Abstract
FLOWERING LOCUS C (FLC) has a key role in the timing of the initiation of flowering in Arabidopsis. FLC binds and represses two genes that promote flowering, FT and SOC1. We show that FLC binds to many other genes, indicating that it has regulatory roles other than the repression of flowering. We identified 505 FLC binding sites, mostly located in the promoter regions of genes and containing at least one CArG box, the motif known to be associated with MADS-box proteins such as FLC. We examined 40 of the target genes, and 20 showed increased transcript levels in an flc mutant compared with the wild type. Five genes showed decreased expression in the mutant, indicating that FLC binding can result in either transcriptional repression or activation. The genes we identified as FLC targets are involved in developmental pathways throughout the life history of the plant, many of which are associated with reproductive development. FLC is also involved in vegetative development, as evidenced by its binding to SPL15, delaying the progression from juvenile to adult phase. Some of the FLC target genes are also bound by two other MADS-box proteins, AP1 and SEP3, suggesting that MADS-box genes may operate in a network of control at different stages of the life cycle, many ultimately contributing to the development of the reproductive phase of the plant.
Keywords: reproductive transition, phase change, environmental response, floral morphology, ChIP sequencing
Encoding a MADS-box transcription factor, FLOWERING LOCUS C (FLC) is a major repressor of flowering in Arabidopsis (1, 2). The regulatory role of FLC in the control of flowering initiation is of special significance in vernalization (2), a period of low temperature that stimulates flowering. Before vernalization, FLC represses the initiation of flowering, preventing the changes that convert the apical meristem to one producing the reproductive structures. After the prolonged period of low temperature, FLC expression is repressed and plants are able to initiate flowering. The repression of FLC is associated with modifications to FLC chromatin, which prevent transcriptional activity of the gene (3, 4). The state of reduced transcriptional activity is maintained through the subsequent cell divisions of the developing plant when growing under normal temperature conditions (5). Two loci regulating FLC are FRIGIDA (FRI) (6) and VERNALIZATION INSENSITIVE 3 (VIN3) (4). FRI is responsible for a high level of production of the FLC protein and VIN3, which is induced by low temperature, reduces FLC transcriptional activity during vernalization. The vernalization process overrides the FRI-mediated control of FLC, resulting in the repression of transcriptional activity and the promotion of flowering initiation (7).
The transition of the vegetative apical meristem to one producing reproductive structures involves the interaction of FLC with a small number of key genes. Three flowering-time genes, FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSSON OF CONSTANS 1 (SOC1), and FLOWERING LOCUS D (FD), have been reported to be targeted by FLC (8, 9). FLC binds to the promoters of SOC1 and FD as well as to the first intron of FT (8, 9). FLC binding prevents the transcriptional activation of these genes, including their induction by daylength controls of flowering time (5).
FLC is expressed in most parts of the plant during all developmental stages (2), suggesting that, in addition to direct and indirect functions in the initiation of flowering and the production of reproductive structures, FLC may have other regulatory roles. FLC has been reported as being involved in the control of temperature-dependent seed germination, which requires other genes active in the flowering pathway, including FT, SOC1, and APETALA1 (AP1) (10). Another regulatory function claimed for FLC is a role in the regulation of the circadian clock (11). How FLC regulates the circadian clock at the molecular level is unclear (12), but it is known that FLC binds weakly to the LATE ELONGATED HYPOCOTYL (LHY) promoter, which may be one of the mechanisms by which FLC affects circadian rhythm (13).
There are more than 100 MADS-box transcription factors in Arabidopsis. Many of these proteins regulate important plant developmental processes (14). MADS-box proteins bind to a consensus sequence, the CArG box, that has the core motif CC(A/T)6GG (15). Some MADS-box proteins function in complexes with other MADS-box proteins (14). FLC interacts with another MADS-box protein, SHORT VEGETATIVE PHASE (SVP), to delay flowering (16), but loss of SVP does not fully suppress the delay of flowering (16), suggesting that FLC may interact with other proteins or is able to act by itself.
We show that FLC binds to more than 500 target sites in the Arabidopsis genome, potentially regulating genes that function in many developmental pathways. We present data showing that, for 40 of the target genes, 25 have expression regulated by FLC. Some FLC targets are also bound by the MADS-box proteins AP1 and SEPALLATA3 (SEP3), suggesting that this class of transcription factor acts in concert to regulate progress through the phases of the plant life cycle.
Results
Genome-Wide FLC Binding Sites.
To explore potential roles of FLC, we identified the genes across the genome that had FLC binding sites. We used ChIP followed by high-throughput sequencing (ChIP-seq) of immunoprecipitated DNA fragments from 12-d-old whole seedlings of Col FRI (a line with a high level of FLC) and compared the result to binding sites in flc-3, a FLC deletion mutant in the Col FRI background (17). Protein–DNA complexes were immunoprecipitated with antiserum raised against the FLC protein without the conserved MADS domain (18). The FLC antiserum recognizes the FLC protein but could interact with the related MADS AFFECTING FLOWERING (MAF) proteins (18). ChIP DNA from both the wild type and the mutant was sequenced with an Illumina Genome Analyzer (GAII).
We obtained ∼4.6 million and 2.9 million 75-bp single-end reads for Col FRI and flc-3, respectively; 50% of the reads mapped to the Arabidopsis Col genome (TAIR9 Build), allowing one mismatch at any position or two mismatches at low quality score positions. The regions enriched for mapped reads in the wild-type FLC-ChIP dataset itself, as well as relative to the mutant ChIP dataset, were identified by the quantitative enrichment of sequence tags (QuEST) algorithm (19) using three levels of stringency parameters (stringent, recommended, and relaxed), which are listed in Table S1. The relaxed data were then separated into two parts according to the q value with 200 binding sites for the first half and 205 sites for the second half. We obtained similar peak regions using two MACS algorithms (20) with the default parameters, except that regions identified by MACS were longer than those from QuEST. Peaks in MACS but not in QuEST were checked manually, based on the read distributions, resulting in seven more peaks being added to the QuEST peaks. There were 505 binding sites identified with higher read numbers in Col FRI wild type than in the flc-3 mutant (Dataset S1).
Majority of FLC Binding Sites Are in Gene Promoters.
Potential FLC target genes were defined as annotated genes containing binding sites within 3 kb upstream of the 5′ end and 1 kb downstream of the 3′ end of annotated genes. Using these criteria, we identified 786 genes as putative FLC targets (Dataset S1) because some binding sites occurred between two possible gene targets. Among the putative FLC targets, we found the known targets SOC1 and FT, but not FD or LHY. We found a low number of signals in both Col FRI and flc-3, which suggested that the FLC antiserum may be recognizing loci bound by some FLC-related MAF proteins.
Of all sites, 52.5% were located in a promoter region or in a promoter and 5′ UTR region of the nearest gene (Fig. S1). When considering the genes on both sides of binding sites, some peaks located in the gene body or in the 3′ end of a gene could also be considered to be in the promoter region of a neighboring gene. We may have underestimated the numbers of sites in promoters by assigning peaks to the nearest gene. We found that 26.5% of all sites were located within the gene body, 14.9% were located at the 3′ end of the gene, and 6.1% were located in intergenic regions (Fig. S1). Of the peaks in the gene body, 17.4% were in exons, 4% were in introns, and 5.1% were in both exons and introns (Fig. S1).
Binding Motifs of the FLC Protein.
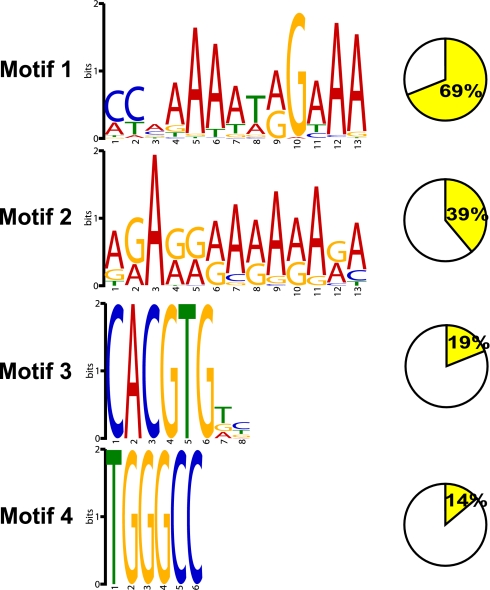
In vitro binding studies have shown that MADS-box proteins bind to a specific DNA motif, the CArG box, with the consensus sequence CC(A/T)6GG (15). CArG-box sequences differ for the various MADS-box proteins. The previously known FLC targets (SOC1 and FT) contain a CArG box in their binding regions (8, 9). To determine the consensus sequence of FLC binding motifs across the genome, we used MEME software (21) to analyze the sequences of the 505 target sites. Of the FLC binding sites, 69% contained at least one CArG-box motif with the core consensus sequence CCAAAAAT(G/A)G and an AAA extension at the 3′ end (Fig. 1). When we used the fuzznuc program (22) to search for the CCWWWWWWRG motif (W = A/T and R = A/G), there was significant enrichment of this motif over randomly selected promoter sequences for the stringent, the recommended, and the first half of the relaxed dataset (Fisher P < 0.0001) (Table S2). The second half of the relaxed dataset is not significantly enriched for this motif (Table S2). When we did the search against CCWWWWWWRGWAA (including the AAA extension) motif, all of the datasets showed an enrichment over the randomly selected promoter sequences (Fisher P < 0.00001). This result suggests that the AAA trinucleotide extension is a functional part of the CArG-box motif in the binding of the FLC protein. When we compared the FLC CArG-box motif against the aggregated DNA motif databases by using TOMTOM (23), we found similarity between the CArG-box motifs of FLC and AGL15 or SEP3, including an AAA extension (Fig. S2). Although the CArG-box motif might be the basic recognition sequence for MADS-box protein binding, other elements might be important. FLC binds to a CArG box 2.7 kb upstream of the translation start of SEP3 (CCCAAAATAGAAA); three other MADS-box flowering-time regulators, SOC1, SVP, and AGAMOUS-LIKE 24 (AGL24), bind to two different CArG-box motifs at 502 bp (CTAAATATGG) and 287 bp (CAATAATTGG) upstream of the translation start in the SEP3 gene (24), consistent with different specificities for the different MADS-box proteins.
Fig. 1.
Analysis of motifs overrepresented in the FLC binding sites. Motifs overrepresented in the FLC binding sites were analyzed by MEME software. Motifs were numbered 1–4 and ordered by the number of occurrences. The percentage of the motif in the binding sites was indicated to the right of each motif.
Besides the CArG-box motif, we found other sequence motifs in the FLC binding regions. A series of GA runs and TGGGCC, which occur in the promoter regions of many genes, are found in 39% and 14% of the FLC binding sites, respectively (Fig. 1); 19% of the binding peaks contain the G-box motif (CACGTG) (Fig. 1), a binding motif for basic helix–loop–helix (bHLH) and basic region leucine zipper (bZIP) transcription factors (25), raising the possibility that these genes might be coregulated by other transcription factors in combination with FLC.
FLC Binding Affects Gene Expression.
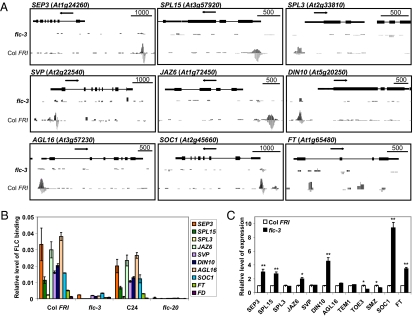
Data from ChIP-PCR experiments on chromatin from Col FRI and C24, as well as from the same ecotypes carrying the flc null mutants flc-3 and flc-20, paralleled the ChIP-seq findings (Fig. 2 A and B and Fig. S3). We selected 20 genes for validation of FLC binding; all correlated with the ChIP-seq data (Fig. 2 A and B and Fig. S3). Both the ChIP-PCR and the ChIP-seq analyses showed that there are different levels of FLC binding to different targets. SEP3, C-REPEAT/DRE BINDING FACTOR 1 (CBF1), JASMONATE-ZIM-DOMAIN PROTEIN 6 (JAZ6), and AGAMOUS-LIKE 16 (AGL16) are bound more strongly than SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 15 (SPL15), DARK INDUCIBLE 10 (DIN10), SVP, and SOC1, whereas FT is bound by FLC at only a low level in both assays (Fig. 2B). There is no significant binding peak at the FD locus in the ChIP-seq analysis; consistent with this finding, the ChIP-PCR result did not show any significant FLC binding at the FD promoter region in wild type relative to the flc mutant (Fig. 2B).
Fig. 2.
Validation and expression analyses of selected FLC target genes. (A) Binding profiles for selected target genes. The TAIR annotation of the genomic locus is shown at the top of each box. The profile in flc-3 is shown in the middle part of each box, and the profile in Col FRI is shown at the bottom of each box. Scale bars indicate sequence lengths, and arrows indicate gene orientation. The scale of the y axis is adjusted for each trace for visual clarity. (B) ChIP-PCR validation for selected FLC target genes. ChIP was done with FLC antiserum in Col FRI and C24 and compared with the corresponding flc mutant: flc-3 and flc-20. Input DNA was used as the reference in the PCR. Error bars represent SEs. (C) Transcriptional responses of the selected genes with loss of flc activity. Actin was used as the internal reference. The expression level of each gene in flc-3 was normalized to the level in Col FRI. Error bars represent SEs. Asterisks indicate a significant change (*P < 0.05; **P < 0.01; Student's t test).
To look at the relationship between gene expression and FLC binding, we checked the expression level of 40 candidate FLC target genes taken from the three levels of binding strengths in Col FRI 12-d-old whole seedlings compared with expression levels in flc-3. Of the 40 genes, 25 showed changed expression in the flc-3 mutant compared with the Col FRI wild type (selected genes are shown in Fig. 2C), whereas 15 remained unchanged in our seedling assay, which is consistent with other data showing that binding of transcription factors does not always induce changes in gene expression (26, 27). It is possible that the genes without expression changes could show changes at other stages of development or in particular tissues. Twenty of the 25 target genes showed increased expression (6 are shown in Fig. 2C: SEP3, SPL15, JAZ6, DIN10, SOC1, and FT). The five genes showing decreased expression in the mutant are SCHLAFMÜTZE (SMZ) and TARGET OF EAT 3 (TOE3), two targets in the AP2 transcription factor family (Fig. 2C); CBF1; CBF3; and At5G25900, involved in the GA response pathway (Fig. S4). This finding suggests that FLC, like AP1 (26), can regulate genes positively or negatively, although the major direction of regulation for AP1 is enhancement of expression, and FLC predominantly acts as a repressor.
FLC Targets Several Genes Involved in Floral Transition Pathways.
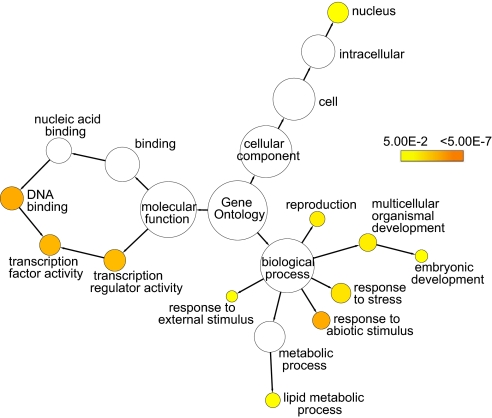
To further understand the nature of the FLC target genes, we analyzed the functional assignment in the Gene Ontology (GO) classifications (Fig. 3). Transcription factors are highly represented by three families, the MADS-box, AP2-EREBP and NAC families (Table S3), implying that the FLC protein is likely to be modulating the activity of a number of transcription factors that regulate important biological processes. The analysis showed that other FLC binding targets are genes that are concerned with response to stress as well as genes involved in reproductive and embryonic development (Fig. 3). In terms of cellular location, there is a high proportion of target genes with nuclear located products (Fig. 3).
Fig. 3.
GO categories enriched in the FLC target genes were analyzed with BiNGO. GOslim categories with significant enrichment in the dataset were highlighted in color, with different colors representing different levels of significance.
We found a number of FLC target genes, in addition to SOC1 and FT, that are involved in the regulation of the floral transition. FLC binds to the promoter of SVP (Fig. 2A), which delays flowering, but the expression of SVP was similar in Col FRI and flc-3 seedlings (Fig. 2C). SVP is expressed most highly in the shoot apical meristem, where it has been shown to interact with FLC (16). This finding may explain why we did not detect any changes in the expression of SVP in flc seedlings.
Two genes, SMZ and TOE3, both in the AP2 transcription factor family, are targeted by FLC. They act redundantly with other members of this family as flowering repressors (28, 29). The expression of TOE3 and SMZ was decreased in the flc mutant (Fig. 2C), suggesting that FLC positively regulates the expression of the genes to repress flowering. FLC also binds to the promoter of TEMPRANILLO1 (TEM1), an AP2-domain transcription factor acting as a flowering repressor (30). TEM1 regulates flowering time by repressing FT expression in leaves by binding to the 5′ UTR region of the FT gene (30).
FLC binds to the 3′ end of FRI (AT4G00650) in a genome region that could also be in the promoter region of the downstream gene AT4G00651. We could not detect any significant changes in the expression of either FRI or AT4G00651 in flc seedlings (Fig. S4), so it is not clear whether this FLC binding plays a role in regulating the FRI gene.
FLC Slows the Progression from Juvenile to Adult Phase.
Our binding data show that FLC targets two SPL genes, SPL15 and SPL3. Both genes are involved in promoting the juvenile-to-adult phase transition of the developing plant (31, 32). The binding signal at the SPL15 locus is much stronger than at the SPL3 locus (Fig. 2B), and SPL15 expression is significantly up-regulated in the flc mutant, indicating that FLC strongly represses the expression of SPL15 (Fig. 2C). There is no significant change of the expression of SPL3 in the flc mutant (Fig. 2C), which suggests that FLC regulates the juvenile-to-adult phase transition mainly by regulating the expression of SPL15.
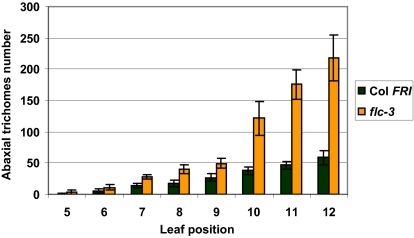
A morphological marker differentiating leaves of the juvenile phase from leaves formed during the adult growth phase is the presence of abaxial trichomes (33). The first leaves that display abaxial trichomes in Col FRI and flc-3 are the fifth or sixth leaves, but the number of trichomes increases more rapidly in flc-3 than in Col FRI. There are ∼40 trichomes on the eighth leaf in flc-3 but only ∼20 in Col FRI; on the 10th leaf, there are ∼120 abaxial trichomes in flc-3 but only ∼40 trichomes in Col FRI (Fig. 4). Leaf shape is also different between flc and wild type, with the mutant showing an earlier transition to the adult shape than wild type (34). These findings are consistent with a recent report from Willmann et al. (35) that FLC delays the progression from juvenile-to-adult phase. Our data indicate that the delay may be a consequence of the binding of FLC to the promoter of SPL15, repressing its activity.
Fig. 4.
Comparison of abaxial trichome numbers between Col FRI and flc-3. Abaxial trichome numbers for Col FRI and flc-3 were counted 4 wk after germination. Six plants were sampled for Col FRI, and 10 plants were sampled for flc-3. Error bars represent SEM.
FLC Is Involved in Floral Pattern Regulation.
Several FLC target genes, including SEP3, are involved in floral morphology regulation. FLC binds directly to the promoter region of SEP3, repressing its expression (Fig. 2) and consequently changing the activity of B and C class homeotic genes (24). This finding suggests that FLC may be involved in floral morphology processes, although there is no floral morphology phenotype in the flc mutant. FLC has an indirect control of floral morphology through its binding to SOC1 and SVP, which express proteins that, together with AGL24, bind to the promoter of SEP3, repressing its action (24). There is no floral morphology phenotype of the single or double mutants of soc1-2, agl24-1, and svp-41 (24). However, the triple mutant soc1-2 agl24-1 svp-41 shows severely abnormal flower organs (24) (Fig. 5F). We found that the flc-3 soc1-2 agl24-1 triple mutant also showed floral defects with abnormal reproductive organs (Fig. 5 B–D). These plants were bushy, with many leafy structures in the inflorescences (Fig. 5B). The flowers had no stamens or petals and showed carpelloid sepals (Fig. 5 C and D). At a later stage, most of the inflorescences developed into small floral buds that did not open and were infertile. Only a few flowers had normal floral organs and produced some seeds. We confirmed that there is no floral phenotype in the soc1-2 agl24-1 double mutant (24) (Fig. 5E), which suggests that the loss of FLC function in the triple mutant contributes to the floral defects. This result indicates that FLC—like SOC1, AGL24, and SVP—is involved in floral pattern regulation, probably through binding and regulation of expression of SEP3.
Fig. 5.
Floral defects of flc-3 soc1-2 agl24-1. (A) Inflorescence apex of wild-type Col. (B) Early stage of flc-3 soc1-2 agl24-1 inflorescence with many leafy structures. (C) Flowers of flc-3 soc1-2 agl24-1 with no petals or stamens. (D) Flower of flc-3 soc1-2 agl24-1 with carpelloid sepals. (E) Inflorescence of soc1-2 agl24-1 with normal floral organs. (F) Inflorescence of soc1-2 agl24-1 svp-41 with abnormal floral organs.
FLC and Environmental Response Pathways.
The GO analysis showed that FLC binding targets included many stress-responsive genes in the cold, light, and some hormone response pathways. The cold response genes CBF1 and CBF3 are regulated by SOC1 (36) as well as by FLC (Fig. S3 and Fig. S4).
There are also a large number of FLC target genes in hormone response pathways, such as the abscisic acid (ABA), jasmonate (JA), ethylene, and auxin pathways. Two genes in the JA signaling pathway, JAZ6 and JAZ9, showed increased expression in flc (Fig. 2C and Fig. S4). Hormone stress, including that generated by environmental challenge, frequently leads to alteration in flowering time, so, in these cases, FLC may play an indirect role in controlling flowering time.
In our data, there are FLC target genes categorized in the gibberellin (GA) pathway, including GA3, a multifunctional cytochrome P450 in GA biosynthesis (37), and the GA INSENSITIVE DWARF 1C (GID1C), which encodes an ortholog of the rice GA receptor gene OsGID1 (38). GA response mutants have been linked with FLC levels because of mutation in the flowering-time gene FPA (39). FLC may act with GA to ensure appropriate initiation of flowering.
It has been reported that FLC plays a role in controlling circadian rhythm (12); there are four major genes related to the circadian clock that are targets of FLC, CIRCADIAN 1 (CIR1), FIONA1 (FIO1), LHY/CCA1-like 1 (LCL1), and CONSTANT-LIKE 1 (COL1). Identification of these targets provides supporting evidence that FLC does have a role in regulating the circadian clock (11).
Discussion
FLC, a key regulatory gene for the initiation of flowering in Arabidopsis (1), codes for a protein that acts as a repressor of flowering through the regulation of the transcriptional activity of FT (8). FT protein is synthesized in leaves and transported to the apical meristem (40), where it interacts with FD to stimulate the activity of AP1, one of the major genes controlling the transition of the apical meristem from vegetative to reproductive mode. FLC also binds to SOC1 (8), which controls the second major gene concerned in the generation of the reproductive meristem, LEAFY. The actions of AP1 and LEAFY promote the development of the inflorescence meristem, which produces flowers. The FLC protein binds to the first intron of FT and to the promoter of SOC1, in each case inhibiting transcriptional activity (8).
FLC mRNA production is itself repressed by exposure of the germinating seed or of seedling growth stages to an extended period of low temperature. The key negative control of FLC transcriptional activity is mediated by the action of a repressor (VIN3) acting through the polycomb group protein complex (PRC2) (4, 41). This interaction of an environmental stimulus with transcriptional activities of a particular gene has provided fertile ground for the understanding of epigenetic activity in the control of patterns of gene action in plants.
In our earlier studies of FLC, we noted that it is expressed widely in the tissues of the plant and also throughout plant development, including the earliest stages of embryo development in the maturing seed (2, 42). We found that FLC activity is required at all stages of embryo development to provide effective regulation of the timing of flowering initiation and particularly of repression of flowering (42), which suggested that the FLC protein is likely to be interacting with genes that are subsequently involved in the formation of the reproductive phase of the apical meristem.
In our present work, we have shown that the FLC protein binds to some 500 sites. Among some of these target genes, we have been able to show a direct effect of FLC binding on the modulation of gene activity. Approximately 50% of the target genes we analyzed for regulation of mRNA production showed a level of transcription different to the level in the flc mutant. This proportion of regulated genes may be a significant underestimate of the number of genes in which FLC plays a regulatory role because we checked only at one stage of plant development; other transcription factors or cofactors produced at particular stages of development and in specific tissues may be required for gene activity.
FLC may exert its repressive function at other stages in the plant life cycle. The control of floral initiation by FLC during vegetative growth is not the only case where FLC acts at an early developmental stage. We have shown that FLC binds to SPL15, a gene that is directly involved in the transition of the developing plant from the juvenile phase producing vegetative structures to the first signs of an adult phase, which ultimately is capable of producing the floral structures (31).
FLC is concerned not only with the initiation of flowering but also with the development of floral architecture through its binding to homeotic genes producing the different floral organs. We have shown that FLC binds to SEP3 and regulates its expression, activating AP3 and AGAMOUS (AG), two of the genes involved in the “ABC” patterning of the whorls of floral organs (24).
Like most MADS-box genes, FLC very likely binds as a dimer to a particular recognition motif (9), the CArG box. In some cases, we know that other MADS-box proteins also bind to the same target gene. We determined the consensus sequence of the FLC CArG box to be CCAAAAAT(A/G)G and noted a highly conserved terminal AAA triplet adjacent to the terminal GG of the CArG box. Two other MADS-box proteins, AGL15 and SEP3, have been reported to have a consensus sequence similar to FLC (27, 43); AGL15 sites frequently have an AAA triplet adjacent to the CArG box and, less frequently, to SEP3 (Fig. S2). The highest probability of binding of FLC to its target genes occurs in those sequences that have the adjacent AAA triplet and where their sequence most closely fits the consensus. However, even for those target sites in the lowest stringency class of binding, the presence of the adjacent AAA triplet enhanced the significance of the binding site over random sequences. In the case of FLC, the AAA triplet appears to expand the length of the functional CArG box.
Many members of the MADS-box gene family have been linked to regulation of genes in metabolic and developmental pathways that relate to the reproductive phase of the life cycle; FLC also fits this pattern. It has a unique position in that, in contrast to other MADS-box genes where there are detailed analyses of their action in regulatory activities, FLC is actively transcribed in most tissues in the plant and from some of the earliest developmental stages of the embryo onwards, and its usual regulatory control is repression of transcriptional activity. Other MADS-box proteins, such as AP1 and SEP3, have transcriptional activity limited to the terminal apex after the floral transition, producing reproductive tissues with regulatory actions being promotive rather than repressive. Also, 28% of FLC targets are also bound by AP1 (26) and 36% by SEP3 (43). SEP3 is one example where FLC binding is repressive and AP1 binding is promotive, which may be indicative of a balancing mechanism of regulatory control where there are both repressive and promotive actions by the different MADS-box proteins.
The fact that FLC is involved in regulating genes in many different gene pathways through the developmental life of the plant could mean that its actions do not always relate to the reproductive phase. However, many pathways, from the earliest activities of FLC in the juvenile-to-adult transition and in response to abiotic stress and hormonal action of various kinds, do relate to the vegetative to reproductive transition. Some FLC binding, such as to genes in the cold acclimation pathway, appear unrelated to the reproductive transition.
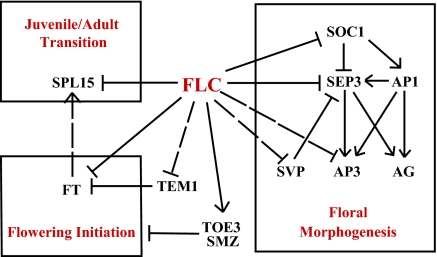
The large number of FLC targets and their locations in many different pathways emphasize the importance of the reproductive transition and the need to have that transition occur at the most appropriate time of the life cycle. The plant has a robust and exquisite set of controls ensuring that the reproductive phase has an optimal chance of success. This strategy is supported by the multiple pathways that regulate the switch to reproductive development; FLC appears to regulate genes involved in all of these pathways (Fig. 6).
Fig. 6.
FLC functions in plant development. FLC is involved in various aspects of plant development, including the juvenile-to-adult transition, flowering initiation, and floral morphogenesis. Arrows indicate gene activation, and blunted lines indicate repression. For FLC binding targets, solid lines indicate confirmed expression changes in the flc seedlings, and broken lines indicate either no expression changes or was not checked in the flc seedlings.
Materials and Methods
Plant Materials, Growing Conditions, and Genotyping.
Arabidopsis thaliana lines Col FRI, C24, flc-3 (in Col FRI), and flc-20 (in C24) were grown on MS medium (Murashige and Skoog medium supplemented with 10 g/L−1 sucrose and 8 g/L−1 agar, pH 5.7) under long-day conditions (16-h light/8-h dark cycle) at 23 °C. Mutants for phenotypic analysis, flc-3, soc1-2, svp-41, and agl24-1, are in Col background and were grown under standard greenhouse conditions (16-h light/8-h dark cycle at 23 °C). The flc-3 soc1-2 agl24-1 triple mutant was obtained from the segregation of flc-3 × soc1-2 agl24-1 svp-41 (kindly provided by Hao Yu, National University of Singapore, Singapore).
ChIP-Seq and Data Analysis.
ChIP was performed as described previously (44) with 28 g of 12-d-old Col FRI or flc-3 (Col FRI) whole seedlings. FLC antiserum was raised against the FLC protein without the conserved MADS domain (7, 9). Then, 32 ng and 20 ng of ChIP DNA were isolated from Col FRI and flc-3 (Col FRI), respectively, and sequenced with an Illumina Genome Analyzer (GAII) on one lane with indexing.
The sequencing reads were mapped to the Arabidopsis genome (TAIR9 build), allowing one mismatch at any position or two mismatches at low quality score positions. The regions enriched for binding sites were first identified by the QuEST algorithm using three levels of stringency parameters and then compared with the regions identified by MACS algorithms. Peaks in MACS but not in QuEST were checked manually and added to the final list.
Motif Analysis.
For motif analysis, we generated a set of peak-associated sequences using QuEST software. MEME software (version 4.5.0) (21) was applied to yield overrepresented motifs in the dataset. For the distribution of a single motif among the sequences, motifs 1 and 2 used 0 or 1 per sequence, whereas motifs 3 and 4 used any number of repetitions.
GO Analysis.
GO enrichment analysis was performed by using the BiNGO 2.3 plug-in (45) in Cytoscape 2.6.3 (46) with the GOslim_plants dataset. To test for enrichment, a hypergeometric test was conducted, and the Benjamini and Hochberg false-discovery rate was calculated. The network of the enriched categories was presented.
Supplementary Material
Acknowledgments
We thank Masumi Robertson for providing the FLC antiserum; Jean Finnegan, Ian Greaves, and Candice Sheldon for discussions; and Hao Yu for providing the mutant seeds.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession no. SRP005412). The track files are available from the authors upon request.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103175108/-/DCSupplemental.
References
- 1.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheldon CC, et al. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastow R, et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 4.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 5.Dennis ES, Peacock WJ. Epigenetic regulation of flowering. Curr Opin Plant Biol. 2007;10:520–527. doi: 10.1016/j.pbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Johanson U, et al. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 7.Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci USA. 2000;97:3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Searle I, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- 10.Chiang GC, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106:11661–11666. doi: 10.1073/pnas.0901367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170:387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards KD, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spensley M, et al. Evolutionarily conserved regulatory motifs in the promoter of the Arabidopsis clock gene LATE ELONGATED HYPOCOTYL. Plant Cell. 2009;21:2606–2623. doi: 10.1105/tpc.109.069898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Folter S, et al. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Folter S, Angenent GC. trans meets cis in MADS science. Trends Plant Sci. 2006;11:224–231. doi: 10.1016/j.tplants.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Li D, et al. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell. 2008;15:110–120. doi: 10.1016/j.devcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson M, Helliwell CA, Dennis ES. Post-translational modifications of the endogenous and transgenic FLC protein in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1859–1866. doi: 10.1093/pcp/pcn167. [DOI] [PubMed] [Google Scholar]
- 19.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 22.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 23.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Xi W, Shen L, Tan C, Yu H. Regulation of floral patterning by flowering time genes. Dev Cell. 2009;16:711–722. doi: 10.1016/j.devcel.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Menkens AE, Cashmore AR. Isolation and characterization of a fourth Arabidopsis thaliana G-box-binding factor, which has similarities to Fos oncoprotein. Proc Natl Acad Sci USA. 1994;91:2522–2526. doi: 10.1073/pnas.91.7.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann K, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell. 2009;21:2563–2577. doi: 10.1105/tpc.109.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009;7:e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid M, et al. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 30.Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- 34.Mentzer L, Yee T, Wang TY, Himelblau E. FLOWERING LOCUS C influences the timing of shoot maturation in Arabidopsis thaliana. Genesis. 2010;48:680–683. doi: 10.1002/dvg.20683. [DOI] [PubMed] [Google Scholar]
- 35.Willmann MR, Poethig RS. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development. 2011;138:677–685. doi: 10.1242/dev.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo E, et al. Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell. 2009;21:3185–3197. doi: 10.1105/tpc.108.063883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helliwell CA, Poole A, Peacock WJ, Dennis ES. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima M, et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- 39.Meier C, et al. Gibberellin response mutants identified by luciferase imaging. Plant J. 2001;25:509–519. doi: 10.1046/j.1365-313x.2001.00980.x. [DOI] [PubMed] [Google Scholar]
- 40.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 41.Wood CC, et al. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheldon CC, et al. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci USA. 2008;105:2214–2219. doi: 10.1073/pnas.0711453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaufmann K, et al. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7:e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng W, et al. Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol. 2007;143:1660–1668. doi: 10.1104/pp.106.095521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 46.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.