Abstract
The identification of genes that participate in melanomagenesis should suggest strategies for developing therapeutic modalities. We used a public array comparative genomic hybridization (CGH) database and real-time quantitative PCR (qPCR) analyses to identify the AMP kinase (AMPK)-related kinase NUAK2 as a candidate gene for melanomagenesis, and we analyzed its functions in melanoma cells. Our analyses had identified a locus at 1q32 where genomic gain is strongly associated with tumor thickness, and we used real-time qPCR analyses and regression analyses to identify NUAK2 as a candidate gene at that locus. Associations of relapse-free survival and overall survival of 92 primary melanoma patients with NUAK2 expression measured using immunohistochemistry were investigated using Kaplan–Meier curves, log rank tests, and Cox regression models. Knockdown of NUAK2 induces senescence and reduces S-phase, decreases migration, and down-regulates expression of mammalian target of rapamycin (mTOR). In vivo analysis demonstrated that knockdown of NUAK2 suppresses melanoma tumor growth in mice. Survival analysis showed that the risk of relapse is greater in acral melanoma patients with high levels of NUAK2 expression than in acral melanoma patients with low levels of NUAK2 expression (hazard ratio = 3.88; 95% confidence interval = 1.44–10.50; P = 0.0075). These data demonstrate that NUAK2 expression is significantly associated with the oncogenic features of melanoma cells and with the survival of acral melanoma patients. NUAK2 may provide a drug target to suppress melanoma progression. This study further supports the importance of NUAK2 in cancer development and tumor progression, while AMPK has antioncogenic properties.
Keywords: sucrose nonfermenting-like kinase, chromosome 1q
The identification of genes that participate in melanomagenesis should suggest strategies for developing effective therapeutic modalities (1, 2). For more than three decades, cytogenetic analyses have been used to identify genes that have an impact on tumorigenesis (3). However, those cytogenetic analyses have had limited success in elucidating such genes in solid tumors such as malignant melanoma because of the complexity of chromosomal and genomic aberrations (4, 5). Recent advances in microarray technologies, including array-based comparative genomic hybridization (CGH), have allowed the genomic characterization of cancer cells in solid tumors (6–10). Gains of chromosome 1q are frequent events in many types of cancers, including breast cancer, medulloblastoma, retinoblastoma, hepatocellular carcinoma, non-small cell lung carcinoma, cervical cancer, and others (11–16). Cytogenetic studies have revealed that melanoma cells have several characteristic abnormalities including a recurring translocation involving chromosomes 1 and 6 that results in gains of chromosome 1q (17, 18). We previously reported a CGH analysis which showed that gains of chromosomes 1q and 6p correlate strongly with the clinical outcome of patients with primary cutaneous melanomas (19).
NUAK2 [also known as “sucrose nonfermenting (SNF1)-like kinase,” SNARK], which resides at 1q32, is a member of the SNF1/AMP kinase (AMPK) family (serine/threonine kinases) that is regulated by the putative tumor suppressor LKB1 (20–23) and also by death receptor signaling through NF-κB (21). AMP-related kinases function as critical sensors coupling cellular energy status to cell growth and proliferation by modulating the cell-cycle machinery and, when deregulated, result in cancer development and tumor progression in several cancers of different cell lineages (24–26). In melanomas, the LKB1–AMPK signaling pathway is deregulated by oncogenic B-RAF and participates in cancer development (27, 28). However, the exact mechanisms by which AMP-related kinases participate in cancer development and tumor progression remain unknown.
In this study, we analyzed data from a public array CGH database and used real-time quantitative PCR (qPCR) analyses to identify NUAK2 as a candidate gene for melanomagenesis. Additional experiments demonstrate that knockdown of NUAK2 induces cellular senescence and decreases the migration of melanoma cells that harbor NUAK2 amplification. We report that the expression level of NUAK2 is significantly associated with the relapse-free survival of acral melanoma patients. Our study highlights the crucial role of NUAK2 in cancer development and in tumor progression, whereas AMPK has antioncogenic properties.
Results
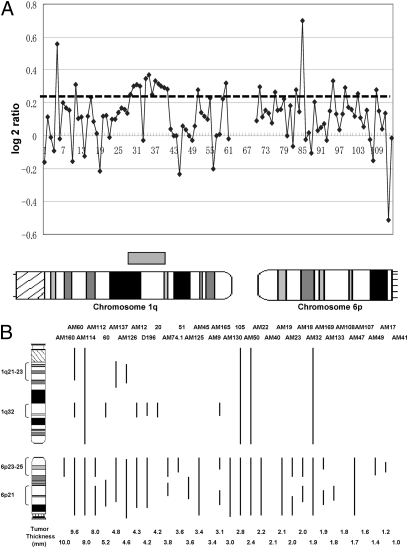
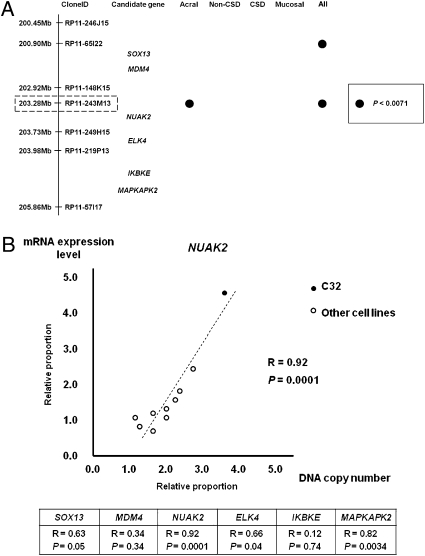
Previous cytogenetic studies reported that gains of chromosomes 1q and 6p are frequent cytogenetic aberrations in primary cutaneous melanomas (17–19). To identify genes that participate in melanomagenesis within those loci, we used a public array database (http://www.ncbi.nlm.nih.gov/geo/; Series GSE2631) to analyze correlations between genomic loci within chromosomes 1q and 6p and tumor thickness. We initially focused on examining gains at the chromosomal level (Fig. 1A). Data from 33 cases of acral melanomas, 34 cases of nonchronic sun-induced damage (non-CSD) melanomas, 28 cases of CSD melanomas, and 15 cases of mucosal melanomas were examined. The four most frequent loci (1q21–23, 1q32, 6p23–25, and 6p21) with gains were identified (Fig. 1B). Analyses of correlations of those loci showed that 1q32 correlated with tumor thickness in acral melanomas (P = 0.017) and in all melanomas (P = 0.003) (SI Appendix, Table S1). Those analyses indicate that, of the loci identified on chromosomes 1q and 6p, 1q32 has the strongest statistical correlation with tumor thickness. To characterize the significantly correlated clone further, we focused on genomic loci from 193.52 Mb (D1S2794, clone ID: RP11-154A22) to 208.18 Mb (D1S205, clone ID: RP11-104A2) within the 1q32 locus, because that locus includes most of the genomic gained clones within the 1q32 locus. The genomic clones RP11-65I22 and RP11-243M13 have statistically significant correlations in all melanomas (P < 0.0071), and the genomic clone RP11-243M13 has the strongest statistical significance in acral melanomas (P = 0.0029). Interestingly, correlations existed in only one subset of melanomas, i.e., acral melanomas (Fig. 2A and SI Appendix, Table S2). Thus, analyses of the public array CGH database suggest that a putative oncogene resides in a genomic locus around clone RP11-243M13.
Fig. 1.
Identification of genomic gains associated with tumor thickness. (A) Analysis of chromosomal gains of 1q and 6p from the array CGH database in acral melanomas (Case No. 20). Each spot represents the log2 ratio value of each clone from the array CGH database. Thresholds for gain are shown with the horizontal dashed line at log2 ratios of 0.25. The x-axis represents each clone on chromosomes 1q and 6p; an ideogram of the chromosomes corresponding to each clone is shown below the panel. The gray box above the chromosomal ideogram represents the chromosomal gain of 1q in this case. (B) Chromosomal gains of 1q and 6p in 33 acral melanomas. Chromosomal gains in each case are depicted by the vertical lines to the right of the chromosomal ideogram. The number of each case is shown above the corresponding vertical line. The tumor thickness of each case is shown below the corresponding vertical line. The four regions in which chromosomal gains of 1q and 6p are most frequent (1q21–23, 1q32, 6p23–25, and 6p21) are depicted to the left of the chromosome ideogram.
Fig. 2.
Identification of candidate genes within the 1q32 locus. (A) Six candidate genes and correlations between genomic clones and tumor thickness in each subset of melanoma. The locus spanning ∼5.0 Mb, with the correlated clone most strongly correlated with tumor thickness (RP11-243M13) in the center, is shown. Six candidate genes are located in this locus. Filled circles represent genomic clones with P values <0.0071 in each subset of melanoma. (B) (Lower) Results of regression analyses of mRNA expression levels and DNA copy numbers obtained by real-time qPCR analyses of six candidate oncogenes within the 1q32 locus are shown in the data table. (Upper) Regression analysis of NUAK2 that has the strongest correlation among the six candidate oncogenes. Each circle represents mRNA expression levels and DNA copy numbers for each cell line. The filled circle indicates mRNA level and DNA copy number of C32 melanoma cells that have a gain within the 1q32 locus; the open circles represent nine other melanoma cell lines listed in SI Appendix, Table S3.
Genomic gain or amplification of oncogenes increases DNA copy number and can up-regulate transcriptional levels of those mRNAs. Thus, we hypothesized that oncogenes might have a strong correlation between DNA copy number and mRNA expression levels. We examined six candidate oncogenes [SOX13, MDM4, NUAK2, ELK4, IKBKE, and MAPKAPK2] within the 1q32 locus spanning ∼5.0 Mb from 200.45 Mb (RP11-246J15) to 205.86 Mb (RP11-57117), where the most strongly correlated clone, RP11-243M13, resided in the center (Fig. 2A). We obtained DNA copy numbers and mRNA expression levels using qPCR in 10 melanoma cell lines (SI Appendix, Table S3). Regression analyses revealed that the NUAK2 DNA copy number had the strongest correlation with increases in its mRNA expression level (P = 0.0001), and those for ELK4 and MAPKAPK2 also were significantly correlated (P = 0.04 and P = 0.0034, respectively) (Fig. 2B). After confirming the decreased protein levels of NUAK2, ELK4, and MAPKAPK2 using siRNA SMARTpools (Thermo Scientific) (SI Appendix, Fig. S1), we found significantly reduced cell numbers following the knockdown of NUAK2 (P = 0.04) but no significant effects following the knockdown of ELK4 or MAPKAPK2 (P = 0.20 and P = 0.80, respectively). Thus, NUAK2 is the most promising gene within the 1q32 locus.
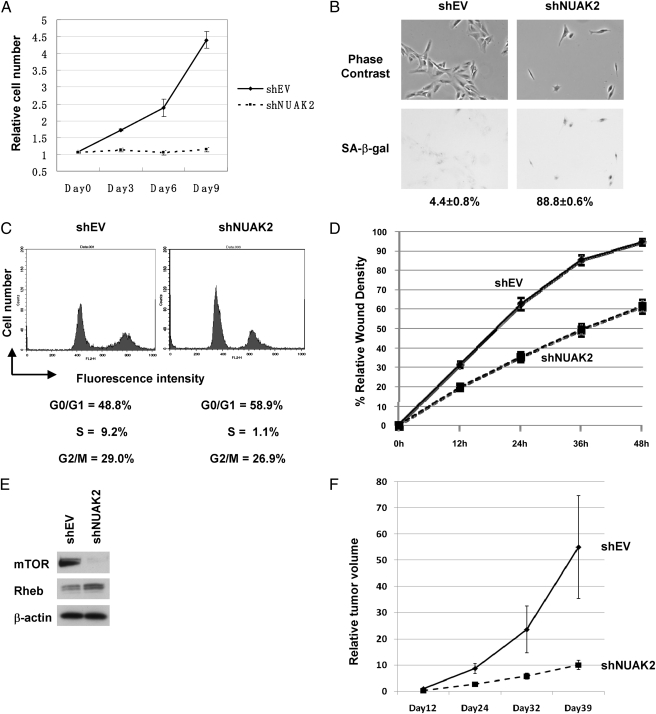
To characterize further the effect of NUAK2 on cell proliferation and migration, we used C32 melanoma cells, which harbor NUAK2 amplification. Knockdown of NUAK2 using a lentivirus containing an shRNA targeting NUAK2 (shNUAK2) caused a significant decrease in cell number (Fig. 3A). In addition, shNUAK2 markedly increased cellular senescence [shown by positive staining with senescence-associated β-galactosidase (SA-β-gal) (Fig. 3B)]. Further analysis revealed that this increased senescence resulted from a significantly decreased S-phase population of cells and decreased levels of cyclin D1, cyclin D3, and cyclin-dependent kinase2 (Fig. 3C and SI Appendix, Fig. S2A). However, knockdown by shNUAK2 had only a marginal effect on apoptosis (SI Appendix, Fig. S2 B and C). Knockdown of NUAK2 significantly impaired the migration of C32 melanoma cells (Fig. 3D and SI Appendix, Fig. S3 A and B). Next, we examined the downstream pathway of NUAK2 and found that knockdown of NUAK2 significantly decreased the expression of mammalian target of rapamycin (mTOR) (Fig. 3E). To validate the effect of NUAK2 in vivo, we examined the tumorigenicity of melanomas in nude mice, where tumor growth was significantly suppressed by knockdown of NUAK2 (Fig. 3F). We also examined the effect of NUAK2 on various melanoma cell lines after characterization of NUAK2 expression and found that NUAK2 dramatically reduces apoptosis in mel18 and in SKMel28 melanoma cells (SI Appendix, Fig. S4 A and B). Taken together, these in vitro and in vivo studies suggest that knockdown of NUAK2 negatively affects the expression of mTOR and that NUAK2 has a significant impact on the proliferation and migration of melanoma cells.
Fig. 3.
Knockdown of NUAK2 in C32 melanoma cells results in reduced proliferation and migration. (A) Cell number analysis of C32 melanoma cells with knockdown of NUAK2. Knockdown of NUAK2 dramatically reduced cell proliferation. (B) SA-β-gal staining in C32 melanoma cells. Images obtained using phase-contrast (Upper) and bright-field (Lower) microscopy are shown. Staining indicates senescent cells. Percentages of positive cells and SE values are given below the images. shEV, empty vector. (C) Cell-cycle profiles of C32 melanoma cells with knockdown of NUAK2. A significant decrease of the S-phase population in cell-cycle profiles is observed. (D) Wound-healing assay using C32 melanoma cells with knockdown of NUAK2. Knockdown of NUAK2 significantly decreased the migration of C32 melanoma cells. (E) Knockdown of NUAK2 by shNUAK2 significantly decreased the expression of mTOR. (F) Tumor growth of C32 melanoma cells infected with shNUAK2 relative to the empty vector. Tumor growth was significantly suppressed by infection of shNUAK2.
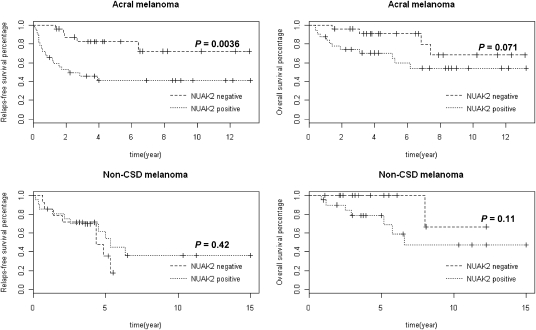
To explore the effects of NUAK2 in the clinical setting, we analyzed the correlations between levels of NUAK2 expression using immunohistochemistry (SI Appendix, Fig. S5 A and B) and clinical parameters, as well as clinical outcome using both univariate and multivariate analyses. Mann–Whitney tests and χ2 tests were performed on 57 acral melanomas and 35 non-CSD melanomas (SI Appendix, Table S4), and the results showed that the NUAK2 expression level is associated with tumor thickness and ulceration in acral melanomas (P = 0.0026 and P = 0.017, respectively) (SI Appendix, Table S5). Log rank tests were used to analyze acral melanomas and non-CSD melanomas to explore differences between the NUAK2-positive and NUAK2-negative groups with respect to relapse-free survival and overall survival (Fig. 4). The relapse-free survival of acral melanoma patients differed significantly between the NUAK2-positive and NUAK2-negative groups (P = 0.0036). Kaplan–Meier survival analysis estimated that the lower-quartile relapse-free survival time in the NUAK2-negative group (77 mo) is longer than in the NUAK2-positive group (6.5 mo). The median survival time (the time when 50% of acral melanoma patients relapsed) was not reached in the NUAK2-negative group. Multivariate Cox regression analysis showed that acral melanoma patients with high levels of NUAK2 expression were more likely to relapse than were acral melanoma patients with low levels of NUAK2 expression (hazard ratio = 3.88; 95% confidence interval = 1.44–10.50; P = 0.0075) (Table 1). The binary status of NUAK2 expression was used as the classifier to classify the acral melanoma patients into either relapse or not-relapse categories. The sensitivity and the specificity of the classifier at three time points (2 y, 3 y, and 4 y) were calculated using R package survivalROC (http://cran.r-project.org/web/packages/survivalROC/index.html, refs. 29 and 30). At the 4-y time point, the sensitivity of the classifier is 80%, and the specificity of the classifier is 60% (SI Appendix, Table S6). Acral melanoma patients also were divided into two sample sets based on tumor thickness; one set consisted of the thinner pTis+pT1/pT2+pT3 sample set, and the other set consisted of the thicker pT4 sample set (defined in SI Appendix, Materials and Methods). A log rank test showed that the NUAK2 expression level is correlated significantly with relapse-free survival in patients with thicker acral melanomas (SI Appendix, Fig. S6; P = 0.048). Thus, these findings suggest that NUAK2 is the gene at chromosome 1q32 that is significantly associated with the clinical survival of acral melanoma patients, particularly those patients with thicker acral melanomas.
Fig. 4.
High expression levels of NUAK2 and its significance on clinical outcome. Kaplan–Meier survival analyses of high expression levels of NUAK2 for relapse-free survival in 57 cases of acral melanoma (Upper Left) and in 35 cases of non-CSD melanoma (Lower Left) and for overall survival in 57 cases of acral melanoma (Upper Right) and in 35 cases of non-CSD melanoma (Lower Right). P values are indicated.
Table 1.
Multivariate Cox regression analysis of high expression levels of NUAK2 in relapse-free survival of acral melanoma patients
| Variable* | Hazard ratio | 95% confidence interval | P value |
| NUAK2 | 3.88 | 1.44–10.50 | 0.0075† |
| Sex | 0.87 | 0.38–1.97 | 0.73 |
| Age | 1.31 | 0.55–3.11 | 0.55 |
*Coding of variables: High expression of NUAK2 was coded as 1 = negative; 2 = positive. Sex was coded as 1 = male; 2 = female. Age was coded as 1 = ≥60 y; 2 = >60 y.
†P < 0.05.
Discussion
Gains and/or amplifications of the long arm of chromosome 1 are among the frequent chromosomal abnormalities in a wide variety of cancers, and gains in the region spanning 1q31–1q32 are the most frequent abnormalities in these loci. The observation that the gain of the 1q32 locus is shared by a variety of cancers emphasizes the importance of this locus for cancer development and tumor progression in general (12). Public databases such as GeneCards (www.genecards.org) show that NUAK2 is highly expressed in various cancers, including cancers of lymphoid tissues, lung, and breast. In this study, we report that high expression levels of NUAK2, which resides at 1q32, have a significant association with clinical relapse in acral melanoma patients, particularly in patients who have thicker acral melanomas. To take advantage of the public array CGH database, we identified a locus that correlates with tumor thickness on chromosomes 1q and 6p and used real-time qPCR analyses to identify NUAK2 as the most promising gene within those loci. Knockdown studies in vitro and in vivo using a lentiviral vector containing an shRNA targeting NUAK2 showed that NUAK2 has a significant impact on the proliferation and migration of melanoma cells that harbor NUAK2 amplification and that the knockdown of NUAK2 down-regulates the expression of mTOR. We used immunohistochemistry and survival analyses to show that acral melanoma patients with high levels of NUAK2 expression are more likely to relapse than are acral melanoma patients with low levels of NUAK2 expression. This result suggests that the gain of NUAK2 at the genomic level induces high expression levels of NUAK2 at the protein level and that high expression of this gene participates in melanomagenesis and the poor relapse-free survival of acral melanoma patients, particularly patients with thicker acral melanomas.
Our study reveals that high expression levels of NUAK2 correlate inversely with clinical outcome, particularly for patients with acral melanomas, and have strikingly different effects in acral and non-CSD melanomas. The microenvironment that affects the differentiation and/or the proliferation of melanocytes in acral areas might be a factor in the discrepancy in the clinical outcomes of patients with acral melanomas and those with non-CSD melanomas. We have shown previously that dickkopf homolog 1 secreted by dermal fibroblasts in the skin affects the differentiation and proliferation of melanocytes in acral areas by mesenchymal–epithelial interactions (31) and that it does so by modulating the Wnt signaling pathway (32). These activated signaling pathways and paracrine factors might participate in the poorer relapse-free survival of patients with acral melanomas. Our study also reveals that high expression levels of NUAK2 correlate inversely with relapse-free survival and have a striking difference between relapse-free and overall survival. Recent studies revealed that NUAK2 participates in cell detachment by regulating the phosphorylation of MLC2 through MYPT1, which is a specific substrate for NUAK2 (23, 33). The mechanisms through which NUAK2 regulates cell migration and invasion need to be explored further, and these effects may contribute to the differences in relapse-free and overall survival.
AMPK-related kinases are modulated by the AMP:ATP ratio and function as sensors in cellular metabolism (24). These sensors are closely connected with cell proliferation and motility (21). However, AMPK-related kinases have different impacts on tumorigenesis; e.g., AMPKα1 and AMPKα2 function as tumor suppressors, whereas NUAK1/AMPK-related kinase5 functions as an oncogene. NUAK2 itself has contradictory functions on tumorigenesis, acting as a tumor suppressor or as an oncogene. One study showed that NUAK2-deficient mice develop colorectal tumors, but other studies have reported that NUAK2 has antiapoptotic properties and promotes motility, invasiveness and cell-to-cell detachment (34). Some important oncogenes in melanomas, such as MITF, have a similar dual role in melanomagenesis and require expression within a limited range to act as an oncogene (35). Previous studies have indicated that NUAK2 might have complicated functions as an oncogene, and tightly regulated gain-of-function studies would be quite valuable to unravel the various functions of NUAK2. Our data also reveal that high expression levels of NUAK2 correlate inversely with clinical outcome, particularly for melanomas arising from palmoplantar-subungual-mucosal areas. Interestingly, lentigines of Peutz–Jeghers syndrome, which is caused by mutation of LKB1, are distributed in palmoplantar-subungual-mucosal areas, and melanomas rarely occur in Peutz–Jeghers syndrome (36, 37). The activity of NUAK2 is reduced in LKB1-deficient cells, and NUAK2-deficient mice develop colorectal tumors (20, 34), but there has been no previous report of its involvement with melanomas. Taken together, these clinical, in vivo, and in vitro observations suggest that the effects of NUAK2 on tumorigenesis might differ depending on cell lineage. Although the exact upstream regulation of NUAK2 still is poorly understood, LKB1 might have a pivotal role in regulating NUAK2, and death receptor signaling also might have an important role in inducing NUAK2 activity to facilitate migration and antiapoptotic properties. Our results suggest that the deregulation of NUAK2 has profound effects on cancer development and tumor progression of melanocytic cells.
In sum, high expression levels of NUAK2 reflect a distinct molecular subset of melanomas with poor clinical outcome, which metastasize quickly and produce ulcerative and thicker tumors. Our data suggest that high expression levels of NUAK2 could have a potential prognostic value for patients with acral melanomas inferred from the detailed association and survival analyses, although further studies are required to establish its medical utility. NUAK2, which resides at the 1q32 locus that is gained and/or amplified in a wide variety of cancers, promotes cancer development by affecting the proliferation and/or migration of cancer cells. Our study emphasizes the crucial role of AMPK-related kinases in cancer development and tumor progression and also suggests that blocking NUAK2 and/or its downstream pathway could be an effective strategy to target acral melanomas.
Experimental Procedures
Details of some of experimental procedures, including statistical analysis of the public database; quantitation of DNA copy numbers and mRNA expression levels; vectors, siRNA transfection, and lentiviral infection; and in vitro assays, are available in SI Appendix.
Tumor Specimens and Cell Lines.
We used clinical specimens of primary cutaneous melanomas. This study was approved by each institution. The clinical features of each case are summarized in SI Appendix, Table S5. Full details are published in SI Appendix.
Immunohistochemical Analyses.
Immunohistochemistry was performed after confirmation of specificities of antibodies. Eccrine and/or sebaceous glands were used as internal positive controls (SI Appendix, Fig. S7 A and B). Full details and the list of antibodies used are published in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. K. Miura (pathologist), T. Yamanaka (dermatologist), and K. Nishida, K. Sakamaki, A. Tamura, N. Ando, and C. Miyagishi (Pathology Staff Members) for their assistance. We also thank D. R. Lowy for valuable comments and suggestions about the study and Drs. Stuart Yuspa and Luowei Li for help with the wound-healing assay. We are grateful to Dr. Richard Simon for his advice and suggestions for improving and clarifying the manuscript. This work was supported in part by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007694108/-/DCSupplemental.
References
- 1.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Treatment for chronic myelogenous leukemia: The long road to imatinib. J Clin Invest. 2007;117:2036–2043. doi: 10.1172/JCI31691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaneko Y, et al. Correlation of karyotype with clinical features in acute lymphoblastic leukemia. Cancer Res. 1982;42:2918–2929. [PubMed] [Google Scholar]
- 4.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 5.Mertens F, Johansson B, Höglund M, Mitelman F. Chromosomal imbalance maps of malignant solid tumors: A cytogenetic survey of 3185 neoplasms. Cancer Res. 1997;57:2765–2780. [PubMed] [Google Scholar]
- 6.Pinkel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 7.Bittner M, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 8.Dissanayake SK, et al. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282:17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr KM, Bittner M, Trent JM. Gene-expression profiling in human cutaneous melanoma. Oncogene. 2003;22:3076–3080. doi: 10.1038/sj.onc.1206448. [DOI] [PubMed] [Google Scholar]
- 10.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 11.Orsetti B, et al. Genetic profiling of chromosome 1 in breast cancer: Mapping of regions of gains and losses and identification of candidate genes on 1q. Br J Cancer. 2006;95:1439–1447. doi: 10.1038/sj.bjc.6603433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–4753. doi: 10.1038/sj.onc.1208641. [DOI] [PubMed] [Google Scholar]
- 13.Gratias S, et al. Genomic gains on chromosome 1q in retinoblastoma: Consequences on gene expression and association with clinical manifestation. Int J Cancer. 2005;116:555–563. doi: 10.1002/ijc.21051. [DOI] [PubMed] [Google Scholar]
- 14.Kim TM, et al. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer. 2008;123:2808–2815. doi: 10.1002/ijc.23901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai AL, et al. Recurrent chromosomal imbalances in nonsmall cell lung carcinoma: The association between 1q amplification and tumor recurrence. Cancer. 2004;100:1918–1927. doi: 10.1002/cncr.20190. [DOI] [PubMed] [Google Scholar]
- 16.Wilting SM, et al. Integrated genomic and transcriptional profiling identifies chromosomal loci with altered gene expression in cervical cancer. Genes Chromosomes Cancer. 2008;47:890–905. doi: 10.1002/gcc.20590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trent JM, Thompson FH, Meyskens FL., Jr Identification of a recurring translocation site involving chromosome 6 in human malignant melanoma. Cancer Res. 1989;49:420–423. [PubMed] [Google Scholar]
- 18.Thompson FH, et al. Cytogenetics of 158 patients with regional or disseminated melanoma. Subset analysis of near-diploid and simple karyotypes. Cancer Genet Cytogenet. 1995;83:93–104. doi: 10.1016/0165-4608(95)00057-v. [DOI] [PubMed] [Google Scholar]
- 19.Namiki T, et al. Genomic alterations in primary cutaneous melanomas detected by metaphase comparative genomic hybridization with laser capture or manual microdissection: 6p gains may predict poor outcome. Cancer Genet Cytogenet. 2005;157:1–11. doi: 10.1016/j.cancergencyto.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Lizcano JM, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legembre P, Schickel R, Barnhart BC, Peter ME. Identification of SNF1/AMP kinase-related kinase as an NF-kappaB-regulated anti-apoptotic kinase involved in CD95-induced motility and invasiveness. J Biol Chem. 2004;279:46742–46747. doi: 10.1074/jbc.M404334200. [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre DL, et al. Identification and characterization of a novel sucrose-non-fermenting protein kinase/AMP-activated protein kinase-related protein kinase, SNARK. Biochem J. 2001;355:297–305. doi: 10.1042/0264-6021:3550297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zagórska A, et al. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci Signal. 2010 doi: 10.1126/scisignal.2000616. 10.1126/scisignal.2000616. [DOI] [PubMed] [Google Scholar]
- 24.Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 25.Martin MJ, Carling D, Marais R. Taking the stress out of melanoma. Cancer Cell. 2009;15:163–164. doi: 10.1016/j.ccr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Ashrafian H. Cancer's sweet tooth: The Janus effect of glucose metabolism in tumorigenesis. Lancet. 2006;367:618–621. doi: 10.1016/S0140-6736(06)68228-7. [DOI] [PubMed] [Google Scholar]
- 27.Zheng B, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell. 2009;33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteve-Puig R, Canals F, Colomé N, Merlino G, Recio JA. Uncoupling of the LKB1-AMPKalpha energy sensor pathway by growth factors and oncogenic BRAF. PLoS ONE. 2009;4:e4771. doi: 10.1371/journal.pone.0004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 30.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi Y, et al. Mesenchymal-epithelial interactions in the skin: Increased expression of dickkopf1 by palmoplantar fibroblasts inhibits melanocyte growth and differentiation. J Cell Biol. 2004;165:275–285. doi: 10.1083/jcb.200311122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi Y, et al. The effects of dickkopf 1 on gene expression and Wnt signaling by melanocytes: Mechanisms underlying its suppression of melanocyte function and proliferation. J Invest Dermatol. 2007;127:1217–1225. doi: 10.1038/sj.jid.5700629. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto H, et al. Identification of a novel substrate for TNFalpha-induced kinase NUAK2. Biochem Biophys Res Commun. 2008;365:541–547. doi: 10.1016/j.bbrc.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Tsuchihara K, et al. Susceptibility of Snark-deficient mice to azoxymethane-induced colorectal tumorigenesis and the formation of aberrant crypt foci. Cancer Sci. 2008;99:677–682. doi: 10.1111/j.1349-7006.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray-Schopfer VC, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 36.Bauer AJ, Stratakis CA. The lentiginoses: Cutaneous markers of systemic disease and a window to new aspects of tumourigenesis. J Med Genet. 2005;42:801–810. doi: 10.1136/jmg.2003.017806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong SS, Rajakulendran S. Peutz-Jeghers syndrome associated with primary malignant melanoma of the rectum. Br J Dermatol. 1996;135:439–442. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.