Abstract
Perrault syndrome is a genetically heterogeneous recessive disorder characterized by ovarian dysgenesis and sensorineural hearing loss. In a nonconsanguineous family with five affected siblings, linkage analysis and genomic sequencing revealed the genetic basis of Perrault syndrome to be compound heterozygosity for mutations in the mitochondrial histidyl tRNA synthetase HARS2 at two highly conserved amino acids, L200V and V368L. The nucleotide substitution creating HARS2 p.L200V also created an alternate splice leading to deletion of 12 codons from the HARS2 message. Affected family members thus carried three mutant HARS2 transcripts. Aminoacylation activity of HARS2 p.V368L and HARS2 p.L200V was reduced and the deletion mutant was not stably expressed in mammalian mitochondria. In yeast, lethality of deletion of the single essential histydyl tRNA synthetase HTS1 was fully rescued by wild-type HTS1 and by HTS1 p.L198V (orthologous to HARS2 p.L200V), partially rescued by HTS1 p.V381L (orthologous to HARS2 p.V368L), and not rescued by the deletion mutant. In Caenorhabditis elegans, reduced expression by RNAi of the single essential histydyl tRNA synthetase hars-1 severely compromised fertility. Together, these data suggest that Perrault syndrome in this family was caused by reduction of HARS2 activity. These results implicate aberrations of mitochondrial translation in mammalian gonadal dysgenesis. More generally, the relationship between HARS2 and Perrault syndrome illustrates how causality may be demonstrated for extremely rare inherited mutations in essential, highly conserved genes.
Perrault syndrome is a rare recessive disorder characterized by ovarian dysgenesis in females and sensorineural hearing loss in females and males (1). Affected females are karyotypically 46XX and infertile; affected males are 46XY and fertile (2). The syndrome is clinically heterogeneous: Some families have only sensorineural hearing loss and female ovarian dysgenesis, whereas others have neurological manifestations, which can include ataxia, limited extraocular movements, nystagmus, ophthalmoplegia, lower limb weakness, mental retardation, sensory polyneuropathy, hypotonia, or poor reflexes (3–5). In addition, families have been reported with either short stature (6, 7) or marfanoid features (8). The syndrome is also genetically heterogeneous. In one family, Perrault syndrome is caused by mutations in HSD17B4, encoding 17β-hydroxysteroid dehydrogenase type 4 (9, 10). In 10 other families diagnosed with Perrault syndrome, HSD17B4 sequences were wild type.
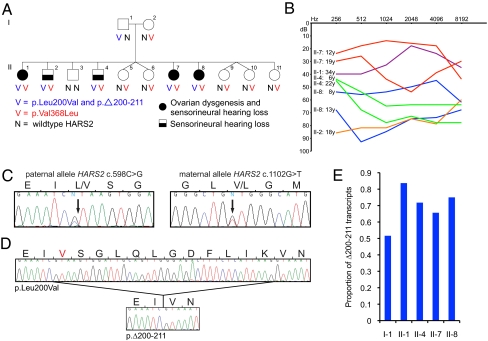
The goal of the present study was to identify and characterize the genetic basis of Perrault syndrome in one of the first reported kindreds (11) (Fig. 1A). In this nonconsanguineous family of mixed European ancestry, affected females II-1, II-7, and II-8 presented with ovarian dysgenesis, with amenorrhea and streak gonads (i.e., nonfunctional gonads composed primarily of fibrous tissue). All had 46XX karyotypes. These three females and two males in the family had sensorineural hearing loss, which was progressive in all five siblings, but varied in age of onset and severity (Fig. 1B). Affected males were fertile, with healthy, hearing children. Both parents had normal hearing. All persons in the family had normal intelligence. Thorough clinical evaluation of the family was provided by Pallister and Opitz (11).
Fig. 1.
Identification of mutations in HARS2 in a family with Perrault syndrome. (A) A nonconsanguineous family of French, Irish, and Scottish ancestry, with Perrault syndrome in 5 of 11 siblings. Affected siblings are compound heterozygotes for mutations in HARS2. (B) Progressive hearing loss in the affected individuals, measured by pure tone audiometry as described by Pallister and Opitz (11). (C) Paternal allele chr5:140,075,395C > G, corresponding to HARS2 c.598C > G, and maternal allele chr5:140,076,926, corresponding to HARS2 c.1102G > T. (D) Sequence from cDNA from lymphoblasts of II-1, indicating that HARS2 c.598C > G yields two transcripts, encoding HARS2 p.L200V and HARS2 p.Δ200–211. (E) The paternal allele HARS2 c.598C > G encodes HARS2 p.Δ200–211 and HARS2 p.L200V. The proportion of HARS2 c.598C > G transcripts encoding HARS2 p.Δ200–211 is significantly lower for the unaffected father I-1 than for his affected children II-1, II-4, II-7, and II-8 (P = 0.009). Transcripts were derived from lymphoblast cDNA. P value for the comparison is based on the Z score for significance of a difference between independent proportions.
Results
Identification of the Gene Responsible for Perrault Syndrome.
Genome-wide linkage analysis, under a model of fully penetrant recessive inheritance, was undertaken for 11 members of the family. Only one chromosomal region, between D5S2115 and D5S436 on chromosome 5q31, yielded a multipoint lod score >3.0. Fine mapping of this region defined a linkage interval of 4.142 Mb bounded by D5S479 and D5S2508 at chromosome 5:136,305,608–140,447,387 (hg19) with lod score Z = 3.10 (Fig. S1). This region harbors 58 genes, all of which were evaluated by Sanger sequencing of exons and flanking regulatory regions. Rare variants of predicted functional effect that cosegregated with the Perrault phenotype were found in only one gene, HARS2, which encodes the mitochondrial histidyl tRNA synthetase. Affected individuals carried a paternally inherited mutation at chr5:140,075,395C > G in HARS2 exon 6, corresponding to HARS2 c.598C > G (p.L200V), and a maternally inherited mutation at chr5:140,076,926G > T in HARS2 exon 10, corresponding to HARS2 c.1102G > T (p.V368L) (Fig. 1 A and C).
To test whether these mutations were the only plausible candidates, the entire 4.142-Mb linkage region was tiled with overlapping cRNA oligonucleotide bait probes. Genomic DNA from affected individual II-1 was hybridized to the baits and sequenced to a median 150-fold coverage, with 97% of targeted bases having >20-fold coverage. HARS2 was the only gene in this region with two variants of predicted functional effect (Table 1). Neither HARS2 c.598C > G nor HARS2 c.1102G > T was present in any of 982 control individuals of Caucasian ancestry or in 960 individuals genotyped in phase 3 of the 1,000 Genomes Project.
Table 1.
Rare variants in the 5q31 linked region in individual II-1 with Perrault syndrome
| SBP | Indel | Total | |
| Rare variants in linked region | 104 | 40 | 144 |
| Nonsense, missense, frameshift, or splice variants | 6 | 0 | 6 |
| Genes with ≥2 nonsense, missense, frameshift, or splice variants | 1 | 0 | 1 |
SBP, single base pair variants; Indel, insertion and deletion variants.
Analysis of HARS2 cDNA from patient-derived lymphoblast cell lines revealed that the paternal mutant allele created an alternative splice site, resulting in an in-frame deletion of the 12 remaining codons in exon 6 (p.Δ200–211) (Fig. 1D). The proportion of transcripts encoding HARS2 p.Δ200–211, among those encoding HARS2 p.L200V or p.Δ200–211, was significantly lower in the father, I-1, compared with his affected children, II-1, II-4, II-7, and II-8 (P = 0.009, Fig. 1E).
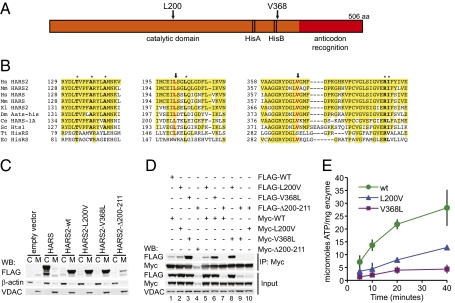
HARS2 encodes a histidyl tRNA synthetase (Fig. 2A) that is predicted to function in mitochondria (12). Aminoacyl tRNA synthetases are highly conserved enzymes that catalyze the covalent linkage of specific amino acids to their cognate tRNAs (13). Aminoacyl tRNA synthetase activity is required in both the cytoplasm and mitochondria for translation of nuclear- and mitochondrially encoded proteins, respectively. In mammals, for most amino acids, these two activities are encoded by separate genes, hence the cytoplasmic HARS and mitochondrial HARS2 histidyl tRNA synthetases. The residues mutated in HARS2 in Perrault syndrome are conserved from humans to yeast, with HARS2 p.L200 also conserved in Thermus thermophilus, and HARS2 p.V368 also conserved in Escherichia coli (Fig. 2B). HARS2 p.V368 is located just C-terminal to the highly conserved HisB region, which is specific to the histidine tRNA synthetases and forms part of the histidine binding pocket (14). HARS2 p.L200V and HARS2 p.V368L were predicted by the SIFT (Sorting Intolerant from Tolerant) algorithm to be not tolerated (P = 0.01 and P = 0.02, respectively) and by PolyPhen2 to be damaging (HumDiv scores 0.988 and 0.998 and HumVar scores 0.970 and 0.996, respectively). In 10 other families with Perrault syndrome, HARS2 sequences were wild type.
Fig. 2.
Effects of the mutations on the HARS2 protein. (A) Schematic of the HARS2 protein. (B) Protein sequence alignment of HARS2 orthologs. Mutated amino acids are in red and indicated by arrows; other amino acids critical to the protein structures of Fig. 3 are in boldface type and marked with asterisks. (C) Analysis by Western blot (WB) of cytoplasmic (C) and mitochondrial (M) extracts of 293T cells transfected with HARS and HARS2 constructs demonstrates that HARS localizes to cytoplasm and HARS2 localizes to mitochondria. HARS2 p.Δ200–211 is poorly expressed relative to HARS2 wild type, HARS2 p.L200V, and HARS2 p.V368L. β-Actin and VDAC are cytoplasmic and mitochondrial controls, respectively, for fractionation and loading. (D) Immunoprecipitates (IP) from mitochondrial extracts of 293T cells transfected with pairs of constructs. Coimmunoprecipitation between Myc- and FLAG-tagged HARS2 p.V368L (lane 3) is stronger than between Myc- and FLAG-tagged HARS2 or HARS2 p.L200V (lanes 1 and 2), indicating that HARS2 p.V368L homodimerizes more efficiently than does HARS2 or HARS2 p.L200V. Coimmunoprecipitation between HARS2 p.V368L and either HARS2 or HARS2 p.L200V (lanes 6 and 8) is stronger than between HARS2 and HARS2 p.L200V (lane 5), indicating that dimerization is increased even when only one monomer in the dimer harbors the p.V368L mutation. HARS2 p.Δ200–211 was expressed at very low levels; no dimerization with this mutant protein was detected (lanes 4, 7, 9, and 10). (E) Measurement of the enzymatic activity, using the pyrophosphate exchange assay, of purified wild-type HARS2 (wt), HARS2 p.L200V, and HARS2 p.V368L indicates that activities of both mutant proteins are reduced relative to wild-type protein, with HARS2 p.V368L more severely affected. Results illustrate a representative experiment, performed in triplicate; error bars indicate SD.
Mitochondrial Localization, Dimerization, and Enzyme Activity of HARS2 and HARS2 Mutants.
To determine whether HARS2 was indeed localized to the mitochondria and whether the Perrault-linked mutations affected protein expression or localization, we expressed wild-type and mutant HARS2 and the cytoplasmic histidyl tRNA synthetase HARS in mammalian cells. C-terminally epitope-tagged proteins were transiently expressed in 293T cells. HARS was highly expressed in the cytoplasm and virtually undetectable in mitochondria; most expression of both wild-type and mutant HARS2 was in mitochondria (Fig. 2C). Wild-type HARS2, HARS2 p.L200V, and HARS2 p.V368L were expressed at similar levels, whereas very little HARS2 p.Δ200–211 was detected, suggesting that the deletion mutant was unstable.
HARS2 functions in vivo as a homodimer (15). To determine whether the mutations affected the ability of HARS2 proteins to dimerize, Myc- and FLAG-tagged wild-type and mutant HARS2 proteins were coexpressed in pairs and immunoprecipitated with an anti-Myc antibody (Fig. 2D). HARS2 p.V368L dimerizes more efficiently than does wild-type HARS2 or HARS2 p.L220V. In contrast, HARS2 p.Δ200–211 is poorly expressed with no dimerization detected.
To determine whether the HARS2 mutations affected enzyme activity, we assayed the activity of purified recombinant wild-type and mutant HARS2 using the pyrophosphate exchange assay, which measures the first step of the aminoacylation reaction (Fig. 2E). Compared with wild-type HARS2, both HARS2 p.L200V and HARS2 p.V368L had decreased activity, with HARS2 p.V368L more severely affected. HARS2 p.Δ200–211 was poorly expressed in bacteria and could not be purified.
Analysis of HARS2 Structure.
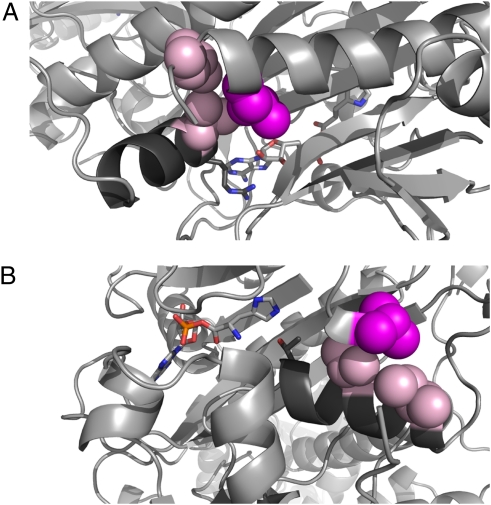
Crystal structures of prokaryotic homologs of vertebrate histidyl tRNA synthetases, in complex with histidyl-adenosine monophosphate (HAM), allowed us to examine the putative structural roles of HARS2 p.L200 and HARS2 p.V368 and to predict consequences of mutations at these sites. Sequences of these regions are conserved among prokaryotic and eukaryotic species (Fig. 2B). The amino acid analogous to HARS2 p.L200 in the T. thermophilus enzyme is involved in packing interactions with two other strongly conserved hydrophobic amino acids, analogous to HARS2 p.L203 and HARS2 p.I390 (Fig. 2B). The amino acid analogous to HARS2 p.I390 is on an α-helix involved in ATP binding that is conserved in all class II tRNA synthetases (15). HARS2 p.R389, the residue adjacent to p.I390, is absolutely conserved and is involved in coordinating the γ-phosphate and positioning the adenine ring of the HAM (16, 17) (Fig. 3A). Substitution of the side chain of HARS2 p.L200 with a shorter valine side chain may destabilize or adjust this packing interaction, allowing movement of the helix containing p.I390 and p.R389, thereby altering binding in the active site. HARS2 p.V368 is located at the end of the highly conserved HisB motif, which is involved in the recognition of histidine (14). Its side chain is also engaged in packing interactions with two other hydrophobic amino acids, analogous to p.A137 and p.A141. These three amino acids are completely conserved in eukaryotes and strongly conserved as methyl-containing hydrophobic residues in prokaryotes (Fig. 2B). HARS2 p.A137 and p.A141 are located on two neighboring turns of an α-helix that begins with the invariant residue p.T133, which makes contact with the histidyl end of the HAM (Fig. 3B). Substitution of HARS2 p.V368 with the longer leucine side chain could cause an adjustment in the packing interaction leading to movement of the helix and p.T133 away from the histidine in the binding pocket. This adjustment could lead to reduction in the binding affinity for the activated histidine.
Fig. 3.
Structures of homologs of HARS2 at the mutant sites. Ribbon diagrams are shown of histidyl tRNA synthetases from (A) T. thermophilus (PDB 1ADY) and (B) E. coli (PDB 1KMM), each in complex with histidyl-adenylate monophosphate (HAM), which is represented by light gray sticks with element coloring in red (oxygen), blue (nitrogen), and orange (phosphorus). Substitution at either L200 or V368 could result in movement of conserved HAM contact residues. Numbers listed in A and B indicate analogous residues in human/prokaryote proteins. (A) Region surrounding residue L151, analogous to HARS2 p.L200. L200/L151 (magenta) contacts L203/L154 and I390/V312 (pink). The helix containing I390/V312 and the conserved R389/R311 residue is shown in dark gray, with R389/R311, which contacts HAM, shown in dark gray sticks with element coloring as above. For clarity, residues 151–160 have been removed from the view. (B) Region surrounding residue V292, analogous to HARS2 p.V368. V368/V292 (magenta) contacts A137/V89 and A141/I93 (pink) on adjacent turns of the α-helix containing the invariant T133/T85 (dark gray sticks with element coloring), which contacts the HAM. For clarity, residues 293–304 have been removed from the view.
Analysis of HTS1 Mutations in Yeast.
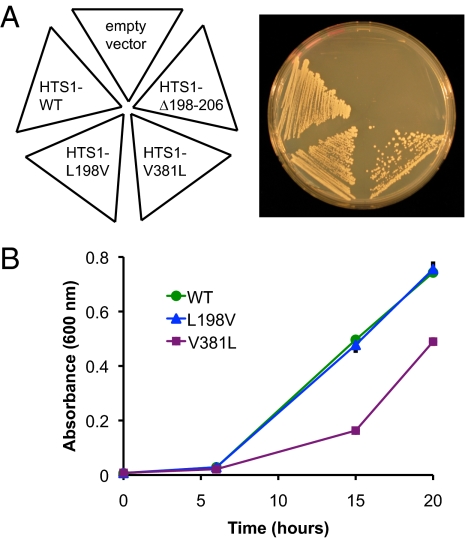
We investigated the effects of the HARS2 mutations in vivo by evaluating the analogous mutations in yeast (Fig. 4). Saccharomyces cerevisiae has a single essential hisitdyl tRNA synthetase gene, HTS1, that provides both mitochondrial and cytoplasmic activity (18). We determined the ability of HTS1 mutations analogous to the HARS2 mutations to complement the lethality of the yeast HTS1 deletion strain (hts1Δ). Growth of cells expressing Hts1 p.L198V (analogous to HARS2 p.L200V) was similar to the growth of cells expressing wild-type Hts1 (Fig. 4A), indicating that Hts1 p.L198V was able to completely rescue the lethality of the hts1Δ strain. In contrast, growth was only partially rescued by Hts1 p.V381L (analogous to HARS2 p.V368L). Hts1 p.Δ198–207 and the empty vector were unable to rescue growth. Similar results were observed when cells from the rescued colonies were grown in liquid media, with wild-type Hts1 and Hts1 p.L198V rescuing growth and Hts1 p.V381L providing partial rescue (Fig. 4B). Consistent with the results of the HARS2 enzyme assay, these results suggest that the Hts1 p.V381L (HARS2 p.V368L) mutation is the more severe of the two missense mutations, whereas Hts1 p.L198V (HARS2 p.L200V) may provide wild-type or near wild-type activity in vivo. In contrast, the failure of HTS1 p.Δ198–207 to rescue hts1Δ and the poor expression of HARS2 p.Δ200–211 in mammalian cells and bacteria suggest that this mutant is unlikely to provide any activity in vivo.
Fig. 4.
Effects on growth in yeast of mutations in HTS1 analogous to mutations in HARS2. HTS1-L198V, corresponding to HARS2 p.L200V, complements loss of HTS1 more effectively than does HTS1-V381L, corresponding to HARS2 p.V368L. HTS1-Δ198–205, corresponding to HARS2 p.Δ200–211, fails to complement loss of HTS1. (A) hts1Δ yeast with a rescuing wild-type copy of HTS1 on a URA3 plasmid were transformed with TRP1 plasmids encoding HTS1 of various genotypes. The resulting strains were streaked on Trp− media containing 5-fluoroorotic acid, to select against the rescuing wild-type HTS1. Growth indicates complementation by wild-type or mutant HTS1 on the TRP plasmid. (B) Growth in liquid culture of the strains from A.
Effects of Down-Regulation of hars-1 on Fertility in C. elegans.
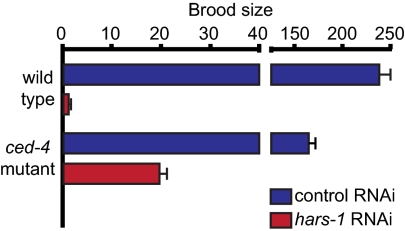
To investigate the impact of reduced histidyl tRNA synthetase activity on a multicellular organism, we exposed wild-type Caenorhabditis elegans to hars-1 RNA interference (RNAi) by feeding (Fig. 5). Hars-1 is the single histidyl tRNA synthetase gene of C. elegans. Embryos from wild-type animals were laid onto bacteria expressing double-stranded RNA and the progeny were exposed to RNAi throughout development. Expression of hars-1 was reduced by at least 50% in L4 larvae exposed to hars-1 RNAi relative to larvae exposed to control RNAi (Fig. S2). Reduced hars-1 expression caused severe gonadal defects including smaller, narrower gonads and the absence of oocytes or fertilized eggs in the majority of animals (Fig. S3). When oocytes or fertilized eggs were present, they were often misshapen. Consistent with these morphological defects, hars-1 RNAi-treated animals were almost completely sterile relative to control RNAi-treated animals (Fig. 5).
Fig. 5.
RNAi of the C. elegans histidyl tRNA synthetase gene hars-1 reduces fertility. hars-1 RNAi decreased the brood size of wild-type animals more dramatically than it did that of ced-4(n1162) animals, which have a defect in apoptosis. Eggs were collected from wild-type N2 or ced-4(n1162) adults on plates seeded with control or hars-1 RNAi bacteria and allowed to develop to the L4 stage. F2 progeny of 14 F1 animals exposed to control RNAi and 48 F1 animals exposed to hars-1 RNAi were counted. A representative experiment is shown. Error bars indicate SD.
Deficient numbers of functional germ cells could be due to reduced production of germ cells, loss of germ cells due to cell death, or both. To investigate this mechanism, we compared the response of wild-type animals and ced-4(n1162) mutant animals, which have a defect in apoptosis, to hars-1 RNAi. When wild-type and ced-4(n1162) animals were treated with control RNAi, ced-4(n1162) animals had a smaller brood size, as expected (19). Treatment with hars-1 RNAi decreased brood size in both wild-type and ced-4(n1162) mutants, but less severely in ced-4(n1162) mutants (Fig. 5). These results indicate that hars-1 activity is required in C. elegans for fertility and proper germ cell development and that the loss of fertility caused by hars-1 RNAi is partially mediated by apoptosis.
We also investigated the consequences of complete loss of hars-1 using the hars-1(tm4074) null allele. Homozygous hars-1(tm4074) animals arrested development as L2 larvae, as determined by overall body size and by extent of gonad development (Fig. S3 and Table S1). The L2 stage is before substantial germ-line expansion. The arrested larvae remained active and apparently healthy for at least several days. In addition, the pseudocoelae of mutant animals contained a large number of granules (Fig. S3) that did not stain with oil red O or Nile red so were unlikely to contain fat or to be lysosomal in origin (20, 21).
Discussion
Aminoacyl-tRNA synthetases are highly conserved proteins that play essential and nonredundant roles in cytoplasmic and mitochondrial protein translation. In mammals, the cytoplasmic and mitochondrial aminoacyl-tRNA synthetases for each amino acid except glycine and lysine are encoded by separate genes. Null mutations in cytoplasmic or bifunctional (i.e., cytoplasmic and mitochondrial) aminoacyl-tRNA synthetases are lethal, both in yeast (22) and in mouse (23). Null mutations in the corresponding mitochondrial enzymes have more variable phenotypes. Those in yeast are unable to grow on glycerol (a nonfermentable carbon source) and have unstable mitochondrial DNA (24). C. elegans with a probable null mutation in mitochondrial lars-2 grow to late larval stage and are sterile and long-lived (25), suggesting a critical role for mitochondria in the development of the germ line. In humans, homoplasmic loss-of-function mutation of the mt-tRNAVal gene is lethal (26). Deleterious mtDNA mutations are rarely homoplasmic, suggesting a threshold of activity that is necessary for life (27).
Mutations in mammalian cytoplasmic and bifunctional aminoacyl-tRNA synthetases share phenotypic and genotypic features. Dominantly inherited missense mutations in tRNA synthetases for alanine (AARS), glycine (GARS), lysine (KARS), and tyrosine (YARS) cause Charcot–Marie–Tooth (CMT) disease, a group of peripheral neuropathies characterized by progressive degeneration of motor and sensory neurons (28–31). Although most CMT-associated mutations result in reduced enzyme activity, this is not always the case, suggesting that the CMT phenotype is not always caused by loss of function (32). In support of this, mice heterozygous for a Gars null mutation have reduced Gars activity but no neurodegeneration, whereas the complex spontaneous mutation Gars p.P278YK causes a dominant peripheral neuropathy phenotype similar to human CMT (23). This observation suggests that the CMT-like phenotype depends on the presence of the mutant protein. The similarity of the phenotype in the AARS, GARS, and YARS cases and the absence of phenotypes characteristic of mitochondrial dysfunction suggest that for individuals with mutations in the two bifunctional enzymes GARS and KARS, the CMT phenotype is primarily due to a defect in cytoplasmic activity.
In contrast to the phenotypes associated with cytoplasmic and bifunctional aminoacyl-tRNA synthetases, phenotypes caused by mutations in the corresponding mitochondrial enzymes are clinically more variable. We have seen that mutations in HARS2 leading to loss of enzyme activity cause progressive sensorineural hearing loss and ovarian dysgenesis. Mutations in the mitochondrial tRNA synthetases for aspartic acid (DARS2), arginine (RARS2), tyrosine (YARS2), and serine (SARS2) have been implicated, respectively, in the following syndromes: leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation (LBSL); pontocerebellar hypoplasia type 6 (PCH6); myopathy, lactic acidosis, and sideroblastic anemia (MLASA); and hyperuricemia, pulmonary hypertension, renal failure in infancy, and alkylosis (HUPRA) (33–36). LBSL and PCH6 involve specific progressive central nervous system dysfunction, whereas the primary symptoms of MLASA are progressive exercise intolerance and sideroblastic anemia. HUPRA is a multisystemic disorder involving progressive renal failure. All of these conditions are recessively inherited and are likely due to reduced synthetase activity. Despite their different clinical presentations, the primary cellular defect resulting from these mutations is probably the same. Skeletal muscle biopsies from patients with RARS2, YARS2, and SARS2 mutations show an overall decrease in the activity of respiratory chain complexes, particularly for those complexes containing mitochondrially encoded subunits (33, 35, 36). For YARS2 this defect was shown to result from a general decrease in mitochondrial translation. We speculate that the Perrault syndrome-associated HARS2 mutations also lead to a general decrease in mitochondrial translation and respiratory chain defects in affected tissues.
Disruption of mitochondrial translation has previously been implicated in progressive sensorineural hearing loss. Mutations in mitochondrial rRNA and tRNA genes, including mt-tRNAHis, have been associated with both syndromic and nonsyndromic hearing loss (27, 37, 38). Mutations in mitochondrial tRNA genes would be predicted to lead to phenotypes similar to those caused by mutations in mitochondrial aminoacyl-tRNA synthetases. In addition to their role in cellular energy production, mitochondria are major producers of reactive oxygen species and play a key role in the initiation of apoptosis. It has been hypothesized that disruptions in oxidative phosphorylation resulting from decreased mitochondrial translation could lead to increased production of reactive oxygen species and the initiation of apoptosis (38). Because the inner ear is very sensitive to these processes, a slight increase in apoptosis could lead to cochlear degeneration (39, 40).
Disruption of mitochondrial translation has not previously been implicated in mammalian gonadal dysgenesis. However, mitochondria-dependent apoptosis is a critical part of gonadal development. Oocyte numbers reach their maximum at midgestation, have decreased by approximately two-thirds at birth, and continue to decrease throughout postnatal life (41, 42). It has been suggested that this extensive apoptosis provides a mechanism for removing cells carrying mtDNA mutations (43) and could also remove cells with poor mitochondrial function as a result of mutations in the nuclear genome or other defects. Thus, there is a critical balance between oocyte proliferation and apoptosis. Disruption of either process could lead to inadequate numbers or complete loss of oocytes. In addition, adequate mitochondrial function is thought to be an important determinant of the developmental competence of human oocytes (44). Loss of activity of C. elegans hars-1 or lars-2, or of other genes critical for mitochondrial function (25, 45, 46), leads to sterility and/or early larval arrest, indicating that a critical role for mitochondria in fertility and early development has been conserved throughout evolution.
The mitochondrial aminoacyl-tRNA synthetases are involved in essential and ubiquitous cellular processes, but the phenotypes caused by mutant alleles vary in tissue specificity and in clinical presentation. This variety may suggest differences among cell types in the resilience of mitochondrial translation given reduction in specific tRNA synthetase activities. Cell types may differ due to higher requirements for respiratory chain complex activity in some tissues, such as skeletal muscle, or to greater sensitivity to mitochondrial dysfunction in other tissues, such as the inner ear and developing ovary. In addition, interaction between a specific mutation and the cellular environment could result in the reduction of enzyme activity to below a critical threshold in particular cells. For example, variability in baseline levels of gene expression or variability in tissue-specific enhancers, splicing factors, or chaperones could affect levels of active tRNA synthetase enzymes in different tissues, making certain tissues more vulnerable to reduced activity. Disorders resulting from mutations in DARS2, RARS2, and HARS2 all involve splicing mutations. Variation in the amount of alternative splicing may modify the overall amount of enzyme activity. In the family harboring the HARS2 mutations, the ratio of HARS2 p.L200V to HARS2 p.Δ200–211 transcripts is higher in the lymphoblasts of unaffected individual I-1 than in those of his affected children. Because HARS2 p.L200V is stable with nearly wild-type activity and HARS2 p.Δ200–211 is unstable, the level of HARS2 activity is likely to be higher in the cells of I-1. Splicing variability, whether due to cis effects, interacting proteins, or stochastic influences, could result in activity falling below a critical threshold in specific cells while adequate activity is maintained in other cells.
With the vast amounts of DNA sequence data now available for genomic analysis, a major challenge to both research and medicine is to determine which of a patient's many genetic changes cause the critical phenotype. If a gene is mutant in only one family, statistical methods are not useful: Causality must be evaluated biologically. In the case of Perrault syndrome and HARS2, conservation analysis, transcript analysis, activity assays of expressed proteins, and functional studies in yeast and C. elegans were combined to demonstrate that missense mutations in HARS2 are responsible for this family's disorder. The hallmarks of Perrault syndrome are progressive sensorineural hearing loss and ovarian dysgenesis, but the disorder is both clinically and genetically heterogeneous. It is possible that some cases of apparently nonsyndromic sensorineural hearing loss may be unrecognized Perrault syndrome, if only males and prepubertal females are evaluated. We have identified mutations in HARS2 and HSD17B4 (10) as causes of Perrault syndrome in two families. However, mutations in these genes do not explain Perrault syndrome in nine other families, indicating that other critical genes remain to be identified.
Methods
Detailed methods are provided in SI Methods.
Subjects and Gene Identification.
The family was referred by the diagnosing physician (J.M.O.). Control subjects were 982 individuals of self-described Caucasian ancestry without deafness or ovarian dysgenesis. Linkage analysis and sequencing of genes in the critical region were carried out as described in SI Methods and Table S2.
Analysis of Alternate Transcripts.
Proportions of transcripts encoding the L200V and Δ200–211 mutant forms of HARS2 were determined by amplifying and sequencing individual clones.
cDNA Cloning and Mutagenesis.
Human HARS and HARS2 cDNA and yeast HTS1 genomic constructs were generated by PCR, cloning, and site-directed mutagenesis.
Transfection, Cell Fractionation, and Western Blotting.
293T cells were transfected with epitope-tagged HARS and HARS2 constructs, followed by isolation of cytoplasmic and mitochondrial fractions. Proteins were analyzed by immunoprecipitation and/or Western blotting.
Purification of Recombinant HARS2 Proteins.
Wild-type and mutant HARS2 were expressed in bacteria and proteins were purified by affinity chromatography.
Pyrophosphate Exchange Assay.
Pyrophosphate exchange assays were performed by a modification of published procedures.
Yeast HTS1 Deletion Mutant and Complementation Assay.
The ORF of HTS1 was deleted by PCR-mediated gene replacement. The haploid hts1Δ strain was isolated after transformation with pRS316-HTS1. The hts1Δ strain containing pRS316-HTS1 was transformed with wild-type or mutant HTS1. Complementation was tested on media containing 5-FOA, to select against pRS316-HTS1, and assessed by growth on plates or in liquid media.
C. elegans Strains and RNAi by Feeding.
Wild-type N2 Bristol and ced-4(n1162) strains were obtained from the Caenorhabditis Genetic Center. hars-1(tm4074) was received from S. Mitani (Tokyo Women's Medical University, Tokyo). C. elegans were maintained using standard methods and RNAi by feeding was performed essentially as described. F2 progeny of individual F1 animals were counted.
Supplementary Material
Acknowledgments
We are grateful to T. Davis and E. First for technical advice. This work was supported by National Institutes of Health/National Institute of Deafness and Communication Disorders Grant R01DC005641, National Institutes of Health/National Institute of Environmental Health Sciences Predoctoral Fellowship F30ES13069, and National Institutes of Health/National Institute of General Medical Sciences Training Grant T32GM07266.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103471108/-/DCSupplemental.
References
- 1.Perrault M, Klotz B, Housset E. [Two cases of Turner syndrome with deaf-mutism in two sisters] Bull Mem Soc Med Hop Paris. 1951;67:79–84. [PubMed] [Google Scholar]
- 2.Christakos AC, Simpson JL, Younger JB, Christian CD. Gonadal dysgenesis as an autosomal recessive condition. Am J Obstet Gynecol. 1969;104:1027–1030. doi: 10.1016/0002-9378(69)90697-8. [DOI] [PubMed] [Google Scholar]
- 3.Amor DJ, Delatycki MB, Gardner RJ, Storey E. New variant of familial cerebellar ataxia with hypergonadotropic hypogonadism and sensorineural deafness. Am J Med Genet. 2001;99:29–33. doi: 10.1002/1096-8628(20010215)99:1<29::aid-ajmg1119>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 4.Marlin S, et al. Perrault syndrome: Report of four new cases, review and exclusion of candidate genes. Am J Med Genet A. 2008;146A:661–664. doi: 10.1002/ajmg.a.32180. [DOI] [PubMed] [Google Scholar]
- 5.Kobe C, Kracht LW, Timmermann L, Bachmann J, Schmidt MC. Perrault syndrome with progressive nervous system involvement. Clin Nucl Med. 2008;33:922–924. doi: 10.1097/rlu.0b013e31818c4e25. [DOI] [PubMed] [Google Scholar]
- 6.Nishi Y, Hamamoto K, Kajiyama M, Kawamura I. The Perrault syndrome: Clinical report and review. Am J Med Genet. 1988;31:623–629. doi: 10.1002/ajmg.1320310317. [DOI] [PubMed] [Google Scholar]
- 7.Nikolaou DS, Winston RM. Sporadic Perrault syndrome. J Obstet Gynaecol. 1999;19:436–437. doi: 10.1080/01443619964896. [DOI] [PubMed] [Google Scholar]
- 8.Jacob JJ, Paul TV, Mathews SS, Thomas N. Perrault syndrome with Marfanoid habitus in two siblings. J Pediatr Adolesc Gynecol. 2007;20:305–308. doi: 10.1016/j.jpag.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Fiumara A, et al. Perrault syndrome: Evidence for progressive nervous system involvement. Am J Med Genet A. 2004;128A:246–249. doi: 10.1002/ajmg.a.20616. [DOI] [PubMed] [Google Scholar]
- 10.Pierce SB, et al. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am J Hum Genet. 2010;87:282–288. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pallister PD, Opitz JM. The Perrault syndrome: Autosomal recessive ovarian dysgenesis with facultative, non-sex-limited sensorineural deafness. Am J Med Genet. 1979;4:239–246. doi: 10.1002/ajmg.1320040306. [DOI] [PubMed] [Google Scholar]
- 12.O'Hanlon TP, Raben N, Miller FW. A novel gene oriented in a head-to-head configuration with the human histidyl-tRNA synthetase (HRS) gene encodes an mRNA that predicts a polypeptide homologous to HRS. Biochem Biophys Res Commun. 1995;210:556–566. doi: 10.1006/bbrc.1995.1696. [DOI] [PubMed] [Google Scholar]
- 13.Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- 14.Arnez JG, et al. Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. EMBO J. 1995;14:4143–4155. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freist W, Verhey JF, Rühlmann A, Gauss DH, Arnez JG. Histidyl-tRNA synthetase. Biol Chem. 1999;380:623–646. doi: 10.1515/BC.1999.079. [DOI] [PubMed] [Google Scholar]
- 16.Arnez JG, Augustine JG, Moras D, Francklyn CS. The first step of aminoacylation at the atomic level in histidyl-tRNA synthetase. Proc Natl Acad Sci USA. 1997;94:7144–7149. doi: 10.1073/pnas.94.14.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aberg A, Yaremchuk A, Tukalo M, Rasmussen B, Cusack S. Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase. Biochemistry. 1997;36:3084–3094. doi: 10.1021/bi9618373. [DOI] [PubMed] [Google Scholar]
- 18.Natsoulis G, Hilger F, Fink GR. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986;46:235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- 19.Hengartner MO. In: C. elegans II. Riddle DL, editor. Plainview, NY: Cold Spring Harbor Lab Press; 1997. pp. 383–415. [Google Scholar]
- 20.Ashrafi K, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 21.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 23.Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 24.Tzagoloff A, Shtanko A. Mitochondrial and cytoplasmic isoleucyl-, glutamyl- and arginyl-tRNA synthetases of yeast are encoded by separate genes. Eur J Biochem. 1995;230:582–586. doi: 10.1111/j.1432-1033.1995.tb20599.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 26.McFarland R, et al. Multiple neonatal deaths due to a homoplasmic mitochondrial DNA mutation. Nat Genet. 2002;30:145–146. doi: 10.1038/ng819. [DOI] [PubMed] [Google Scholar]
- 27.Kokotas H, Petersen MB, Willems PJ. Mitochondrial deafness. Clin Genet. 2007;71:379–391. doi: 10.1111/j.1399-0004.2007.00800.x. [DOI] [PubMed] [Google Scholar]
- 28.Antonellis A, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordanova A, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 30.Latour P, et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet. 2010;86:77–82. doi: 10.1016/j.ajhg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaughlin HM, et al. NISC Comparative Sequencing Program. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antonellis A, et al. Functional analyses of glycyl-tRNA synthetase mutations suggest a key role for tRNA-charging enzymes in peripheral axons. J Neurosci. 2006;26:10397–10406. doi: 10.1523/JNEUROSCI.1671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edvardson S, et al. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheper GC, et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet. 2007;39:534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 35.Riley LG, et al. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia—MLASA syndrome. Am J Hum Genet. 2010;87:52–59. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belostotsky R, et al. Mutations in the mitochondrial seryl-tRNA synthetase cause hyperuricemia, pulmonary hypertension, renal failure in infancy and alkalosis, HUPRA syndrome. Am J Hum Genet. 2011;88:193–200. doi: 10.1016/j.ajhg.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crimi M, et al. A mitochondrial tRNA(His) gene mutation causing pigmentary retinopathy and neurosensorial deafness. Neurology. 2003;60:1200–1203. doi: 10.1212/01.wnl.0000055865.30580.39. [DOI] [PubMed] [Google Scholar]
- 38.Guan MX. Molecular pathogenetic mechanism of maternally inherited deafness. Ann N Y Acad Sci. 2004;1011:259–271. doi: 10.1007/978-3-662-41088-2_25. [DOI] [PubMed] [Google Scholar]
- 39.Someya S, Tanokura M, Weindruch R, Prolla TA, Yamasoba T. Effects of caloric restriction on age-related hearing loss in rodents and rhesus monkeys. Curr Aging Sci. 2010;3:20–25. doi: 10.2174/1874609811003010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh T, et al. Genomic duplication and overexpression of TJP2/ZO-2 leads to altered expression of apoptosis genes in progressive nonsyndromic hearing loss DFNA51. Am J Hum Genet. 2010;87:101–109. doi: 10.1016/j.ajhg.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morita Y, Tilly JL. Oocyte apoptosis: Like sand through an hourglass. Dev Biol. 1999;213:1–17. doi: 10.1006/dbio.1999.9344. [DOI] [PubMed] [Google Scholar]
- 42.Vaskivuo TE, et al. Survival of human ovarian follicles from fetal to adult life: Apoptosis, apoptosis-related proteins, and transcription factor GATA-4. J Clin Endocrinol Metab. 2001;86:3421–3429. doi: 10.1210/jcem.86.7.7679. [DOI] [PubMed] [Google Scholar]
- 43.Krakauer DC, Mira A. Mitochondria and germ-cell death. Nature. 1999;400:125–126. doi: 10.1038/22026. [DOI] [PubMed] [Google Scholar]
- 44.Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion. 2010 doi: 10.1016/j.mito.2010.09.012. 10.1016/j.mito.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Tsang WY, Lemire BD. The role of mitochondria in the life of the nematode, Caenorhabditis elegans. Biochim Biophys Acta. 2003;1638:91–105. doi: 10.1016/s0925-4439(03)00079-6. [DOI] [PubMed] [Google Scholar]
- 46.Curran SP, Leverich EP, Koehler CM, Larsen PL. Defective mitochondrial protein translocation precludes normal Caenorhabditis elegans development. J Biol Chem. 2004;279:54655–54662. doi: 10.1074/jbc.M409618200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.