Abstract
The hippocampus is a key brain region involved in both short- and long-term memory processes and may play critical roles in drug-associated learning and addiction. Using whole genome sequencing of mRNA transcripts (RNA-Seq) and immunoprecipitation-enriched genomic DNA (ChIP-Seq) coupled with histone H3 lysine 4 trimethylation (H3K4me3), we found extensive hippocampal gene expression changes common to both cocaine-addicted and alcoholic individuals that may reflect neuronal adaptations common to both addictions. However, we also observed functional changes that were related only to long-term cocaine exposure, particularly the inhibition of mitochondrial inner membrane functions related to oxidative phosphorylation and energy metabolism, which has also been observed previously in neurodegenerative diseases. Cocaine- and alcohol-related histone H3K4me3 changes highly overlapped, but greater effects were detected under cocaine exposure. There was no direct correlation, however, between either cocaine- or alcohol- related histone H3k4me3 and gene expression changes at an individual gene level, indicating that transcriptional regulation as well as drug-related gene expression changes are outcomes of a complex gene-regulatory process that includes multifaceted histone modifications.
Keywords: drug addiction, histone methylation
Knowledge of the cellular and molecular adaptations underlying addiction to cocaine and alcohol has primarily been gained from animal models of acute and chronic drug exposure, from which specific molecular pathways to addiction have been identified (1, 2). Studies on human postmortem brain (3) have yielded data that are complementary and tend to validate animal models of exposure. Addiction-associated molecular alterations observed in animal and human studies are diverse. Clearly, cellular and molecular responses to addictive drugs depend on the type and timing of exposure, the timing of observations within the progression to addiction, and the brain regions and cells in which observations are made. Specific changes in signal transduction pathways, including in the transcription factor ΔFosB and the cAMP-response element binding protein (CREB), are likely to be more prominent in early stages of drug-induced neuroadaptation (4). Longstanding adaptations may be marked by changes in expression of genes involved in the regulation of cellular functions including ion transport, chromosome remodeling, stress and immune response, cell adhesion, cell cycle, apoptosis, protein and lipid metabolism, and mitochondrial functions (3). Later changes may be more closely relevant to addictive behaviors and long-lasting vulnerability to relapse.
The hippocampus is also a brain region critically involved in addiction. Most studies have focused on the mesolimbic system, in which medium-sized spiny neurons in the dorsal striatum and nucleus accumbens mediate dopaminergic, glutamatergic, and GABAergic neurotransmission (5) and are key in drug-reward and drug-seeking behavior. However, hippocampal functions related to short- and long-term memory processes and involving synaptic plasticity are necessary for drug-associated learning and memory. The hippocampus directly projects excitatory efferents to the nucleus accumbens in the mesolimbic system and can also activate dopaminergic neurons of the ventral tegmental area, explaining its involvement in cue-conditioned dopamine release. Cocaine modulates hippocampal functions such as long-term potentiation (LTP) (6), raising the possibility that long-term drug exposure may impair adaptive plasticity relevant to learning.
By using a massively parallel sequencing approach, which enables whole genome views of RNA transcription and epigenetic state, we surveyed genome-wide changes in gene expression and histone methylation modification in postmortem hippocampal tissue from individuals chronically exposed to cocaine or alcohol, and from carefully matched drug-free controls. Based on their mechanisms of action, and, to some extent, published literature, we hypothesized that this approach would reveal changes that were substance-specific and other changes that were common to general processes of addiction.
Results
Global Change in mRNA Expression in Chronic Cocaine and Alcohol Exposure.
The hippocampal mRNA transcriptome was sequenced in 24 age-, ethnicity-, and postmortem interval-matched men: eight chronic cocaine addicts, eight alcoholics, and eight drug-free control subjects (SI Appendix, Table S1). RNA expression was detected for 16,008 RefSeq genes by using the sequencing reads pooled from all individuals. The expression of each gene was thereby quantified for each individual within a consistent physical framework, defined by pooled analysis of expression signals and by National Center for Biotechnology Information gene annotations that allowed signals from multiple exons to be combined into a measure of average sequencing read counts for each gene. Gene expression was highly correlated among individual samples (SI Appendix, Fig. S1A), indicating reproducibility and quality of RNA-Seq results. The signal distribution (SI Appendix, Fig. S1B) of all expressed transcripts was also consistent with previous studies of gene expression in the brain (7). However, the depth of sequencing coverage enabled the detection, and quantification of more transcripts of low-abundance compared with microarray-based analysis.
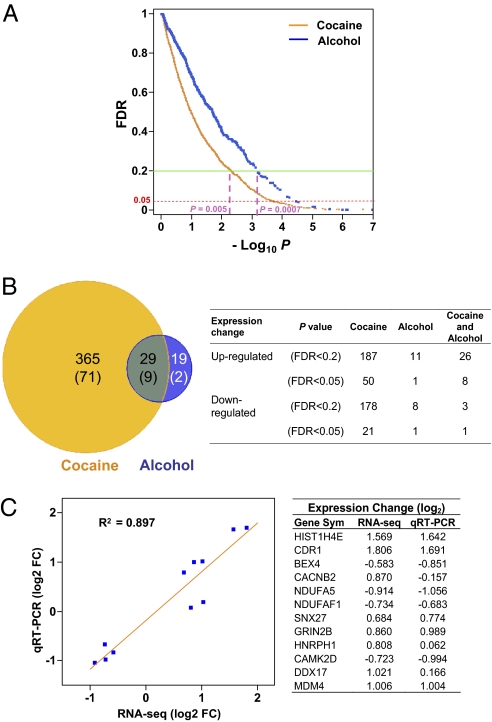
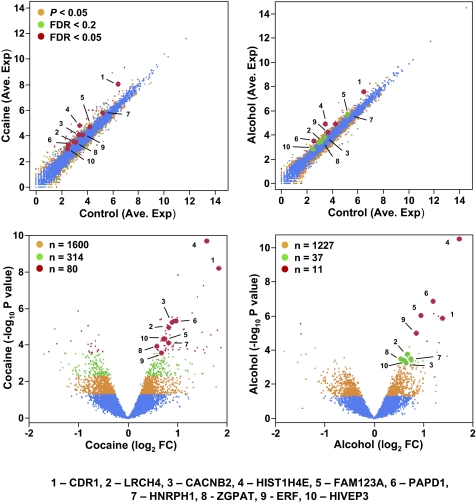
Among the 16,008 hippocampal expressed genes, at an uncorrected P value lower than 0.05, a total of 1,994 were differentially expressed in chronic cocaine-exposed individuals, and 1,275 were differentially expressed in the chronic alcohol-exposed individuals. However, to compensate for multiple testing leading to false-positive findings at any significance threshold, we calculated a false discovery rate (FDR; Benjamini–Hochberg) q value for each gene and applied it for genome-wide corrections. As seen in Fig. 1A, by using a relatively stringent FDR cutoff of less than 0.2, which includes the genes with the maximum probability of false discovery of less than 20% (P < 0.005 in cocaine group and P < 0.0007 in alcohol group) and many of those with a lower FDR probability, there were a total of 394 differentially expressed genes in the cocaine-addicted subjects and 48 in the alcoholic subjects (Fig. 1B and SI Appendix, Tables S2 and S3). The use of an FDR lower than 0.05 identified 80 genes in the cocaine addicts and 11 in the alcoholics (Fig. 1B). The numbers of genes that are either up-regulated or down-regulated in the cocaine addicts and alcoholics are summarized in Fig. 1B. Gene expression changes detected by RNA-Seq were also validated by quantitative RT-PCR for 11 transcripts in eight control and eight cocaine-addicted individual samples. There was a strong correlation (R2 = 0.90) between the RNA-Seq and quantitative RT-PCR measurement (Fig. 1C and SI Appendix, Fig. S2), indicating overall reliability of the RNA-Seq results. The scatterplots of relative expression levels (average normalized sequencing read counts), degree (fold) of change, and P values (both uncorrected and FDR cutoffs) of all 16,008 expressed genes in both cocaine and alcohol groups are also shown in Fig. 2.
Fig. 1.
Differentially expressed genes in the hippocampus of cocaine- and alcohol-addicted individuals, as detected by RNA-Seq. (A) Scatterplot of −log10 P (uncorrected P value) versus FDR q value for all 16,008 expressed genes. FDR thresholds of 0.2 and 0.05 are marked, as well as corresponding uncorrected P values at an FDR of 0.2. (B) Genes differentially expressed (FDR < 0.2) in cocaine only, alcohol only, and common to both. Numbers of genes with an FDR lower than 0.05 are shown in parentheses. Right: Number of genes that are up- or down-regulated in cocaine only, alcohol only, and common to both. (C) Correlation of gene expression changes detected by RNA-Seq and quantitative RT-PCR. Validation was performed for 11 genes in eight cocaine addicts and eight control subjects. The gene symbols and fold-change (log2 FC) for each gene are listed on the right.
Fig. 2.
Transcripts detected in the hippocampus by RNA-Seq and changes of expression in cocaine- or alcohol-addicted individuals. Upper: Scatterplots of expression levels of all 16,008 gene transcripts in control versus cocaine- (Left) or alcohol-addicted (Right) individuals. Lower: Volcano plots providing P values and fold change for all 16,008 gene transcripts in cocaine- (Left) and alcohol-addicted (Right) individuals. Genes that are differentially expressed at uncorrected P < 0.05, FDR lower than 0.2, or FDR lower than 0.05 are indicated by three-level color coding. Ten protein-coding genes that are among the most highly differentially expressed in both the cocaine and alcohol-addicted hippocampus are highlighted in red, and their gene symbols are listed.
Genes Affected by both Cocaine and Alcohol Exposure.
Although it is apparent that long-term cocaine exposure resulted in wider and more severe effects on gene expression in the hippocampus (Fig. 1), we also observed a significant overlap (P < 4.04 × 10−7) of differentially expressed genes between cocaine addiction and alcoholism. Under a FDR lower than 0.2, a total of 29 genes showed expression changes in both the cocaine-exposed and alcohol-exposed hippocampus (Fig. 1 A and B). Furthermore, in each case, the change was in the same direction [nominal likelihood, (1/2)29]. The details of these 29 genes are listed in Table 1. Examining the 11 genes whose expression in alcoholics was altered with a FDR lower than 0.05, nine were also common in the cocaine group. These convergent alterations in gene expression indicate sharing of processes in cocaine- and alcohol-related hippocampal neural adaptation. However, it is also apparent that there was a more severe, or more consistent, long-term impact in the cocaine-addicted individuals, as indicated by the disparity in the number of genes affected (Fig. 1B).
Table 1.
Genes with significant differential expression (FDR < 0.2) observed in both chronic cocaine- and alcohol-addicted individuals
| Cocaine |
Alcohol |
||||||||
| Gene symbol | Gene name | Log FC | P Value | FDR | Rank* | Log FC | P Value | FDR | Rank* |
| HIST1H4E | Histone cluster 1, h4e | 1.57 | 1.6 × 10−10 | 0.000 | 1 | 1.70 | 2.5 × 10−11 | 0.000 | 1 |
| RN7SK | RNA, 7SK small nuclear | 2.42 | 5.3 × 10−10 | 0.000 | 2 | 1.31 | 3.6 × 10−5 | 0.044 | 11 |
| CDR1 | Cerebellar degeneration-related protein 1, 34 kDa | 1.81 | 5.2 × 10−9 | 0.000 | 3 | 1.36 | 1.1 × 10−6 | 0.004 | 6 |
| SNORD89 | Small nucleolar RNA, C/D box 89 | 1.53 | 5.6 × 10−8 | 0.000 | 5 | 1.30 | 1.1 × 10−6 | 0.004 | 5 |
| SNORA73A | Small nucleolar RNA, H/ACA box 73A | 1.41 | 2.2 × 10−7 | 0.001 | 6 | 0.81 | 6.5 × 10−4 | 0.190 | 48 |
| SCARNA17 | Small Cajal body-specific RNA 17 | 1.30 | 1.6 × 10−6 | 0.004 | 8 | 1.08 | 3.0 × 10−5 | 0.042 | 10 |
| PAPD1 | Mitochondrial poly(A) polymerase | 0.95 | 4.1 × 10−6 | 0.004 | 14 | 1.17 | 1.1 × 10−7 | 0.001 | 2 |
| CACNB2 | Calcium channel, voltage-dependent, β2 subunit | 0.87 | 4.8 × 10−6 | 0.005 | 15 | 0.61 | 4.8 × 10−4 | 0.171 | 38 |
| LRCH4 | Leucine-rich repeats and calponin homology (CH) domain containing 4 | 0.80 | 8.8 × 10−6 | 0.008 | 18 | 0.65 | 1.5 × 10−4 | 0.112 | 16 |
| SNORD42A | Small nucleolar RNA, C/D box 42A | −1.47 | 1.2 × 10−5 | 0.010 | 19 | −1.48 | 1.1 × 10−5 | 0.018 | 9 |
| SNORA47 | Small nucleolar RNA, H/ACA box 47 | 1.82 | 1.5 × 10−5 | 0.010 | 22 | 1.48 | 2.3 × 10−4 | 0.140 | 21 |
| LENG8 | Leukocyte receptor cluster (LRC) member 8 | 0.77 | 2.3 × 10−5 | 0.014 | 26 | 0.72 | 6.2 × 10−5 | 0.066 | 12 |
| FAM123A | Family with sequence similarity 123A | 0.72 | 3.9 × 10−5 | 0.019 | 30 | 0.92 | 8.0 × 10−7 | 0.004 | 4 |
| HIVEP3 | HIV type I enhancer binding protein 3 | 0.70 | 3.9 × 10−5 | 0.019 | 31 | 0.59 | 3.3 × 10−4 | 0.163 | 26 |
| HNRPH1 | Heterogeneous nuclear ribonucleoprotein H1 | 0.81 | 6.5 × 10−5 | 0.026 | 38 | 0.72 | 2.7 × 10−4 | 0.141 | 25 |
| ZGPAT | Zinc finger, CCCH-type with G patch domain | 0.55 | 9.2 × 10−5 | 0.033 | 43 | 0.51 | 2.7 × 10−4 | 0.141 | 24 |
| ERF | Ets2 repressor factor | 0.65 | 2.2 × 10−4 | 0.046 | 73 | 0.82 | 8.7 × 10−6 | 0.018 | 8 |
| SNORD116-29 | Small nucleolar RNA, C/D box 116–29 | −1.68 | 3.0 × 10−4 | 0.053 | 89 | −1.82 | 1.2 × 10−4 | 0.103 | 14 |
| C9orf139 | Chromosome 9 ORF 139 | 0.90 | 4.4 × 10−4 | 0.068 | 103 | 0.89 | 5.2 × 10−4 | 0.176 | 41 |
| C9orf3 | Aminopeptidase O | 0.70 | 6.5 × 10−4 | 0.084 | 119 | 0.73 | 4.0 × 10−4 | 0.171 | 29 |
| KCNA2 | Potassium voltage-gated channel, shaker-related subfamily, member 2 | 0.67 | 1.1 × 10−3 | 0.106 | 153 | 0.74 | 3.7 × 10−4 | 0.171 | 27 |
| EXOC6B | Exocyst complex component 6B | 0.51 | 1.4 × 10−3 | 0.124 | 180 | 0.55 | 6.4 × 10−4 | 0.190 | 45 |
| CENTB5 | Arfgap with coiled-coil, ankyrin repeat and PH domains 3 | 0.54 | 1.8 × 10−3 | 0.139 | 198 | 0.61 | 4.7 × 10−4 | 0.171 | 37 |
| TAOK2 | TAO kinase 2 | 0.56 | 1.8 × 10−3 | 0.139 | 201 | 0.68 | 2.4 × 10−4 | 0.141 | 22 |
| TNRC6C | Trinucleotide repeat containing 6C | 0.59 | 2.8 × 10−3 | 0.170 | 255 | 0.69 | 6.5 × 10−4 | 0.190 | 47 |
| ADAMTS4 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 | 0.65 | 2.9 × 10−3 | 0.173 | 263 | 0.79 | 4.3 × 10−4 | 0.171 | 33 |
| MSH4 | Muts homologue 4 (Escherichia coli) | 0.79 | 3.5 × 10−3 | 0.180 | 302 | 0.99 | 4.2 × 10−4 | 0.171 | 31 |
| C16orf72 | Chromosome 16 ORF 72 | 0.54 | 4.3 × 10−3 | 0.189 | 358 | 0.66 | 6.0 × 10−4 | 0.186 | 44 |
| CCR5 | Chemokine (C-C motif) receptor 5 | −0.79 | 4.7 × 10−3 | 0.194 | 386 | −1.19 | 7.4 × 10−5 | 0.070 | 13 |
FC, fold change.
*Rank in each group according to P value.
Some of the protein-coding genes with expression changes common to both drugs are highlighted in Fig. 2. It is noticeable that these genes play important roles in neuronal functions. Briefly, CDR1 is a cerebellar degeneration-related protein, LRCH4 is a leucine-rich repeat-containing neuronal protein, CACNB2 is a subunit of voltage-gated calcium channel and is involved in neuronal functions, and FAM123A (AMER2) is a member of the gene family involved in neurogenesis (8), suggesting that they all play critical roles in brain functions. HIST1H4E encodes a member of the histone H4 family, a critical component of the nucleosome. PAPD1 is a mitochrondrial poly(A) polymerase, and HNRPH1 is a heterogeneous nuclear ribonucleoprotein, both being involved in RNA processing and stability. ZGPAT, ERF, and HIVEP3 (ZAS3) are transcription regulators. ZGPAT is a zinc finger transcription factor protein. ERF contains a highly conserved DNA-binding domain, the ETS domain, and was implicated in RAS/ERK signaling and the transcriptional regulation of c-Myc. HIVEP3 is also a zinc finger protein, binding to the κB motif and gene promoter and enhancers. HIVEP3 has also been linked to Parkinson disease (9).
Chronic Cocaine Exposure Inhibits Mitochondrial Inner Membrane Oxidative Phosphorylation.
We subjected the differentially expressed genes with FDR less than 0.2 to Gene Ontology (GO) and pathway analysis to detect molecular and cellular functional domains impacted by chronic cocaine or alcohol exposure (Table 2). We observed a great effect of long-term cocaine exposure on genes involved in mitochondrial inner membrane functions and oxidative phosphorylation. Interestingly, the cocaine-affected genes were common to those observed in some of the neurodegenerative diseases that also involve mitochondrial inner membrane genes responsible for ATP synthesis, maintenance of mitochondrial membrane potential, and defense against oxidative stress (SI Appendix, Table S4). To objectively assess the impact of chronic cocaine exposure on mitochondrial ATP synthesis, we examined all 90 genes encoding components of oxidative phosphorylation, At an uncorrected P value lower than 0.05, 32 of the 90 genes (compared with five expected randomly) were differentially expressed (SI Appendix, Table S5). All were down-regulated. Furthermore, 74 of the 90 genes (including the 32 genes that were significantly down-regulated) displayed reduced expression levels in cocaine addicts. This down-regulation of mitochondrial inner membrane genes was not observed in the alcoholics. In that group, there were only three genes that showed significant expression changes (uncorrected P < 0.05). These results strongly indicate that the inhibition of the mitochondrial inner membrane genes in cocaine addiction is a robust phenomenon and a specific effect of chronic cocaine exposure, with potential negative implications for brain energy metabolism and diverse brain functions that depend on it. These findings are also highly consistent with brain imaging studies that have revealed negative effects of cocaine on brain glucose metabolism (10–13).
Table 2.
Summary of GO and pathway analysis
| Cocaine |
Alcohol |
|||
| GO term | Count | P value | Count | P value |
| Mitochondrial inner membrane (GO:0005743) | 18 | 0.00015 | — | — |
| Huntington disease (hsa05016) | 13 | 0.00027 | — | — |
| Alzheimer disease (hsa05010) | 11 | 0.00169 | — | — |
| Parkinson disease (hsa05012) | 9 | 0.00441 | — | — |
| Oxidative phosphorylation (hsa00190) | 8 | 0.01654 | — | — |
| Negative regulation of gene expression (GO:0010629) | 22 | 0.00086 | — | — |
| Gene silencing (GO:0016458) | 6 | 0.00455 | — | — |
| Covalent chromatin modification (GO:0016569) | 8 | 0.01154 | — | — |
| RNA-binding (SP_PIR_KEYWORDS) | 18 | 0.02450 | 5 | 0.019506 |
| Extracellular matrix part (GO:0044420) | 7 | 0.03018 | — | — |
Chronic Cocaine Exposure Alters Expression of Genes Involved in Regulation of Transcription and Chromatin Modification.
Chronic cocaine exposure also altered the expression of genes that are responsible for the regulation of transcription, gene silencing, and chromatin modification as revealed by GO analysis (using FDR < 0.2 as threshold for individual genes; Table 2 and SI Appendix, Table S6). The genes in these several GO categories overlap highly. Several had been previously implicated in cocaine addiction, such as DNMT3a, a DNA methyltransferase that was recently reported to play an important role in regulating cocaine response and spine plasticity in the nucleus accumbens in the rat (14), and HDAC2, a histone deacetylase found to be involved in cocaine-induced transcription changes in rat nucleus accumbens and cocaine-seeking behavior (15). Although more significant impact was observed only in the cocaine addicts, there was also substantial overlap in altered expression of genes in the alcoholics at an FDR lower than 0.02 or an uncorrected P value lower than 0.05 (SI Appendix, Table S6). In addition, we also observed convergent evidence that chronic cocaine exposure, and, to a lesser degree, alcohol exposure, alters expression of genes involved in RNA processing, including significant alteration in the expression of genes encoding RNA-binding and processing proteins (Table 2 and SI Appendix, Table S7) and enrichment of differentially expressed small nucleolar RNA genes, which are involved in both ribosomal RNA and mRNA processing (16). Among 102 small nucleolar RNA genes expressed in hippocampus, 15 were significantly changed (FDR < 0.2) in cocaine addicts (compared with 394 of 16,008 for all of the expressed genes at an FDR < 0.2), and five were significantly changed in the alcoholics (48 of 16,008 for all genes; SI Appendix, Table S8). Overall, these results reveal pronounced and persistent effects of long-term cocaine exposure and some common molecular changes induced by chronic alcohol exposure on gene expression regulation and RNA processing.
Other Genes and Molecular Networks Affected by Chronic Cocaine or Alcohol.
Genes involved in ECM functions were also differentially expressed in the cocaine addicts (Table 2 and SI Appendix, Table S9), as reported in other studies (3). We also observed cocaine-depressed transcript levels for all five members of the BEX gene family (BEX 1–5), which encodes brain expressed, X-linked proteins (Table 2 and SI Appendix, Table S10) and are thought to mediate neurotrophin signaling and neuronal differentiation (17). This inhibition of the whole of BEX gene family was observed only in cocaine-exposed individuals. We also observed expression changes of genes involved in LTP (SI Appendix, Table S11) in the cocaine addicts. There were a total of 10 differentially expressed genes (FDR < 0.02 or uncorrected P < 0.05) in the Kyoto Encyclopedia of Genes and Genomes LTP pathway, indicating perturbation of LTP and alteration of hippocampal plasticity by long-term cocaine exposure.
Although gene expression changes in the alcoholics were not as extensive as in the cocaine addicts, detailed examination of the results, in some cases using a less stringent statistical cutoff (P < 0.01), also revealed some potentially important insights to the molecular adaptation to chronic alcohol exposure. The changes included genes critical in neural transmission and related brain functions (SI Appendix, Table S3) such as GRIN2B, GABA, MAOA, SEMA7A, CREB5, HDAC2. These changes are convergent with previous findings (18, 19).
Global Correlation of Histone H3K4Me3 with Gene Expression.
To investigate potential epigenetic effects of chronic cocaine and alcohol exposure on chromatin histone modification, we measured hippocampal histone H3 lysine 4 trimethylation. A total of 27,569 H3K4me3 peaks were detected across the genome. A pattern of colocalization with gene promoters was revealed (SI Appendix, Fig. S3A). Among these, 13,113 peaks (48%) were mapped to promoter regions of annotated genes [<1.5 kb from known transcription start sites (TSSs)] for which mRNA expression was also detected by RNA-Seq (SI Appendix, Fig. S3B). A total of 1,413 peaks (5%) were located near the promoters of genes where mRNA expression was not detected, and 13,043 peaks (47%) were in regions more than 1.5 kb away from a known TSS. H3K4Me3 was detected in the promoter regions of 82% of expressed genes, whereas only 11% of nonexpressed genes had H3K4me3 modification (SI Appendix, Fig. S3C). There were also large differences in H3K4me3 signal levels among the peaks detected in expressed genes, nonexpressed genes, and nongenic regions (SI Appendix, Fig. S3D). Expressed genes had, on average, threefold higher signals compared with the nonexpressed genes, and sixfold higher signals compared with the peaks in nongenic regions. In the expressed genes, H3K4Me3 peak size correlated with level of expression (SI Appendix, Fig. S3E); however, many genes that displayed high levels of H3K4 trimethylation at the promoters showed low levels of expression, indicating that H3K4 trimethylation alone is insufficient for activation of expression. These results positively associate H3K4Me3 with gene expression in hippocampus and are consistent with previous findings (20).
Cocaine- and Alcohol-Related H3K4me3 Changes at Gene Promoters.
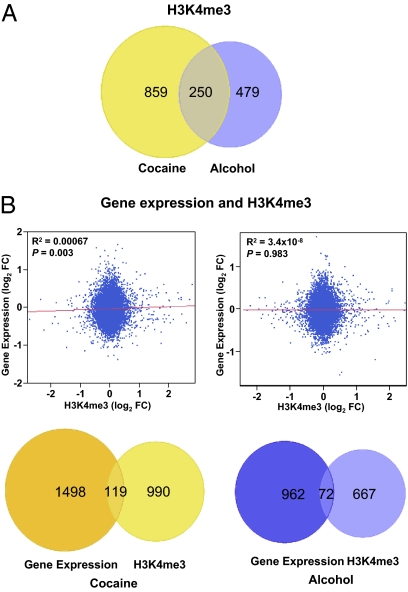
Chronic cocaine exposure resulted in genome-wide changes in H3K4Me3 at gene promoters. Changes in H3K4me3 signal levels were detected in the promoters of 1,115 expressed genes (uncorrected P < 0.05; Fig. 3A and SI Appendix, Fig. S4). By using a relatively more stringent test with an FDR lower than 0.2, significant changes were detected in 108 expressed genes, and five of these were below an FDR of 0.05 (SI Appendix, Table S12). In the alcoholic subjects, there were H3K4me3 signal changes at the promoters of 729 expressed genes (uncorrected P < 0.05; Fig. 3A and SI Appendix, Fig. S4). However, none of these changes withstood an FDR of lower than 0.2 as the cutoff (SI Appendix, Table S13 lists the signal changes with P < 0.005). The smaller number of histone H3K4me3 changes compared with the observed gene expression changes (Fig. 1B) suggests that drug exposure has only a modest effect on histone H3K4me3 in the hippocampus. However, it cannot be ruled out that the apparent limited drug effect might be a result of technical limitations of ChIP to detect more subtle changes. Similar to the observed gene expression changes, we detected a greater number of H3K4me3 changes in cocaine addicts (Fig. 3A), and there was significant overlap of the changes between the cocaine and alcohol exposure groups (Fig. 3A). Among the 729 expressed genes with promoter H3K4me3 changes in the alcoholics, more than one third (n = 250) also showed changes in the cocaine addicts (P < 1 × 10−10), suggesting shared molecular changes in histone modification following long-term cocaine and alcohol exposure.
Fig. 3.
Cocaine- and alcohol-related histone H3K4me3 changes and their correlation with gene expression changes in hippocampus. (A) Number of promoters with H3K4me3 changes (uncorrected P < 0.05) at expressed genes in cocaine- or alcohol-addicted individuals. A significant overlap between the cocaine- and alcohol-addicted groups was observed. (B) A trend of correlation of H3K4me3 change with transcriptional changes among all 13,113 peaks mapped to the hippocampal expressed genes was detected in the cocaine group (Upper, Left), but no correlation was detected in the alcohol group (Upper, Right). The numbers of genes with significant expression changes and H3K4me3 changes are also shown (Lower). No significant overlap was detected between RNA or histone changes in cocaine (Left) or alcohol addiction (Right).
Cocaine- and Alcohol-Related H3K4me3 Changes Do Not Directly Correlate with Expression Changes at Individual Genes.
Globally, among all 13,113 histone H3K4me3 peaks mapped to the promoters of hippcampal expressed genes, we observed a trend of correlation between cocaine-related H3K4me3 and expression changes (P = 0.003, Fig. 3B, Upper Left). However, between the genes with significant H3K4me3 changes or expression changes, there was no significant overlap (Fig. 3B, Lower Left). This indicates that the alteration of histone H3K4me3 resulted from long-term cocaine exposure is only strong enough to be reflected as subtle changes of expression in many genes, but a majority of these individual changes are not great enough to be statistically significant. In fact, it would be unlikely to observe consistent correlation between H3K4me3 and gene expression changes at individual genes, as H3K4me3 is only one of the many histone modifications involved in chromatin remodeling, and gene expression is determined by multilevel and multidimensional regulatory processes. In alcoholics, we did not observe global correlation or individual locus overlap between H3K4me3 and expression changes (Fig. 3B, Right), which again suggests a relative mild effect of alcohol exposure on histone H3 lysine 4 trimethylation. The individual loci that displayed up- or down-regulation of both H3K4me3 and RNA transcripts (uncorrected P < 0.05) in either the cocaine or alcohol group can be found in SI Appendix, Tables S14 and S15.
Discussion
We obtained genome-wide views of gene expression and histone H3 lysine 4 trimethylation in the hippocampus of individuals chronically exposed to cocaine or alcohol compared with drug-free controls. We applied a relatively stringent analytic approach to avoid spurious findings commonly associated with multiple testing. In both cocaine addicts and alcoholics, transcriptional changes significantly overlapped, and many of the commonly affected genes are critical for brain functions. There was also significant overlap between cocaine- and alcohol-related histone H3K4me3 changes. In comparison with alcohol exposure, cocaine exposure led to a stronger shift in hippocampal mRNA transcription, as manifested by the larger number of differentially expressed genes and by changes in molecular and cellular functions defined by GO. More extensive histone alteration was also observed in cocaine addicts. These results may reflect the stronger impact of cocaine on brain functions, as well as divergent impact on different systems. However, the relatively smaller changes observed in alcoholics may be partially related to the fact that, unlike cocaine addicts, whose deaths were directly attributable to drug intoxication, the alcoholic individuals died of various causes. This could increase within-group heterogeneity and make alcohol-related changes more difficult to uncover. Although cocaine- or alcohol-induced histone H3K4me3 changes were not directly predictive of gene expression at the individual gene promoter level, the observed histone H3K4me3 profiles still provide valuable insight into the addiction-induced alteration of histone structure.
Mitochondrial inner membrane dysfunctions are known to play critical roles in neurodegenerative diseases such as Huntington disease, Alzheimer disease, and Parkinson disease (21). The main dysfunctions involved in the pathological processes are suppressed oxidative stress defense, altered membrane ion permeability, and, particularly, depleted ATP synthesis resulting from the inhibition of components of the oxidative phosphorylation pathway. Via brain imaging such as autoradiography and PET, negative effects of cocaine on glucose metabolism and energy state have been repeatedly observed in various brain regions, including hippocampal formation, in humans (10), nonhuman primates (11), mice (13), and rats (12). Prolonged cocaine exposure also lead to more widespread and more severe inhibition of glucose metabolism in various brain regions (12). Strikingly, we observed here that more than one third of the genes responsible for ATP synthesis through glucose metabolism were suppressed (uncorrected P < 0.05) in transcription, and 82% of these genes showed the same trend. ATP provides the energy source required by various cellular biochemical reactions, and brain functions are particularly vulnerable to disruption of mitochondrial energy metabolism. In addition, we also observed significant overexpression (FDR = 0.015) of the ionotropic glutamate receptor gene, GRIN2B, which may result in dysregulation of calcium influx and mitochondrial calcium overload leading to apoptosis. These observed gene expression changes may represent the molecular adaptations of both neurons and the astrocytes and other glial cells that account for a substantial portion of brain energy metabolism (22). The impact of cocaine exposure on mitochondrial functions reflects changes in expression of nuclear chromosomal genes, but it is unknown whether these changes resulted from direct toxic effects on mitochondrial inner membrane components or more indirect effects of mitochondrial toxicity, or on neural networks. Interestingly, changes in expression of genes related to mitochondrial functions were also observed in nicotine-exposed rats (23). However, we did not observe similar changes after alcohol exposure. This may indicate that changes in genes involved in cell energy metabolism are substance-specific, but other factors might have confounded our ability to detect such changes in the alcoholic subjects.
The molecular and cellular changes in response to drug exposure are likely to depend on the stages of the addictive process. Animal studies of drug exposure effects usually involve relatively short courses of drug exposure, and, in contrast, the investigation of molecular changes in human subjects usually relies on postmortem brain tissue from individuals with prolonged heavy exposures. It is likely that, in the early stages of adaptation to cocaine exposure, brain region and signal transduction-specific changes are more visible. In the late stages of the addiction, long-term allostatic processes may become prominent. One well known transcript induced by cocaine and other drugs is CART (cocaine- and amphetamine-regulated transcript; gene symbol CARTPT). After acute exposure to cocaine and other addictive drugs, CART mRNA levels were usually found to be increased in the striatum (24, 25), and the transcript is induced through dopaminergic transmission (26). However, we did not observe significant changes of the CART transcript in the hippocampus of chronic cocaine addicts. Some other molecules identified in animal studies and considered to be important in mediating cocaine-induced changes are CREB1, Δ-FosB (FOSB), two NAD-dependent deacetylases, namely sirtuin1 (Sirt1) and sirtuin 2 (Sirt2) (1, 4), and DNMT3a, a DNA methyltransferase recently found to play key roles in regulating cocaine response and spine plasticity (14). We observed some consistent changes in our chronic cocaine abusers. These include elevated levels of the DNMT3a transcript (Table 4). We also observed altered expression of CREB1 (uncorrected P < 0.05) and the histone deacetylases HDAC2 (P = 0.0007, FDR = 0.09 in cocaine addicts and P = 0.007 in alcoholics), and HDAC4 (uncorrected P < 0.05 in both cocaine and alcohol addicts). However, FOSB, SIRT1, and SIRT2 expression were not altered in the hippocampus of the chronic cocaine abusers. Those differences from previous observations can be stage-specific and also brain region-specific. For example, the cocaine-induced Δ-FosB and sirtuin elevation was observed in the striatum (1).
We profiled the genome-wide distribution of histone H3K4me3 modifications in the human hippocampus and the cocaine and alcohol-induced changes in H3K4me3 distribution. The observation that the majority of the H3K4me3 peaks are located near known gene promoters and the correlation between H3K4me3 modification and gene activation are consistent with previous findings (20). However, it is of great interest to note that many H3K4me3 peaks (47%) were located away from promoters of known protein-coding genes. Although we cannot exclude the possibility that some non-H3K4me3 signals resulted from cross-reactivity, given that, in the known gene regions, H3K4me3 peaks are always positioned near a TSS, the level of cross-reactivity was low in the H3K4me3 ChIP we performed. Therefore, it is likely that many of the peaks that are apparently located far from a known TSS mark promoters of unannotated genes, including noncoding RNA genes. As H3K4me3 is a relatively well studied histone modification known to be linked to gene activation, we intended to examine the effect of cocaine and alcohol addiction on H3K4me3 modification and its relation to changes in gene expression. Although we cannot exclude the possibility that sensitivity of the ChIP assay may be a factor, it appears that histone H3 lysine 4 trimethylation is relatively stable following cocaine and, especially, alcohol addiction. However, we did observe significant overlap between cocaine- and alcohol-related changes, reflecting shared epigenetic alteration related to chromatin remodeling in the brain. It is also apparent that in the majority of the expressed genes, the changes of H3K4me3 modification were not directly associated with gene expression changes. This points to the complexity of gene regulation by multifaceted histone modification (20, 27) and the limited effect of one type of chromatin modification on global gene expression. Similarly, studies have also shown a lack of locus overlap between cocaine-induced methylation and acetylation changes in mice (1). In addition to histone modifications, gene expression is also regulated by many components of the complex transcriptional machinery and also involves other mechanisms such as DNA methylation. Nonetheless, our results reveal genome-wide alteration of histone H3K4 trimethylation resulting from long-term cocaine and alcohol exposure, and accompanying large-scale changes in gene expression that implicate several functional pathways in substance-shared and substance-specific fashion.
Materials and Methods
The materials and methods used in the present study are described in detail in SI Appendix, SI Materials and Methods. Postmortem brain tissue was provided by the University of Miami Brain Bank. Double-stranded cDNA libraries were synthesized from fragmented mRNA. Histone H3K4me3-specific chromatin DNA was enriched by ChIP assays. Sequencing was performed on the Illumina Genome Analyzer IIx. Details of data analysis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M.-A. Enoch for critical reading of the manuscript, and E. Moore for technical support. The acquisition of brain specimens was supported by Public Health Service Grant DA06227-18.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. SRA029279 and SRA029275).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018514108/-/DCSupplemental.
References
- 1.Renthal W, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang GC, et al. Acute administration of cocaine reduces metabotropic glutamate receptor 8 protein expression in the rat striatum in vivo. Neurosci Lett. 2009;449:224–227. doi: 10.1016/j.neulet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mash DC, et al. Gene expression in human hippocampus from cocaine abusers identifies genes which regulate extracellular matrix remodeling. PLoS ONE. 2007;2:e1187. doi: 10.1371/journal.pone.0001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 5.Hyman SE, Malenka RC. Addiction and the brain: The neurobiology of compulsion and its persistence. Nat Rev Neurosci. 2001;2:695–703. doi: 10.1038/35094560. [DOI] [PubMed] [Google Scholar]
- 6.Thompson AM, Swant J, Wagner JJ. Cocaine-induced modulation of long-term potentiation in the CA1 region of rat hippocampus. Neuropharmacology. 2005;49:185–194. doi: 10.1016/j.neuropharm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Carninci P. Constructing the landscape of the mammalian transcriptome. J Exp Biol. 2007;210:1497–1506. doi: 10.1242/jeb.000406. [DOI] [PubMed] [Google Scholar]
- 8.Comai G, Boutet A, Neirijnck Y, Schedl A. Expression patterns of the Wtx/Amer gene family during mouse embryonic development. Dev Dyn. 2010;239:1867–1878. doi: 10.1002/dvdy.22313. [DOI] [PubMed] [Google Scholar]
- 9.Li YJ, et al. Investigation of the PARK10 gene in Parkinson disease. Ann Hum Genet. 2007;71:639–647. doi: 10.1111/j.1469-1809.2007.00353.x. [DOI] [PubMed] [Google Scholar]
- 10.London ED, et al. Cocaine-induced reduction of glucose utilization in human brain. A study using positron emission tomography and [fluorine 18]-fluorodeoxyglucose. Arch Gen Psychiatry. 1990;47:567–574. doi: 10.1001/archpsyc.1990.01810180067010. [DOI] [PubMed] [Google Scholar]
- 11.Lyons D, Friedman DP, Nader MA, Porrino LJ. Cocaine alters cerebral metabolism within the ventral striatum and limbic cortex of monkeys. J Neurosci. 1996;16:1230–1238. doi: 10.1523/JNEUROSCI.16-03-01230.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macey DJ, Rice WN, Freedland CS, Whitlow CT, Porrino LJ. Patterns of functional activity associated with cocaine self-administration in the rat change over time. Psychopharmacology (Berl) 2004;172:384–392. doi: 10.1007/s00213-003-1676-7. [DOI] [PubMed] [Google Scholar]
- 13.Thanos PK, Michaelides M, Benveniste H, Wang GJ, Volkow ND. The effects of cocaine on regional brain glucose metabolism is attenuated in dopamine transporter knockout mice. Synapse. 2008;62:319–324. doi: 10.1002/syn.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrasekar V, Dreyer JL. The brain-specific neural zinc finger transcription factor 2b (NZF-2b/7ZFMyt1) suppresses cocaine self-administration in rats. Front Behav Neurosci. 2010;4:14. doi: 10.3389/fnbeh.2010.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kishore S, Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 17.Vilar M, et al. Bex1, a novel interactor of the p75 neurotrophin receptor, links neurotrophin signaling to the cell cycle. EMBO J. 2006;25:1219–1230. doi: 10.1038/sj.emboj.7601017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devaud LL, Smith FD, Grayson DR, Morrow AL. Chronic ethanol consumption differentially alters the expression of gamma-aminobutyric acidA receptor subunit mRNAs in rat cerebral cortex: competitive, quantitative reverse transcriptase-polymerase chain reaction analysis. Mol Pharmacol. 1995;48:861–868. [PubMed] [Google Scholar]
- 19.Hu XJ, Ticku MK. Chronic ethanol treatment upregulates the NMDA receptor function and binding in mammalian cortical neurons. Brain Res Mol Brain Res. 1995;30:347–356. doi: 10.1016/0169-328x(95)00019-o. [DOI] [PubMed] [Google Scholar]
- 20.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertz L, Peng L, Dienel GA. Energy metabolism in astrocytes: High rate of oxidative metabolism and spatiotemporal dependence on glycolysis/glycogenolysis. J Cereb Blood Flow Metab. 2007;27:219–249. doi: 10.1038/sj.jcbfm.9600343. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Kim JM, Donovan DM, Becker KG, Li MD. Significant modulation of mitochondrial electron transport system by nicotine in various rat brain regions. Mitochondrion. 2009;9:186–195. doi: 10.1016/j.mito.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglass J, McKinzie AA, Couceyro P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. J Neurosci. 1995;15:2471–2481. doi: 10.1523/JNEUROSCI.15-03-02471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salinas A, Wilde JD, Maldve RE. Ethanol enhancement of cocaine- and amphetamine-regulated transcript mRNA and peptide expression in the nucleus accumbens. J Neurochem. 2006;97:408–415. doi: 10.1111/j.1471-4159.2006.03745.x. [DOI] [PubMed] [Google Scholar]
- 26.Rogge G, Jones D, Hubert GW, Lin Y, Kuhar MJ. CART peptides: regulators p;of body weight, reward and other functions. Nat Rev Neurosci. 2010;9:218–758. doi: 10.1038/nrn2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.