organization and Development of the Embryo. New Haven, CT: Yale Univ Press; 1969. pp. 44–66. [Google Scholar]
- 22.Dodge J. The Fine Structure of Algal Cells. New York: Academic Press; 1973. [Google Scholar]
- 23.Rumpho M, Dastoor FP, Manhart JR, Lee J. The kleptoplast. In: Wise R, Hoober J, editors. The Structure and Function of Plastids (Advances in Photosynthesis and Respiration) Vol. 23. Dordrecht, The Netherlands: Springer; 2006. pp. 451–473. [Google Scholar]
- 24.Hirose E, Maruyama T, Cheng L, Lewin R. Intracellular symbiosis of a photosynthetic prokaryote, Prochloron sp., in a colonial ascidian. Invertebr Biol. 1996;115:343–348. [Google Scholar]
- 25.Stat M, Carter D, Hoegh-Guldberg O. The evolutionary history of Symbiodinium and scleractinium hosts—symbiosis, diversity, and the effect of climate change. Perspect Plant Ecol Evol Syst. 2006;8:23–43. [Google Scholar]
- 26.Muscatine L, Pool RR, Trench RK. Symbiosis of algae and invertebrates: Aspects of the symbiont surface and the host-symbiont interface. Trans Am Microsc Soc. 1975;94:450–469. [PubMed] [Google Scholar]
- 27.Pardy R. In: Phycozoans, Phycozoology, Phycozoologists? Algal Symbiosis: A Continuum of Interaction Strategies. Goff L, editor. Cambridge, UK: Cambridge Univ Press; 1983. pp. 5–18. [Google Scholar]
- 28.Hammen C, Hutchison V. Carbon dioxide assimilation in the symbiosis of the salamander Ambystoma maculatum and the algae Oophila amblystomatis. Life Sci. 1962;1:527–532. [Google Scholar]
- 29.Rumpho ME, et al. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc Natl Acad Sci USA. 2008;105:17867–17871. doi: 10.1073/pnas.0804968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wägele H, et al. Transcriptomic evidence that longevity of acquired plastids in the photosynthetic slugs Elysia timida and Plakobranchus ocellatus does not entail lateral transfer of algal nuclear genes. Mol Biol Evol. 2011;28:699–706. doi: 10.1093/molbev/msq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesneau A, et al. Transgenesis procedures in Xenopus. Biol Cell. 2008;100:503–521. doi: 10.1042/BC20070148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karakashian S. Morphological plasticity and the evolution of algal symbionts. Ann N Y Acad Sci. 1970;175:474–487. [Google Scholar]
- 33.van Doorn WG, Yoshimoto K. Role of chloroplasts and other plastids in ageing and death of plants and animals: A tale of Vishnu and Shiva. Ageing Res Rev. 2010;9:117–130. doi: 10.1016/j.arr.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Epel D, Gilbert SF. Ecological Developmental Biology: Integrating Epigenetics, Medicine, and Evolution. Sunderland, MA: Sinauer Associates; 2008. [Google Scholar]
- 35.Shudert E. Nonmotile coccoid and colonial green algae. In: Wehr TD, Sheath RG, editors. Freshwater Algae of North America. New York: Academic Press; 2003. pp. 253–307. [Google Scholar]
- 36.Charlemagne J. Antibody diversity in amphibians. Noninbred axolotls used the same unique heavy chain and a limited number of light chains for their anti-2,4-dinitrophenyl antibody responses. Eur J Immunol. 1987;17:421–424. doi: 10.1002/eji.1830170319. [DOI] [PubMed] [Google Scholar]
- 37.Mescher AL, Neff AW. Limb regeneration in amphibians: Immunological considerations. Sci World J. 2006;6(Suppl 1):1–11. doi: 10.1100/tsw.2006.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behrens S, et al. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl Environ Microbiol. 2003;69:1748–1758. doi: 10.1128/AEM.69.3.1748-1758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worden AZ, Chisholm SW, Binder BJ. In situ hybridization of Prochlorococcus and Synechococcus (marine cyanobacteria) spp. with rRNA-targeted peptide nucleic acid probes. Appl Environ Microbiol. 2000;66:284–289. doi: 10.1128/aem.66.1.284-289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelzer E, et al. Tissue specific regulation of VEGF expression during bone development requires Cbfa1/Runx2. Mech Dev. 2001;106:97–106. doi: 10.1016/s0925-4773(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 42.Fuchs BM, Glöckner FO, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.