Abstract
The Holliday junction (HJ), a cross-shaped structure that physically links the two DNA helices, is a key intermediate in homologous recombination, DNA repair, and replication. Several helicase-like proteins are known to bind HJs and promote their branch migration (BM) by translocating along DNA at the expense of ATP hydrolysis. Surprisingly, the bacterial recombinase protein RecA and its eukaryotic homologue Rad51 also promote BM of HJs despite the fact they do not bind HJs preferentially and do not translocate along DNA. RecA/Rad51 plays a key role in DNA double-stranded break repair and homologous recombination. RecA/Rad51 binds to ssDNA and forms contiguous filaments that promote the search for homologous DNA sequences and DNA strand exchange. The mechanism of BM promoted by RecA/RAD51 is unknown. Here, we demonstrate that cycles of RecA/Rad51 polymerization and dissociation coupled with ATP hydrolysis drives the BM of HJs.
Homologous recombination (HR) is the process responsible for maintaining genome stability in all living organisms; it is particularly important for repairing DNA double-strand breaks (1–5). The process of HR involves the enzymatic degradation of broken dsDNA ends into resected DNA duplex with protruding ssDNA tails (6, 7). Following resection, the central protein of HR, the prokaryotic (bacterial recombinase protein) RecA or its eukaryotic homolog Rad51, is loaded onto the ssDNA tails and forms a contiguous nucleoprotein filament (8, 9). Although RecA and Rad51 share only ∼30% sequence homology, the filaments they form and the conformational changes they induce in DNA are nearly identical (10).
The RecA/Rad51-ssDNA filament possesses the unique ability to search for homologous dsDNA sequences and promote DNA strand exchange: an invasion of ssDNA into homologous DNA duplex that results in the displacement of the identical ssDNA from the duplex and formation of a joint molecule (JM) (Fig. S1A) (11). Besides its essential DNA strand-exchange activity RecA/RAD51 recombinases have an ability to extend JMs by a kinetically distinct process known as heteroduplex extension or three-strand branch migration (BM), in which one DNA strand is progressively exchanged for another (11, 12). When the extending heteroduplex reaches the ssDNA-dsDNA junction on the invading DNA strand, the 3-stranded JM is converted into a 4-stranded Holliday junction (HJ). Then, specialized DNA translocating proteins, like Escherichia coli RuvAB, bind to HJs and promote their migration by four-strand BM (otherwise referred to as the BM of HJs) (13). These DNA translocases are capable of bypassing sequence heterologies and disrupting nucleoprotein complexes encountered during BM. Finally, DNA repair synthesis by DNA polymerases takes place on JMs followed by their resolution.
Surprisingly, the RecA/Rad51 recombinases can also promote four-strand BM (Fig. S1A) (14, 15). This activity may play a significant role in the initial stages of recombination in vivo by helping to form and stabilize HJs, and therefore it is important to understand the mechanism by which these recombinases promote BM of HJs. It is commonly thought that the RecA/Rad51 nucleoprotein filament can carry out both DNA strand exchange and BM of HJs through similar mechanisms. However, there are significant differences between these two activities of RecA/RAD51. Most importantly, BM of HJs requires ATP hydrolysis, whereas DNA strand exchange does not (11, 16).
We hypothesized that DNA strand exchange and BM of HJs represent two fundamentally different activities of RecA/Rad51. While DNA strand exchange is promoted by an established RecA/Rad51-ssDNA filament (Fig. 1A), the BM of HJs is propelled by the polar polymerization of RecA/Rad51 monomers on DNA (Fig. 1B). Here we present data that supports our hypothesis. Our results demonstrate that RecA/RAD51 drives BM by a unique mechanism that involves cycles of protein polymerization/dissociation directed towards HJs. Our work provides a solution to the long-standing puzzle of why ATP hydrolysis is required for BM of HJs: we show that BM depends on dissociation of RecA/RAD51 from DNA, which in turn depends on ATP hydrolysis.
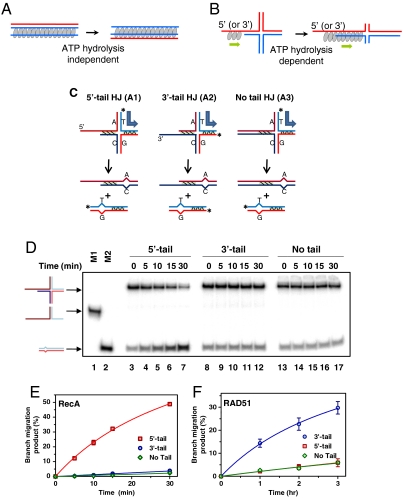
Fig. 1.
RecA and RAD51 require a polarity-specific ssDNA tail to mediate BM. (A) The 3-strand DNA strand exchange occurs with one strand of duplex DNA switching to pair with the ssDNA residing within the stable contiguous filament formed by RecA/Rad51 recombinase (gray ovals) on ssDNA. (B) In contrast, we hypothesize here that the 4-strand reaction (BM of HJs) is driven by polar polymerization of the RecA/Rad51 that proceeds towards the cross-over point. (C) The HJ substrates and the products of their BM. Blue block arrow denotes the direction of BM; asterisk denotes the 32P label; and hatched shading denotes regions of heterology. (D) BM was initiated by adding RecA (3 μM) to the different HJs (A1, A2, and A3, Table S1) (32 nM). After incubation for the indicated periods of time the DNA products were analyzed by electrophoresis in 8% polyacrylamide gels. Lane 1 and 2 contain migration markers: the fork intermediate (M1), and the BM product (M2), respectively. A faint band is a product of incomplete annealing during substrate preparation. (E, F) The kinetics of BM promoted by RecA and RAD51. Data were fit to a one-site binding hyperbola curve. Error bars (small for the RecA reaction) represent standard error of the mean (SEM).
Results
RecA and Rad51 Promote BM in Opposite Directions, Consistent with the Polarities of Their Polymerization on ssDNA.
To test our hypothesis that BM of HJs is driven by the polar polymerization of RecA/Rad51 monomers on DNA we examined the BM activity of RecA and Rad51 on synthetic HJs (17) containing ssDNA tails (Fig. 1C). Because of RecA’s strong preference for ssDNA, filament formation can only initiate on the ssDNA tails, and then, due to the intrinsic polarity of RecA polymerization (18), the filament proceeds into the dsDNA region in the 5′ to 3′ direction. We found that RecA indeed promotes BM of HJ-like substrates containing 5′-ssDNA tails (Table S1), consistent with the polymerization of RecA towards the junction (Fig. 1 D and E). In contrast, RecA does not promote the BM of substrates that contain either 3′-ssDNA tails or lack ssDNA tails; where RecA polymerization proceeds away from the junction or does not initiate, respectively. Using the same set of DNA substrates, we next tested the BM activity of human RAD51. It was previously shown that at elevated salt concentrations, RAD51, similar to RecA, shows a strong binding preference for ssDNA over dsDNA (19). Indeed, we found that at 350–400 mM NaCl, RAD51 promotes the BM of HJs; the rate of BM was approximately ninefold slower than that of the RecA-driven reaction (Fig. 1F). However, the polarity of Rad51 BM was opposite to that of RecA.
While the 5′ to 3′ polarity of RecA polymerization on ssDNA is well documented, evidence for the 3′ to 5′ polarity of Rad51 polymerization is compelling, but indirect (15, 20–22). To provide direct evidence of RAD51 polymerization polarity, we tested the ability of RecA/RAD51 to protect tailed DNA substrates with opposite polarities against cleavage with the restriction endonuclease BamHI. We found that while RecA preferentially protected tailed DNA substrates containing 5′ tails; RAD51 protected those with 3′ tails, which is consistent with a 3′ to 5′ polarity of polymerization (Fig. S2). Thus, the observed polarity of HJ BM by RAD51 and RecA is consistent with their polymerization polarity towards the HJ. Similar to RecA/Rad51-promoted BM on plasmid-based DNA substrates (11, 15, 16), BM of synthetic HJs required ATP hydrolysis and was inhibited in the presence of nonhydrolysable ATP analogs, or when RecA/RAD51 ATPase-deficient mutants were used (Fig. S3 A and B).
DNA Strand Exchange and BM Occur under Differing Conditions.
RecA binding and polymerization on ssDNA occurs at low Mg2+ concentrations (< 1.0 mM free ion) (23). In contrast, DNA strand exchange requires significantly higher Mg2+ during the synaptic step of the reaction that presumably allows dsDNA to bind to the RecA secondary site (24). Therefore, if DNA strand exchange and BM are mechanistically identical, then BM would require high Mg2+ concentrations. On the other hand, if BM is driven by protein polymerization on DNA, we would expect the reaction to occur at low Mg2+ concentrations. Using synthetic DNA substrates we found that RecA promotes BM at Mg2+ as low as 0.39 mM (free ion) (Fig. S1 B and C). In contrast, but as expected, DNA strand exchange promoted by RecA on DNA substrates of similar length and base composition required significantly higher Mg2+. Similarly, RAD51 forms filaments on ssDNA in the presence of Mg2+, but requires Ca2+ or other stimulatory agents for DNA strand exchange (22, 25, 26). Here, we demonstrated that RAD51 promoted BM in the presence of Mg2+ (Fig. S1D), whereas Ca2+ inhibited its BM activity. Thus, both RecA and RAD51 promote BM under conditions that support protein polymerization on DNA, but not DNA strand exchange.
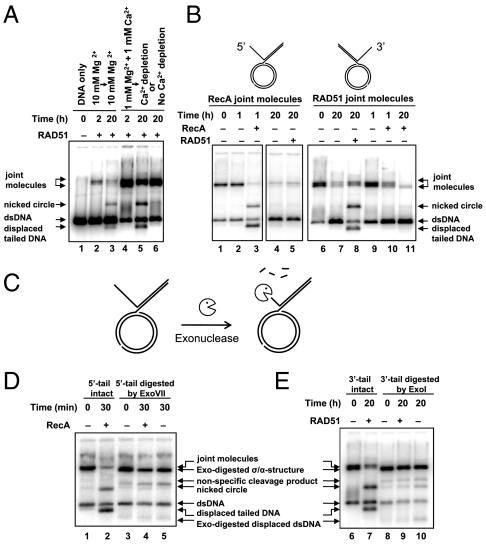
We next wanted to ascertain whether DNA strand exchange and BM of HJs by RecA/Rad51 on plasmid-based DNA substrates (14, 15) also require different reaction conditions. DNA strand exchange was carried out between circular Bluescript pBSK (+) plasmid dsDNA containing an ssDNA region (gapped DNA) and homologous linear dsDNA (Fig. S1A). DNA strand exchange initiates within the ssDNA region of the gapped DNA and results in formation of JMs. As DNA strand exchange proceeds beyond the ssDNA region, the HJ is formed and BM leads to nicked circle and tailed DNA products. For RAD51, JM formation was stimulated by Ca2+ (Fig. 2A, lanes 2 and 4), whereas BM required Mg2+ and was inhibited by Ca2+ (Fig. 2A, lanes 3, 5, and 6). It was previously shown that Ca2+ promotes formation of a stable ATP-bound RAD51-ssDNA filament (high affinity DNA binding state) by inhibiting the RAD51 ATPase; whereas in the presence of Mg2+, RAD51 forms a filament that is prone to dissociation because of a buildup of protein-bound ADP (25, 27). For RecA, high Mg2+ (15 mM) and low Mg2+ (4 mM) concentrations were optimal for the initial DNA strand exchange and for subsequent BM, respectively. Thus, for both RecA and Rad51, DNA strand exchange required a stable filament, but BM needed a filament prone to dissociation/reassociation.
Fig. 2.
The displaced ssDNA strand is essential for BM of plasmid-based DNA substrates. (A) Ca2+ stimulates JM formation, but inhibits BM by RAD51 in strand exchange between 32P-labeled pBSK (+) linear dsDNA (lane 1) and gapped DNA. Reaction scheme is shown in Fig. S1A. The reaction was carried out either in the presence of Mg2+ (lanes 2 and 3), or Ca2+ and Mg2+ (lane 4) followed (lane 5) or not (lane 6) by Ca2+ depletion. The arrows emphasize the continuation of the initial reaction with or without changes to the divalent metal ion concentration. (B) RecA and Rad51 promote BM when added to deproteinized JM. Purified RecA-generated JMs (lane1) were incubated with RecA (lane 3), RAD51 (lane 5) or without protein (lanes 2 and 4). RAD51-generated JMs (lane 6) were incubated with RAD51 (lane 8), RecA (lanes 10 and 11), or without protein (lanes 7 and 9) (C) Scheme of ssDNA tail removal by an exonuclease. (D, E) Removal of the ssDNA tail prevents BM. RecA-generated JMs (lane1) were either mixed with RecA (lane 2) or treated with Exo VII (lane 3,) followed by incubation with RecA (lane 4) or without protein (lane 5). RAD51-generated JMs (lane 6) were either mixed with RAD51 (lane 7) or treated with Exo I (lane 8) followed by incubation with RAD51 (lane 9) or without protein (lane 10). Note, loss of JMs in the absence of RecA/RAD51 was due to spontaneous BM.
The Displaced ssDNA Strand Is Essential for BM on JMs.
We hypothesized that RecA/RAD51 may drive BM by polymerizing on the displaced ssDNA strand of the JM (σ- or α-structures) produced during the initial DNA strand-exchange step (Fig. S1A). To test this hypothesis, we generated JMs by carrying out RecA and RAD51-promoted DNA strand exchange followed by JM deproteinization and purification (Fig. 2B, lane 1 and 6), and then used these JMs as substrates for BM. Addition of RecA to RecA-generated JMs containing the 5′-ssDNA displaced strand (28) led to the appearance of BM products (Fig. 2B, lane 3). In contrast, even after 20 h of incubation, RAD51 produced no BM products on the RecA -generated JM, in which the displaced ssDNA strand polarity directs RAD51 polymerization away from the HJ (Fig. 2B, lane 5). Conversely, RAD51, but not RecA, promoted BM on purified Rad51-generated JM containing a 3′-ssDNA displaced strand (Fig. 2B, lanes 8, 10, 11), consistent with the 3′ to 5′ polarity of RAD51 polymerization on ssDNA. Importantly, reinitiated BM on purified JMs promoted by RecA and RAD51 showed the same requirements as the standard 4-strand BM occurring on gapped DNA, indicating identical mechanisms for these reactions; both required ATP or dATP hydrolysis, Mg2+ and, in the case of RAD51, absence of Ca2+ (Fig. 2A; Fig. S3 C and D; Fig. S1E). The overall rate of BM was approximately 11-fold higher for RecA than for RAD51 (Fig. S4 A and B). Further, we demonstrated that removal of the displaced ssDNA of JMs by Exonuclease VII or Exonuclease I (for RecA- and RAD51- generated JM, respectively, as described in SI Text) abolished BM by RecA or RAD51 (Fig. 2 C–E; Fig. S4C). Using purified plasmid-based JMs we found that RecA/RAD51 promotes BM in the presence of SSB/RPA, ubiquitous single-strand DNA binding proteins (Fig. S4 F and G). This result has important biological implications demonstrating that RecA/RAD51 BM of HJs can occur in the cell, where SSB/RPA is abundant. This result is also consistent with a mechanism of BM that involves polymerization of RecA/RAD51 on the displaced ssDNA strand; it was shown that under similar conditions RecA and RAD51 can displace SSB/RPA from ssDNA, when SSB/RPA is added to ssDNA prior or at the same time as RecA/RAD51 (29, 30).
RecA/RAD51 Binding to the Junction Is Insufficient to Drive BM.
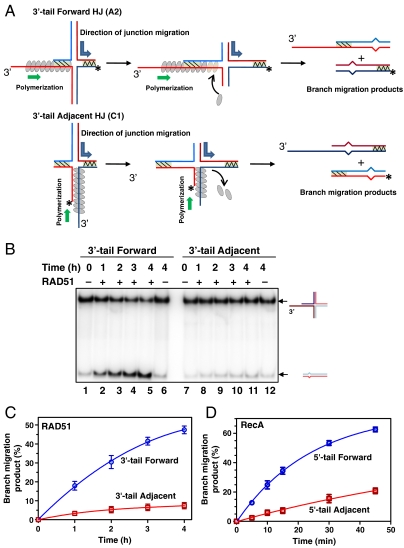
We surmise that the role of the ssDNA displaced strand was not to simply provide a gateway for protein loading, as at low salt concentrations RAD51 binds efficiently to both ssDNA and dsDNA, but does not promote BM (Fig. S4 D and E). The polarity of the displaced ssDNA strand must support RecA/RAD51 polymerization towards the HJ in order for BM to proceed. We suggested that RecA/RAD51 polymerization towards HJs may render BM unidirectional by either triggering a conformational change of the HJ into the open state that is necessary and sufficient for BM (31, 32) or, in addition, by applying a force to the HJ that drives BM forward. We tested these possibilities by designing a pair of synthetic HJ substrates (denoted as “Forward” and “Adjacent”) (Fig. 3A) in which the ssDNA tails were placed on different DNA strands in such a way that it permitted the RecA/RAD51 polymerization toward the HJs on both substrates allowing a conformational change to occur, but the protein would polymerize in the direction that was either the same as that of BM (on the Forward substrate), or opposite to BM (on the Adjacent one). We found that the rate of BM on the Forward DNA substrates was significantly higher than on the Adjacent DNA substrates for both RAD51 and RecA (Fig. 3 B–D). This result indicates that RecA/RAD51 polymerization towards the HJs either generates a force that drives BM or prevents BM, a normally random walk process, from going backward. The latter model is similar to the “Brownian” ratchet mechanism which was recently proposed for helicase-driven BM (33).
Fig. 3.
RAD51 and RecA drive BM preferentially in the direction of protein polymerization. (A) Forward and Adjacent HJ were designed to permit RAD51 (gray ovals) polymerization either in the same direction as BM, or the direction that opposes BM. The direction of BM and protein polymerization is denoted by the blue and green arrows, respectively. Shaded regions denote heterologous DNA arms. (B) The kinetics of RAD51 (2.6 μM) BM on Forward (A2) and Adjacent HJs (C1) (32 nM). (C) Data in (B) presented as a graph.
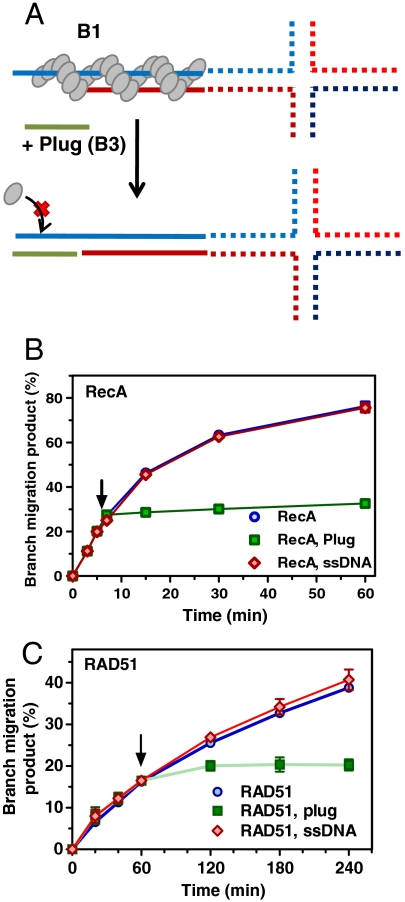
BM Requires Multiple Cycles of RecA/RAD51 Polymerization/Dissociation.
The central question of this work was to understand why the RecA/RAD51-dependent BM of HJs requires ATP hydrolysis. Based on the fact that ATP hydrolysis drives RecA/RAD51 dissociation from DNA (11) and our observation that BM occurs under conditions that favor protein dissociation, e.g., high salt, low Mg2+, absence of Ca2+, we surmised that multiple rounds of protein polymerization and dissociation on DNA are required for BM. To test this hypothesis, we carried out an experiment in which BM was initiated normally by adding RecA/RAD51 to tailed HJs, but new cycles of protein binding were prevented by elimination of ssDNA tails during an outgoing reaction (Fig. 4A). For this purpose, we used an ssDNA-plug complementary to the ssDNA tail, which after annealing would prevent new RecA/RAD51 binding without affecting the protein already bound to the HJ substrate. We found that addition of the ssDNA plug in either RecA- or RAD51-driven reactions led to an almost instant halt of BM, whereas a noncomplementary ssDNA had no effect on BM (Fig. 4 B and C). These results indicate that a single protein polymerization event on DNA is not sufficient for HJ BM. To rule out the possibility that a fraction of DNA molecules remained protein-free during outgoing BM, and addition of the ssDNA-plug simply blocked BM of these DNA molecules, we increased RecA/RAD51 concentration two and threefold. Importantly, this increase had no effect on the ssDNA-plug dependent inhibition of outgoing BM (Fig. S5 A–D). By tracking RecA ATP hydrolysis during BM we found that DNA binding was saturating and that saturation was achieved 2 min after addition of RecA, i.e., prior to ssDNA-plug addition (Fig. S5 E and F). We concluded that multiple rounds of RecA/RAD51 DNA binding and dissociation are required for the BM of HJs.
Fig. 4.
An ongoing BM can be halted by an ssDNA-plug that blocks binding of RecA/Rad51 to HJ. (A) The experimental scheme. (B) BM initiated on 5′-tailed HJs (B7) (33 nM) by RecA (3 μM) and was either carried out uninterrupted for 60 min, or after 11 min ssDNA (33 nM) that was complementary to the 5′-tail ssDNA (B3, denoted as plug) or noncomplementary (B4) was added and the incubation was continued for 49 min. (C) BM initiated on 3′-tailed HJs (A2) (32 nM) by RAD51 (3 μM) was either carried out uninterrupted for 4 h, or after 1 h an ssDNA plug (C2) (48 nM) or nonhomologous ssDNA (C3) (48 nM) were added and the incubation was continued for 3 h. Error bars represent SEM.
Discussion
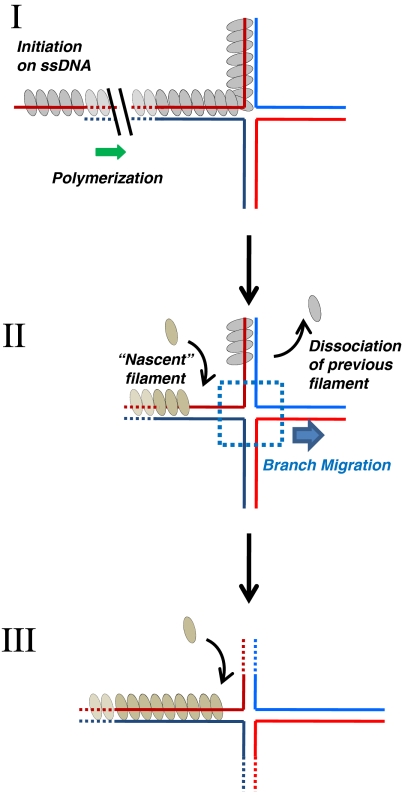
RecA/RAD51 plays the essential role in HR of promoting the search for homology and JM formation (1, 2, 4). In vitro, RecA/RAD51 possesses two activities: DNA strand exchange between homologous ssDNA and dsDNA molecules; and BM of HJs. While the mechanism of DNA strand exchange is fairly well understood, the mechanism of the BM has remained obscure in spite of extensive studies (11, 14–16). Here we demonstrate that RecA/RAD51 promotes BM by a unique mechanism that depends on polymerization and dissociation of RecA/RAD51 on DNA. Also, our results show that RecA and RAD51 polymerize on ssDNA with opposite polarities and promote BM in opposite directions. Previously, the role for RecA polymerization in DNA strand exchange was envisioned by S. Kowalczykowski, who proposed that cycles of RecA polymerization/dissociation would ensure formation of a contiguous nucleoprotein filament that is important for efficient heteroduplex extension on long (plasmid-size) DNA substrates (11). We show that for both RecA and RAD51, DNA strand exchange and BM require different filament conformations indicating these are two separate and distinct activities. DNA strand exchange requires formation of a stable high affinity DNA binding state filament, which occurs in the presence of ATP either at elevated Mg2+ levels or in the presence of Ca2+ for RecA and RAD51, respectively. In contrast, BM requires formation of a dynamic filament that is more prone to dissociation. Given our current results, we propose the following model to explain the mechanism of RecA/RAD51-promoted BM of HJs. This mechanism involves multiple rounds of RecA/RAD51 polymerization on DNA and an ATP hydrolysis-dependent dissociation. The RecA/RAD51 filament dynamics include binding of protein monomers to the growing filament end and dissociation from the opposite end. We suggest that RecA/RAD51 filament formation on DNA, its subsequent dissociation and formation of a “nascent” filament creates a transient protein-free gap within which the HJ can move spontaneously in a random walk fashion (Fig. 5). But because the nascent filament will restrict the backward movement of the HJ, net movement of the HJ will only occur when the junction migrates away from the nascent filament. Because RecA and RAD51 association rates are faster than the dissociation rates, 10- and fivefold respectively (27, 34, 35), the nascent filament will likely capture the HJ before completion of the “old” filament dissociation. At this point of capture, BM is halted and will not resume until the dissociation of the nascent filament occurs. Then polymerization of the next filament begins and the cycle repeats. Therefore the unidirectional progression of the BM reaction comes from multiple rounds of ATP hydrolysis-dependent protein polymerization/dissociation. In addition, RecA/RAD51 polymerization on the displaced DNA strand may impose a direct force on HJs, and/or generate a torsional stress in DNA that facilitates the BM of HJs. Our model rationalizes the linkage between the polarity of BM and the direction of protein polymerization on DNA (which is the same as the direction of protein dissociation from DNA). By linking BM with RecA/RAD51 dissociation from DNA it also provides the mechanistic basis for the dependence of BM on ATP hydrolysis, a thorny question in the HR field of study. Thus, our results clarified the function of the ATPase activity of RecA/RAD51.
Fig. 5.
Proposed mechanism of the recombinase-driven BM. RecA/RAD51 polymerizes on DNA in a polarity-specific direction, capturing the HJ (I). BM proceeds once the first filament dissociates and before polymerization of the nascent filament reaches the junction (II). Formation of the next filament will prevent BM going backward ensuring its unidirectionality (III).
RecA/RAD51 polymerization/dissociation on the displaced ssDNA strand may also contribute to the “standard” DNA strand exchange on plasmid-size DNA substrates, which was shown to involve two kinetically distinct steps: DNA strand exchange and heteroduplex extension, with only the latter being dependent on ATP hydrolysis (11). Our results provide rationale for this observation; heteroduplex extension may be driven by RecA/RAD51 polymerization on the displaced ssDNA strand (Fig. S6). Thus, the RecA/RAD51 activity known as “DNA strand exchange” is constituted, in fact, by two mechanistically distinct activities: (i) canonical DNA strand exchange (also called DNA strand invasion) that is promoted by a stable filament and (ii) BM that requires cycles of protein polymerization and ATP hydrolysis-dependent dissociation.
In vivo, at the first stage of JM formation, initial D-loops are formed by the DNA strand exchange activity of RecA/RAD51. For RecA (which is better studied than RAD51), the length of these initial D-loops was estimated to be 300–500 bp (12). This length is smaller than the average length of ssDNA gaps generated in E. coli after DNA damage (800 nucleotides) (36). At the second stage, RecA/RAD51 may extend the initial D-loops through the BM mechanism described above (Fig. S7). This extension may facilitate capture of the 3′-ssDNA end for priming of DNA synthesis, and contribute to HJ formation. It is important to note that all RecA/RAD51 ATPase mutants, which are deficient in BM, show various degree of deficiency in HR and DNA repair in E. coli, S. cerevisiae, and mammalian cells (37–39).
When compared to specialized helicase-family BM proteins, the BM activity of RecA/RAD51 is relatively slow, e.g., 3–4 bp/s for RecA vs. 10–50 bp/s for the canonical BM enzyme RuvAB (Table S2). The fact that the rate of RAD51 BM is approximately 11-fold slower than RecA may reflect an increased role for specialized BM proteins in eukaryotes, or a need for stimulatory factors (15). However, the conservation of BM activity of the recombinase from bacteria to humans indicates its significance. We believe this activity is especially important during the initial stage of HR when JMs are the least stable. Then following this initial stage of BM, more potent DNA translocating BM proteins, e.g., Rad54 or RuvAB, will gain access to these JMs and either dissociate or expand them further depending on the stage and pathway of HR (40) (Fig. S7).
Materials and Methods
Proteins and DNA.
Human RAD51, RAD51K133R, RAD51K133A, and RPA were purified as described (25). E. coli RecA was a generous gift of E. Golub (Yale University). SSB, Shrimp alkaline phosphatase, and Exonuclease VII were from USB Corp., and Terminal deoxynucleotidyl transferase, restriction endonucleases, Exonuclease I, and T4 polynucleotide kinase were from New England Biolabs. Oligonucleotides were from IDT, Inc. (Table S1). Synthetic HJ DNA substrates (Table S1) were prepared as described (17, 41). Gapped DNA was prepared by annealing the pBSK (+) XhoI-AlwNI fragment (2,065 bp) to pBSK (+) ssDNA and purified as described (41).
BM Assay on Oligonucleotide-Based Substrates.
RecA-mediated BM was performed by incubation of RecA (in the indicated concentrations) with 32P-labeled synthetic HJs (32 nM, molecules) in a BM buffer containing 35 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 2 mM DTT, 3 mM ATP, 10 mM creatine phosphate, 10 units/mL creatine phosphokinase, and 100 μg/mL BSA at 37 °C.
The RAD51-mediated BM was performed by incubating RAD51 (2.6 μM for 3′-tail forward, 3′-tail adjacent and 5′-tail HJ, and 3 μM for no tail HJ) with 32P-HJs (32 nM) in a buffer containing 25 mM Tris-acetate (pH 7.5), 10 mM magnesium acetate, 2 mM DTT, 400 mM NaCl, 2 mM ATP (or other indicated cofactor), 15 mM phosphocreatine, 10 units/mL creatine phosphokinase, and 100 μg/mL BSA at 37 °C. The DNA products of RAD51- and RecA-mediated BM were deproteinized by treatment with stop buffer containing 1.4% SDS, 0.96 mg/mL proteinase K, 7.5% glycerol, and 0.015% bromphenol blue for 5 min at 22 °C. Reactions shown in Fig. 1D were terminated by addition of 1 μg of poly dT319 followed by a 5-min incubation at 37 °C and by addition of stop buffer. The DNA products were analyzed by electrophoresis in 8% polyacrylamide gels (29∶1), as described in SI Text.
BM on Plasmid-Based DNA Substrates by RAD51.
Nucleoprotein filaments were formed by incubating RAD51 protein (5 μM) with pBSK (+) gapped DNA (20 μM, nt) (4-stranded reaction) in buffer containing 25 mM Tris acetate, pH 7.5, 2 mM ATP, 275 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1 mM DTT, and 100 μg/mL BSA for 10 min at 37 °C. RPA (0.4 μM) was added to the nucleoprotein filaments followed by a 10-min incubation. DNA strand-exchange reaction was initiated by addition of 5′-labeled linear pBSK (+) dsDNA (20 μM, nt). BM of nondeproteinized JMs (σ and α-structures) was initiated by Ca2+ depletion with 1.2 mM EGTA.
Deproteinized JMs were prepared as described in refs. 17, 41. BM of deproteinized JMs (0.32 nM, molecules) was carried out in buffer containing 30 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 350 mM NaCl, 2 mM DTT, 2 mM ATP, 8 mM phosphocreatine, and 8 units/mL creatine phosphokinase at 37 °C. RAD51 (10 μM) was added to the reaction mixtures. When indicated, RPA (0.4 μM or 55 nM) was added to the reaction. The DNA products were deproteinized by treatment with stop buffer for 15 min at 37 °C and analyzed in 1.5% agarose gels. RecA-mediated BM was performed as described in SI Text.
Supplementary Material
Acknowledgments.
We thank M. Bouchard and S. Kowalczykowski for the comments and discussion. This work was supported by the National Institutes of Health (NIH) Grant CA100839, and the Leukemia and Lymphoma Society Scholar Award 1054-09 (to A.V.M.) and NIH Grant F31 AG033484-01 (to M.J.R.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016072108/-/DCSupplemental.
References
- 1.Kowalczykowski SC. Structural biology: snapshots of DNA repair. Nature. 2008;453:463–466. doi: 10.1038/453463a. [DOI] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 4.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop DK. Multiple mechanisms of meiotic recombination. Cell. 2006;127:1095–1097. doi: 10.1016/j.cell.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 6.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 9.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 11.Kowalczykowski SC. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu Rev Biophys Biophys Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- 12.Cox MM, Lehman IR. RecA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc Natl Acad Sci USA. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons CA, Stasiak A, West SC. The E. coli RuvAB proteins branch migrate Holliday junctions through heterologous DNA sequences in a reaction facilitated by SSB. EMBO J. 1995;14:5736–5744. doi: 10.1002/j.1460-2075.1995.tb00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conley EC, West SC. Homologous pairing and the formation of nascent synaptic intermediates between regions of duplex DNA by RecA protein. Cell. 1989;56:987–995. doi: 10.1016/0092-8674(89)90632-6. [DOI] [PubMed] [Google Scholar]
- 15.Murayama Y, Kurokawa Y, Mayanagi K, Iwasaki H. Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases. Nature. 2008;451:1018–1021. doi: 10.1038/nature06609. [DOI] [PubMed] [Google Scholar]
- 16.Cox MM. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 17.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 18.Register JC, III, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 19.Liu Y, et al. Conformational changes modulate the activity of human RAD51 protein. J Mol Biol. 2004;337:817–827. doi: 10.1016/j.jmb.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 22.Baumann P, West SC. The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the RecA protein of Escherichia coli: Characterization of ATP hydrolysis. J Biol Chem. 1981;256:8829–8834. [PubMed] [Google Scholar]
- 24.Chow SA, Radding CM. Ionic inhibition of formation of RecA nucleoprotein networks blocks homologous pairing. Proc Natl Acad Sci USA. 1985;82:5646–5650. doi: 10.1073/pnas.82.17.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 27.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn R, Cunningham RP, DasGupta C, Radding CM. Polarity of heteroduplex formation promoted by Escherichia coli RecA protein. Proc Natl Acad Sci USA. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J Mol Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- 30.Sigurdsson S, et al. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karymov M, Daniel D, Sankey OF, Lyubchenko YL. Holliday junction dynamics and branch migration: single-molecule analysis. Proc Natl Acad Sci USA. 2005;102:8186–8191. doi: 10.1073/pnas.0407210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilley DM. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 33.Rasnik I, et al. Branch migration enzyme as a Brownian ratchet. EMBO J. 2008;27:1727–1735. doi: 10.1038/emboj.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 35.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 36.Iyer VN, Rupp WD. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochim Biophys Acta. 1971;228:117–126. doi: 10.1016/0005-2787(71)90551-x. [DOI] [PubMed] [Google Scholar]
- 37.Konola JT, Logan KM, Knight KL. Functional characterization of residues in the P-loop motif of the RecA protein ATP binding site. J Mol Biol. 1994;237:20–34. doi: 10.1006/jmbi.1994.1206. [DOI] [PubMed] [Google Scholar]
- 38.Morgan EA, Shah N, Symington LS. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Mol Cell Biol. 2002;22:6336–6343. doi: 10.1128/MCB.22.18.6336-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark JM, et al. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 40.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat Struct Mol Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 41.Rossi MJ, Mazina OM, Bugreev DV, Mazin AV. Analyzing the branch migration activities of eukaryotic proteins. Methods. 2010;51:336–346. doi: 10.1016/j.ymeth.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.