Abstract
A-kinase anchoring protein 79 (AKAP79) is a human anchoring protein that organizes cAMP-dependent protein kinase (PKA), Ca2+/calmodulin (CaM)-dependent protein phosphatase (PP2B), and protein kinase C (PKC) for phosphoregulation of synaptic signaling. Quantitative biochemical analyses of selected AKAP79 complexes have determined the quaternary structure of these signaling complexes. We show that AKAP79 dimerizes, and we demonstrate that, upon addition of a lysine-reactive cross-linker, parallel homomeric dimers are stabilized through K328–K328 and K333–K333 cross-links. An assembly of greater complexity comprising AKAP79, PP2B, a type II regulatory subunit fragment (RII 1–45) of PKA, and CaM was reconstituted in vitro. Using native MS, we determined the molecular mass of this complex as 466 kDa. This indicates that dimeric AKAP79 coordinates two RII 1–45 homodimers, four PP2B heterodimers, and two CaM molecules. Binding of Ca2+/CaM to AKAP79 stabilizes the complex by generating a second interface for PP2B. This leads to activation of the anchored phosphatases. Our architectural model reveals how dimeric AKAP79 concentrates pockets of second messenger responsive enzyme activities at the plasma membrane.
Following its discovery, cyclic AMP (cAMP) was thought to be freely diffusible within the cell (1). Subsequently, it was demonstrated that utilization of this second messenger is more sophisticated. For example, treatment of cardiac myocytes with norepinephrine, but not prostaglandin E1, stimulates contraction and glycogen metabolism, despite the fact that both hormones elevate cAMP to activate cAMP-dependent protein kinase (PKA) (2). It is now recognized that intracellular cAMP levels fluctuate in cellular microdomains where cAMP effector proteins such as PKA, Epac guanine nucleotide exchange factors, and cyclic nucleotide-gated ion channels reside (3–5). Likewise, other second messengers such as Ca2+ and certain phospholipids operate in cellular microdomains where they interface with their own effector proteins (6).
A-kinase anchoring proteins (AKAPs) organize responses to these second messengers. AKAP79 is a prototypical AKAP, exemplifying the three properties that are characteristic of the family: (i) an amphipathic α-helix (residues 387–406) that binds to the D/D domain (residues 1–45) of PKA RII subunits (7); (ii) a subcellular localization signal, in its case, three tandem membrane-binding basic regions (MBBRs) that bind to PIP2 (8); and (iii) the ability to interact with multiple signaling molecules. AKAP79 targets PKA, the Ca2+-dependent Ser/Thr protein phosphatase PP2B (calcineurin) (9), and protein kinase C (PKC) (10) to substrates including transmembrane receptors and ion channels. A more comprehensive list of AKAP79 binding partners is shown in Fig. S1. Ancillary protein–lipid and protein–protein interactions serve to localize AKAP79 with particular membrane substrates (8). For example, a leucine zipper-like motif in AKAP79 associates with the cytoplasmic tail of L-type Ca2+ channels, enabling channel regulation by PKA and PP2B (11). AKAP79 is also a principal AKAP in the postsynaptic density, where it coordinates phosphoregulation of GluR1 receptors. Consequently, mice deficient in AKAP150 exhibit impaired synaptic plasticity and learning deficits (12). AKAP79 also targets PKC to KCNQ2 channels for receptor-induced suppression of the M current (13), a property that provides AKAP150 knockout animals with a measure of resistance to seizures induced by the nonselective muscarinic agonist pilocarpine (12).
Prior to this study, our understanding of the molecular basis of the AKAP79 signaling complex was limited to structures of AKAP-derived peptides in complex with D/D domains (14, 15). Here, we determine the stoichiometry of a core AKAP79 complex by mass spectrometry (MS) of intact protein assemblies (16, 17), and we map a parallel dimer interface by chemical cross-linking. Each protomer of AKAP79 is capable of binding two PP2B heterodimers, a PKA regulatory homodimer and one CaM. Each kinase/phosphatase pair is anchored in a symmetrical manner. Finally, we show that binding of Ca2+/CaM to AKAP79 generates an additional PP2B interaction site on the anchoring protein to activate the tethered phosphatase.
Results
AKAP79 Dimerizes.
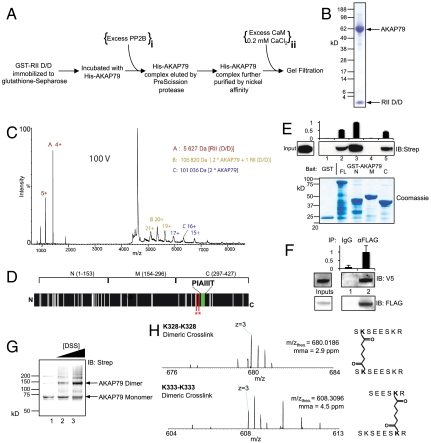
We developed a multistep purification system for reconstitution of AKAP79 complexes in vitro (Fig. 1A). A GST fusion protein with the D/D domain of RIIα (residues 1–45, hereafter abbreviated D/D) was used to purify AKAP79, which was expressed with an N-terminal His6 tag in Sf21 insect cells. AKAP79 and associated proteins were released from glutathione sepharose resin by incubation with PreScission protease that specifically cleaved the link between GST and the D/D domain. A nickel affinity step was included to further enrich His6-tagged AKAP79 and associated proteins before final purification by gel filtration. AKAP79–D/D eluted from a Superdex S200 gel filtration column with an elution volume of 66.6 mL (Fig. S2A), which, according to gel filtration standards, corresponds to a protein of approximately 200 kDa. This suggests that AKAP79 oligomerizes. Peak fractions (Coomassie-stained, Fig. 1B) were pooled and concentrated for analysis by native MS. MS of purified AKAP79–D/D at minimal activation revealed a series of AKAP79–D/D oligomers (Fig. S3A). Fig. 1C shows the tandem MS (MSMS) spectrum of AKAP79–D/D where ions were selected at 4,500 m/z. Upon activation, this complex loses a D/D subunit to form a stripped complex of 106,820 Da [2 ∗ AKAP79 + 1 ∗ D/D] (Fig. 1C, gold). Upon additional activation, a further D/D subunit is lost to form a 101,036-Da complex consisting of the AKAP79 dimer (Fig. 1C, blue). Therefore, the original precursor ion is assigned unambiguously to the 25+ charge state of a complex consisting of [2 ∗ AKAP79 + 2 ∗ D/D], supporting AKAP79 dimerization. MS of the AKAP79–D/D complex was also performed under denaturing conditions. A more precise mass of AKAP79 was calculated by this method at 50,461 ± 18 Da (Fig. S3B). This measurement suggests that the anchoring protein is not posttranslationally modified when expressed in insect cells. Therefore, the small deviations from theoretical masses measured for AKAP79–D/D complexes (Table S1) are likely a result of adduct formation. MSMS of AKAP79–D/D ions selected at 3,640 m/z was also performed, supporting the established ratio of one AKAP79 to two D/D (Fig. S3C).
Fig. 1.
AKAP79 purification and basic stoichiometry. (A) Purification protocol for the AKAP79–D/D complex. The positions of two additional steps sufficient for AKDPC purification are indicated. (B) Coomassie-stained 4–12% SDS-PAGE showing 10 μg purified AKAP79–D/D. (C) MSMS spectrum of AKAP79–D/D. 106,820 Da [2 ∗ AKAP79 + 1 ∗ D/D] (gold) and 101,036 Da [2 ∗ AKAP79] (blue) dissociation products are indicated. Spectra are shown at a collision cell voltage of 100 V. (D) Topology diagram illustrating the location of 42 lysine residues (white bars) in AKAP79 and the boundaries of three constructs. The locations of dimeric cross-linking sites (red) and the PIAIIIT motif (green) are indicated. (E) Anti-strep immunoblot (IB) of Strep–AKAP79 pulled-down by GST, GST–AKAP79, or GST fused to the three regions of AKAP79 as depicted in D. Relative densitometry analysis (n = 3, Top) and pull-down Coomassie (Bottom) are shown. (F) Control immunoglobulin (IgG) or anti-FLAG immunoprecipitates from HEK293 cells coexpressing FLAG–AKAP79 and V5–AKAP79 were immunoblotted as indicated. Relative densitometry analysis (n = 3, Top). (G) Anti-Strep IB shows Strep–AKAP79 dimer cross-linking with 0 (lane 1), 50 (lane 2) and 500 (lane 3) μM DSS, indicated by anti-strep immunoblotting. (H) MS spectra showing peptides generated by trypsinization of cross-linked Strep–AKAP79. The spectra correspond to K328–K328 (Top) and K333–K333 (Bottom) dimeric cross-linked peptides.
To map the dimerization interface, three separate regions of AKAP79 (N, M, and C, Fig. 1D) were fused to GST. Pull-down of Strep-tagged AKAP79, purified from Hi5 insect cells, was performed with GST–AKAP79 and related fragments (Fig. 1E). Strep–AKAP79 interacted with full-length AKAP79 (Fig. 1E, lane 2). However, Strep–AKAP79 also bound to N- and C-terminal regions of the anchoring protein Fig. 1E, lanes 3 and 5). In contrast, protein complexes were not formed with GST alone or the middle region of the anchoring protein (Fig. 1E, lanes 1 and 4). This indicates N- and C-terminal regions of AKAP79 participate in oligomerization. AKAP79 oligomerization was validated in a cellular setting by coexpressing FLAG-tagged AKAP79 and V5-tagged AKAP79 in HEK293 cells (Fig. 1F). V5–AKAP79 was detected in anti-FLAG immune complexes (Fig. 1F, lane 2), but not in control IgG immunoprecipitates (Fig. 1F, lane 1).
More sophisticated chemical cross-linking approaches were used to map the AKAP79 dimerization interface. Addition of the lysine-specific cross-linker disuccinimidyl suberate (DSS) to purified Strep–AKAP79 produced a species of approximately twice the apparent molecular weight, as detected by immunoblotting (Fig. 1G). Low levels of trimeric and higher oligomeric species of AKAP79 were also stabilized by addition of DSS (Fig. 1G). Detection of the 150-kDa species was more prominent with 500-μM DSS (Fig. 1G, lanes 2 and 3; Coomassie staining is shown in Fig. S3D). For mapping studies, Strep–AKAP79 was treated with DSS, and trypsinized peptides were separated by liquid chromatography. MS was used to search for peptides containing cross-linked lysine residues using the xQuest algorithm (18). Two peptides containing homodimeric cross-links corresponding to either K328–K328 or K333–K333 conjugates were identified (Fig. 1H, Fig. S4, and Table S2). The detection of symmetrical cross-linked pairs suggests that the AKAP79 dimer is aligned in parallel. DSS contains amine-reactive NHS esters at each end of an 11.4-Å spacer, so residues 328–333 on each protomer are proximal to each other (Fig. 1D, asterisks). Taken together, the results in Fig. 1 suggest that AKAP79 exists as a dimer with P2 symmetry, with a principal dimerization site located close to the PIAIIIT motif.
The AKAP79–PKA–PP2B–CaM Complex Has 2∶4∶4∶2 Stoichiometry.
Although AKAP79 is named for its ability to anchor PKA, it also targets PP2B and PKC. PKC binds to a pseudosubstrate sequence (residues 32–52) adjacent to the membrane-binding regions of AKAP79 (19). In contrast, the PP2B–AKAP79 interface is less well defined. Although the amino terminus of the anchoring protein can modulate phosphatase activity (9), residues 315–360, which includes the PIAIIIT motif, are important for PP2B binding (20). Other studies have shown that a related PP2B anchoring peptide (PVIVIT) binds to strand β14 of the phosphatase catalytic domain on the face opposite to the active site (21).
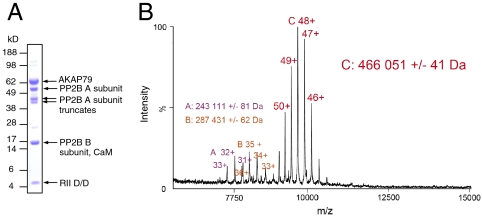
To investigate the molecular basis of phosphatase anchoring, we reconstituted an AKAP79 complex containing PP2B heterodimers. Two steps were added to the basic purification protocol: an excess of purified PP2B was added prior to the nickel affinity step (Fig. 1A, i), and an excess of purified CaM was added in the presence of 0.2 mM CaCl2 prior to gel filtration (Fig. 1A, ii). Fractions that eluted from a Superdex S200 gel filtration column between 64–68 mL were pooled (Fig. 2A and Fig. S2B) and concentrated prior to analysis by MS. PP2B contains labile regulatory elements C-terminal to the phosphatase domain—the bands between 38 and 49 kDa in Fig. 2A were confirmed to be PP2B truncation products by MS analysis of tryptic digests.
Fig. 2.
MS of the AKDPC complex. (A) Fractions 64–68 mL of the gel filtration elution (Fig. S2B) were pooled, concentrated, separated by 4–12% SDS-PAGE, and Coomassie-stained to show the proteins in the AKDPC complex. (B) MS spectrum of the AKDPC complex from A.
We applied nanoelectrospray MS to the AKAP79–PP2B–D/D–CaM(Ca2+) complex (hereafter “AKDPC”—AKAP79, PKAD/D, PP2B, CaM) in order to determine its subunit stoichiometry. The AKDPC complex was buffer exchanged into 50 mM ammonium acetate pH7.5 + 0.1 M CaCl2 and transferred successfully into the gas phase. The MS spectrum of the complex is shown in Fig. 2B. The prevalent series of peaks at the highest m/z range, with charge states between 9,000 to 10,500 m/z, was measured as 466,051 Da (Fig. 2B). The spectrum contains two other series of greater than 200 kDa, around 8,000 m/z, measured at 243,339 and 287,865 Da, that are not readily interpreted and likely contain truncated or modified forms of one or more proteins.
To evaluate possible assignments of the 466,051-Da AKDPC complex, we calculated the masses of all potential combinations of interactions between the different subunits based on the masses predicted by their amino acid sequences (listed in Table S1), using software developed in-house (22). We reasoned that AKAP79 forms a dimer associated with 4 ∗ D/D on the basis of our earlier experiments. Assignments within 0.5% of the measured mass are listed in Table S3. The optimal assignment of a complex, with a predicted mass of 465,854 Da, is 2 ∗ [AKAP79 + 2 ∗ PP2B(A + B subunits) + 2 ∗ D/D + CaM] (Table 1). To account for this stoichiometry, it is likely that the complex is symmetrical, with each protomer of the AKAP79 dimer binding two PP2B heterodimers, a D/D dimer and one CaM.
Table 1.
Stoichiometry of the AKDPC complex
| Protein | Predicted mass, Da | Copy number |
| RIIα D/D | 5,624 | 4 |
| PP2B A subunit | 58,070 | 4 |
| PP2B B subunit (5–170) | 18,915 | 4 |
| AKAP79 | 50,460 | 2 |
| CaM | 17,249 | 2 |
| Predicted total mass of | ||
| 2 ∗ [AKAP79 + 2 ∗ PP2B(A + B) + 2 ∗ D/D + CaM] | 465,854 Da | |
The optimal subunit assignment for the AKDPC complex is shown.
PP2B Engages the N Terminus of AKAP79 in a Ca2+/CaM-Dependent Manner.
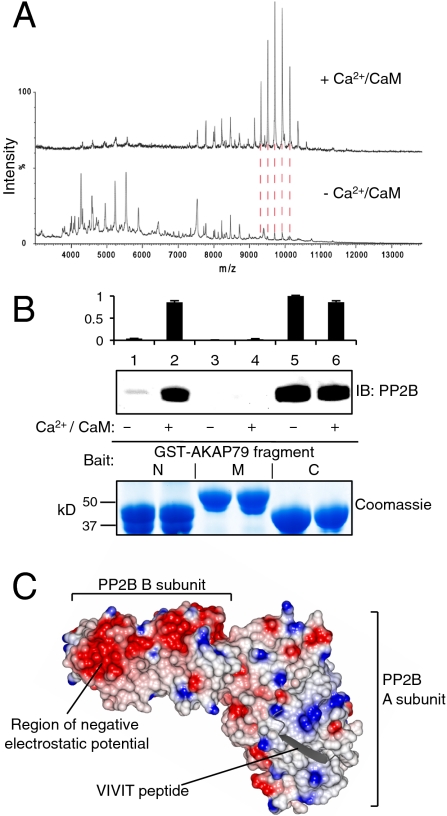
MS spectra of AKAP79–PP2B–D/D complexes are shown under two conditions (Fig. 3A). In the presence of Ca2+/CaM, the 466-kDa complex is prominent (Fig. 3A, Upper). Yet, in the absence of Ca2+/CaM, a relatively high proportion of single subunits and low molecular mass complexes are observed (Fig. 3A, Lower). This indicates that Ca2+/CaM stabilizes the AKAP79–PP2B–D/D complex.
Fig. 3.
Identification of a Ca2+/CaM-sensitive PP2B binding site in AKAP79. (A) MS spectrum of AKDPC complexes isolated by gel filtration in both the presence (Upper) and absence (Lower) of Ca2+/CaM. Red dashed lines highlight peak alignment. (B) Anti-FLAG IB of PP2B–FLAG pulled-down with GST fusion proteins containing indicated AKAP79 fragments in the presence or absence of Ca2+/CaM. Relative densitometry analyis (n = 3, Top) and pull-down Coomassie (Bottom) are shown. (C) PP2B electrostatic surface. Areas of positive and negative electrostatic potential are colored red and blue, respectively. VIVIT peptide is in gray.
Given the ability of Ca2+/CaM to stabilize the 466-kDa complex, we revisited PP2B mapping using GST–AKAP79 fusion proteins. Pull-down of purified FLAG-tagged PP2B (Fig. S5A) was performed with each AKAP79 fragment. Surprisingly, the N-terminal fragment of AKAP79 interacted with PP2B in a Ca2+/CaM-dependent manner (Fig. 3B, lanes 1 and 2). The middle region of AKAP79 did not interact with the phosphatase (Fig. 3B, lanes 3 and 4). However, PP2B bound to the C-terminal fragment of AKAP79 in a Ca2+/CaM-independent manner, as would be expected given that this fusion protein encompasses the PIAIIIT motif (Fig. 3B, lanes 5 and 6). Taken together, these results demonstrate that Ca2+ and CaM are both required for PP2B interaction with the N-terminal region of AKAP79 (Fig. S5C).
By analogy to the crystal structure of PP2B in complex with the PVIVIT anchoring peptide (21), we can offer two feasible explanations for the Ca2+/CaM-dependent association of PP2B with AKAP79. First, CaM binding exposes an additional surface on the anchoring protein (aside from the PIAIIIT motif) to permit bipartite binding with PP2B. A likely candidate is a polybasic region between residues 38–131 of AKAP79 (Fig. S1). This positively charged surface could electrostatically interact with a reciprocal region of negative charge on the phosphatase (Fig. 3C). Second, Ca2+/CaM may act as a molecular bridge to stabilize the AKAP79–PP2B interface.
Mechanics of AKAP79-Anchored PP2B Activation by CaM.
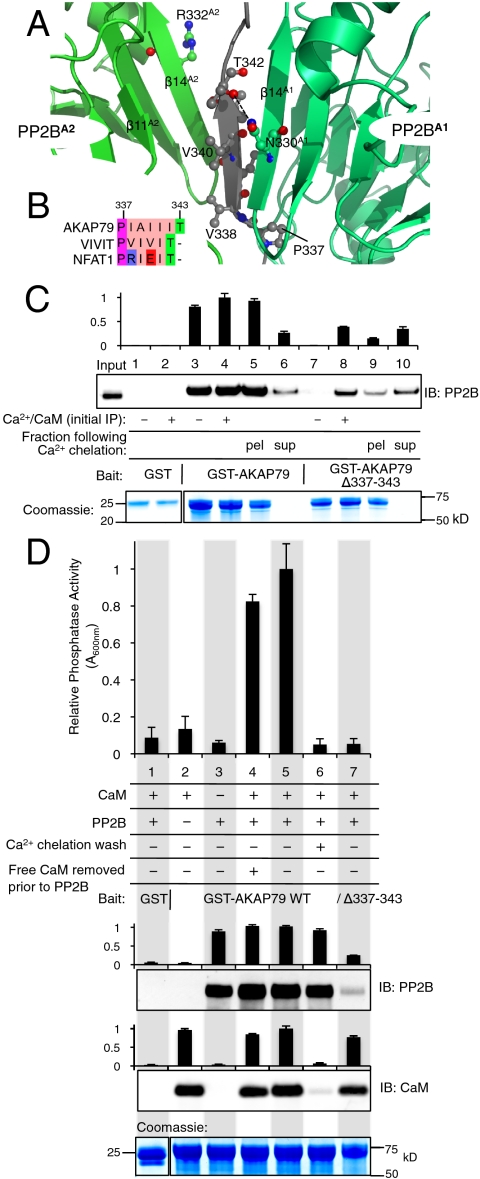
The PP2B–PVIVIT complex crystal structure (20) also offers a structural explanation for the unexpected ratio of one AKAP79 to two PP2B heterodimers. The conformation of the PVIVIT peptide is a parallel β-sheet with hydrophobic side chains alternately protruding on either face. Hence, the same β-sheet can simultaneously contact two A subunits of PP2B (denoted A1 and A2, Fig. 4A). It would appear that the PIAIIIT motif in AKAP79 adopts a similar conformation and retains the ability to concurrently and symmetrically engage two molecules of PP2B. On the contrary, a corresponding region (PRIEIT) in nuclear factor of activated T-cells (NFAT) that lacks hydrophobic residues on one face only associates with a single molecule of the phosphatase (Fig. 4B). Despite the convenience of such a relevant protein–protein interface, we could not exclude the possibility of an asymmetrical binding mechanism in which one PP2B binding event is Ca2+/CaM-dependent and the other phosphatase apoenzyme tonically associates with the PIAIIIT motif.
Fig. 4.
Molecular basis of PP2B anchoring by AKAP79. (A) Close-up view of the interface between the peptide PVIVIT (gray) and two copies of the PP2B A subunit, denoted A1 (dark green) and A2 (light green) in the crystal structure (Protein Data Bank ID code 2P6B) (21). PVIVIT residues are labeled according to the inset alignment (B) with homologous sequences in AKAP79 and NFAT1. (C) Anti-FLAG IB of PP2B–FLAG pulled-down by GST–AKAP79 or GST–AKAP79Δ337–343. EDTA/EGTA-mediated release of PP2B–FLAG is shown by IB and quantified by densitometry (n = 3, Top). Pull-down with GST, GST–AKAP79 and GST–AKAP79Δ337–343 was performed in the presence or absence of Ca2+/CaM as indicated (lanes 1–4, 7, and 8). Lanes 5 and 6, and 9 and 10 show release of PP2B–FLAG from GST–AKAP79 and GST–AKAP79Δ337–343 pull-downs (Ca2+/CaM) by addition of 3 mM EDTA and 3 mM EGTA (pellet vs. supernatant). Pull-down Coomassie is shown (Bottom). (D) The untagged PP2B activity associated with GST, GST–AKAP79 and GST–AKAP79Δ337–343 was monitored under various conditions that modified the Ca2+ and CaM concentrations before or after anchoring of the phosphatase to AKAP79. Phosphatase activity was measured spectrometrically by phosphate release from the phosphopeptide RRA(pT)VA (Top, n = 3). Anti-PP2B and anti-CaM immunoblots for the GST pull-downs are shown and quantified by densitometry (Middle). Pull-down Coomassie is shown (Bottom).
The next phase of this work was to delineate between the symmetrical and asymmetrical anchoring models (see Fig. S6). According to the symmetrical model, all PP2B remains associated with AKAP79 (Fig. S6A). Yet, in an asymmetrical configuration, deactivation of Ca2+/CaM leads to the release of one PP2B apoenzyme per AKAP79 protomer (Fig. S6 B and C). In order to distinguish between these models, we probed for PP2B in the fraction released from AKAP79 after chelating Ca2+ with 3 mM EDTA and 3 mM EGTA by immunoblot (Fig. 4C). As a prelude to these studies, control experiments confirmed that PP2B did not bind to GST regardless of Ca2+/CaM (Fig. 4C, lanes 1 and 2). Moreover, full-length GST–AKAP79 bound PP2B in the absence of Ca2+/CaM (Fig. 4C, lane 1). As anticipated, a GST–AKAP79Δ337–343 mutant that lacks the PIAIIIT motif was unable to interact with the phosphatase (Fig. 4C, lane 7). This implies that PP2B constitutively binds the PIAIIIT motif.
Next, we probed PP2B interaction with GST–AKAP79 and the GST–AKAP79Δ337–343 mutant upon the chelation of Ca2+. PP2B bound to both AKAP79 forms in the presence of Ca2+/CaM (Fig. 4C, lanes 4 and 8). Yet, upon removal of Ca2+ with EDTA/EGTA, the phosphatase was only released from the AKAP79Δ337–343 mutant (Fig. 4C, lane 10). These results support the symmetrical binding model for PP2B–AKAP79 interaction.
There are six potential CaM binding sites in the AKDPC complex, yet only two copies of CaM are present (Fig. 2B). Therefore, it was necessary to investigate the mechanism of anchored PP2B activation by CaM. Purified PP2B was incubated with GST–AKAP79 in the presence of an excess of CaM supplemented with 3 mM CaCl2 (Fig. S5B and Fig. 4D). Phosphatase activity was measured spectrophotometrically using the phospho-substrate peptide RRA(pT)VA. Phosphatase activity was negligible when CaM or PP2B were omitted from the reaction mixture (Fig. 4D, lanes 2 and 3). Likewise, there was no detectable phosphatase activity in pull-downs with either GST or an AKAP79 mutant lacking the PIAIIIT motif (Fig. 4D, lanes 1 and 7). However, robust phosphatase activity was measured when PP2B and an excess of Ca2+/CaM were incubated with AKAP79 (Fig. 4D, lane 5). PP2B activity was sustained when excess Ca2+/CaM was removed prior to the addition of the phosphatase (Fig. 4D, lane 4). All AKAP79-associated phosphatase activity was suppressed when CaM was released by Ca2+ chelation in 3 mM EGTA + 3 mM EDTA (Fig. 4D, lane 6). Taken together, these results imply that CaM associated with AKAP79 is able to activate anchored PP2B within the same signaling complex.
Discussion
A combination of protein chemistry and MS approaches has enabled us to determine the quaternary structure of an AKAP signaling complex. Our studies have uncovered three unexpected biochemical properties that change our understanding of how this signaling complex operates. First, AKAP79 is a dimer. Second, each AKAP79 protomer has the capacity to tether a single PKA holoenzyme and two PP2B heterodimers. Third, binding of Ca2+/CaM reorganizes each half of the AKAP79 complex to activate the anchored phosphatases. This latter property may be particularly relevant to excitatory synaptic transmission because the phosphoregulation of glutamate receptor ion channels is governed by calcium influx across the postsynaptic membrane (23). In a broader context, the assembly of multiple kinases and phosphatases in this manner provides a means to generate concentrated pockets of second messenger responsive enzyme activity on the inner face of the plasma membrane.
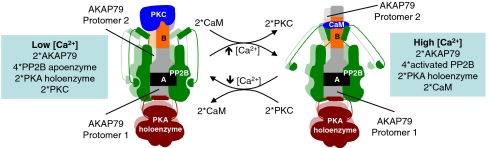
A model for the molecular architecture and [Ca2+]-dependent dynamics of this AKAP79 signaling complex is illustrated in Fig. 5. Two AKAP79 protomers are arranged in parallel through contacts that include a region in the vicinity of K328 and K333. This site is located immediately upstream of the PIAIIIT motifs that are responsible for anchoring PP2B (20). Thus, we propose that under conditions of low cellular [Ca2+], each AKAP protomer anchors two inactive PP2B apoenzymes, principally through the PIAIIIT motif. A rise in intracellular [Ca2+] leads to binding of one molecule of CaM to polybasic regions located between residues 31–52 of each AKAP79 protomer (24). This both releases associated PKC and exposes a second interface that promotes the activation of anchored PP2B. This would drive the AKAP79 complex into a mode that would favor the Ca2+-dependent dephosphorylation of synaptic substrates. Because Ca2+ levels fluctuate at synapses, it is likely that this CaM-dependent activation of anchored PP2B is transient. Thus, we imagine that decreases in cellular [Ca2+] promotes a release of CaM and the concomitant reassociation of PKC. These mutually exclusive binding events will return the AKAP79 signaling complex to a ground state with basal phosphatase activity (Fig. 5).
Fig. 5.
Model of AKAP79 anchoring dynamics. A proposed model for the architecture and dynamics of the core AKAP79 signaling complex, in conditions of both high and low [Ca2+] is illustrated. The second AKAP79 protomer and associated proteins are in the background.
Our updated model of AKAP79 architecture is principally derived from the data in this study, but remains consistent with earlier observations by other investigators. For example, in the PP2B–PVIVIT peptide crystal structure, the authors demonstrated a stoichiometric ratio of two PP2B molecules per phosphatase targeting region of the transcription factor NFAT (21). Hogan and coworkers considered this to be an artifactual consequence of crystal packing (21). However, their empirical findings are upheld by our study because we provide evidence for another example where two molecules of the phosphatase simultaneously interact with a single PP2B-targeting protein. Furthermore, the data presented in Figs. 3 and 4 provide mechanistic details that explain previous mapping studies of PP2B binding to an amino terminal region of the anchoring protein (9).
An accepted role for anchoring proteins is to spatially direct a given cellular signal toward compartmentalized substrates. One concept introduced in this study is the notion that Ca2+ and cAMP responsive AKAP79 binding partners are attached to different regions of the anchoring protein. Our data predicts that PP2B and PKC are anchored in immediate proximity to the membrane (the entry point of Ca2+, the second messenger to which they respond). This configuration would make it possible for the myristoyl groups on the B subunits of anchored PP2B heterodimers (25) to act in concert with the MBBRs of AKAP79 to secure the complex to plasma membrane (Fig. 5 Left). However, one must also recognize that additional protein-–protein interactions must participate in microcompartmentalization of the signaling network, because AKAP79 also interacts with membrane-associated guanylate kinases (MAGUKs) that are attached to the cytoplasmic tails of NMDA receptors (26). On the other hand, the anchoring sites for PKA are predicted to be located further from the membrane (Fig. 5). AKAP79 also interacts directly with type V/VI adenylyl cyclases (27). The active site of adenylyl cyclase isoforms is located on an intracellular loop that protrudes into the cytoplasm (28). This could potentially create a microenvironment in which enzymes that both generate and respond to cAMP are clustered by AKAP79 yet still retain access to the cytoplasm.
In conclusion, we anticipate that approaches described in this report will be applicable to the study of other AKAP complexes. Many AKAPs support intricate signaling networks that incorporate multiple protein components (5, 29). Conventional structural analyses have been hampered by the apparent flexibility of AKAPs and phosphatase-targeting subunits, which often lack predicted secondary and higher structure (30). The only existing crystal structure of a phosphatase-scaffold complex to date is of the PP1 catalytic subunit and the myosin phosphatase targeting subunit, in which a remarkable 2,560 Å2 of surface are buried (31). Likewise, it appears that the PP2B–AKAP79 interaction involves more contacts than were previously anticipated. We now know that this includes CaM-regulated sites in the amino terminal regions of the anchoring protein and the C-terminal PIAIIIT motif. It will be interesting to explore whether this structural information will aid the design of pharmacological agents that selectively modulate CaM-sensitive action of the anchored phosphatase.
Experimental Procedures
Expression and Purification of AKAP79 Complexes.
Human PKA RIIα(1–45), mouse CaM and a bicistron containing human PP2B, adapted from ref. 32, were each cloned into pGEX6P1 for expression in Escherichia coli BL21 cells. AKAP79 was expressed in Sf21 insect cells using the Bac-to-Bac Baculovirus System (Invitrogen). Bacterial lysates were prepared in 25 mM Tris pH 7.5, 0.3–0.5 M NaCl, 2 mM DTT, 0.5 mM EDTA, one EDTA-free protease inhibitor tablet/100 mL (Roche), 0.05 mg/mL DNase 1, 0.1 mg/mL lysozyme. AKAP79 lysate was prepared by homogenization of frozen pellet in 25 mM Tris pH 7.5, 0.3 M NaCl, 2 mM DTT, 0.5 mM EDTA, one EDTA-free protease inhibitor tablet/100 mL. AKAP79 complexes were released from glutathione sepharose beads by incubation with PreScission protease (GE Healthcare). Eluates were desalted into nickel buffer A (25 mM Tris pH 8, 0.3 M NaCl, 15 mM imidazole) prior to incubation with nickel-NTA beads (Qiagen) and subsequently eluted in 25 mM Tris pH 7.5, 0.3 M NaCl, 300 mM imidazole. Superdex S200 (GE Healthcare) gel filtration was performed in 20 mM Na Hepes pH 7.5, 200 mM NaCl, supplemented with excess CaM and 0.2 mM CaCl2 in the case of AKDPC gel filtration.
Chemical Cross-Linking and Cross-Linked Lysine Identification.
Strep–AKAP79, purified from Hi5 insect cells, was incubated at 0.5 mg/mL in 20 mM HEPES, 250 mM NaCl (pH 7.5) for 30 min with between 0–1 mM DSS (Sigma). Strep–AKAP79 cross-linked with 1 mM DSS was trypsinized, and the peptides were separated by liquid chromatography using a nanoAcquity UPLC (Waters). MS was performed using a dual linear ion trap Fourier transform ion cyclotron resonance mass spectrometer, the Velos-FT (SI Methods). Data was searched using xQuest software. All hits were filtered using accurate mass tolerance of 10 ppm for precursor m/z and a tolerance of 0.1 Da for MSMS data. The only cross-linked peptide candidates possible in the sample based on accurate mass measurements (Table S2) coincided with identifications obtained using xQuest (Fig. S4).
Protein Coimmunoprecipitation in HEK293 Cells.
Two pcDNA3.1 vectors were constructed enabling transient expression of AKAP79, with either a FLAG or V5 tag at its N terminus, in HEK293 cells. The cells were harvested 2 d after transfection in 50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM benzamidine, 2 μg/mL LeuPeptin, and FLAG–AKAP79 was immunoprecipitated with either 4 μg mouse anti-FLAG antibody or 4 μg mouse IgG (Sigma). FLAG–AKAP79 was detected with αFLAG–HRP conjugate (Sigma); V5–AKAP79 was detected with αV5–HRP conjugate (Invitrogen).
Pull-Down Assays.
In each pull-down experiment, 40 μL of glutathione sepharose beads saturated with the appropriate GST fusion protein were used to pull-down 2 μg PP2B–FLAG in 200 μL radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM benzamidine, 2 μg/mL (LeuPeptin)]. When testing the effect of Ca2+/CaM, 3 mM CaCl2 and 10 μg purified CaM was included. After 2 h, the beads were washed in 5 × 1 mL RIPA buffer. RIPA buffer was supplemented with 3 mM CaCl2 when testing the effect of Ca2+/CaM on pull-down. Beads were resuspended in SDS-PAGE loading buffer for immunoblotting. For EDTA-mediated release experiments, 50 μL of RIPA buffer supplemented with 3 mM EDTA and 3 mM EGTA was added, and after 30 min the supernatant fraction was separated from the sepharose resin by centrifugation in a spin column.
Pull-Down Protein Phosphatase Assays.
In the default protocol, pull-down of 2 μg high-purity PP2B (Fig. S5B) was attempted with 40 μL of glutathione sepharose beads saturated with GST fusion protein in 250 μL of RIPA buffer supplemented with 3 mM CaCl2 and 25 μg purified CaM. Following 2 h of incubation, pull-downs were washed in 4 × 1 mL RIPA supplemented with 3 mM CaCl2. In one case, CaM was incubated with beads bearing GST–AKAP79FL for 2 h, and CaM that had not associated with AKAP79 was removed by washing with 4 × 1 mL RIPA (+3 mM CaCl2) prior to the PP2B incubation step (Fig. 4D, lane 4). The beads were subsequently washed for a second time in the normal way with 4 × 1 mL RIPA (+3 mM CaCl2) to remove unbound PP2B. In another case, a single wash step with 1 mL of RIPA buffer supplemented with 3 mM EDTA and 3 mM EGTA was included following the final 4 × 1 mL washes in RIPA +3 mM CaCl2 (Fig. 4D, lane 4). In every case, the beads were subject to a final wash in 50 mM imidazole pH 7.2, 50 mM MgCl2, 5 mM NiCl2, 1 mM CaCl2prior to phosphatase assay. Phosphatase activity was determined by quantifying phosphate release following 30-min incubation of pull-downs with 100 μM RRA(pT)VA phosphopeptide (Promega Phosphatase Assay System) in 50 mM imidazole pH 7.2, 50 mM MgCl2, 5 mM NiCl2, 1 mM CaCl2.
Electrospray MS.
Prior to MS analysis, AKAP79 complexes were dialysed into 50 mM ammonium acetate pH 7.5 ± 0.1 M CaCl2. Mass spectra were obtained on a Q-Tof 2 or a Q-Tof 2B (both Waters/Micromass UK Ltd.), modified for high-mass operation (33), using a previously described protocol to preserve noncovalent interactions (34).
Structural representation.
Cartoon representations were generated using PyMol (DeLano Scientific), and the electrostatic surface of PP2B was rendered using CCP4MG (35).
Supplementary Material
Acknowledgments.
The authors thank L. Langeberg for critical evaluation of this work, M. Milnes for assistance in preparation of the manuscript, and the Institute of Cancer Research baculovirus facility for technical assistance. M.G.G. is a Sir Henry Wellcome Postdoctoral Research Fellow. C.V.R. is a Royal Society Professor. The research was funded through a Cancer Research UK grant to D.B.; National Institutes of Health Grant 5R01GM048231 to J.D.S.; National Institutes of Health Grants 7S10RR025107, 5R01GM086688, and 5R01RR023334 to J.E.B.; National Institutes of Health T32 Training Grant GM07750 to P.J.N.; the University of Washington Proteomics Resource (UWPR95794); and was supported by the PROSPECTS grant within the Research Framework Program of the European Commission to C.V.R.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014400108/-/DCSupplemental.
References
- 1.Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–1091. [PubMed] [Google Scholar]
- 2.Keely SL. Activation of cAMP-dependent protein kinase without a corresponding increase in phosphorylase activity. Res Commun Chem Pathol Pharmacol. 1977;18:283–290. [PubMed] [Google Scholar]
- 3.Bacskai BJ, et al. Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science. 1993;260:222–226. doi: 10.1126/science.7682336. [DOI] [PubMed] [Google Scholar]
- 4.Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 5.Dodge-Kafka KL, et al. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parekh AB. Ca2+ microdomains near plasma membrane Ca2+ channels: Impact on cell function. J Physiol. 2008;586:3043–3054. doi: 10.1113/jphysiol.2008.153460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr DW, Stofko-Hahn RE, Fraser ID, Cone RD, Scott JD. Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J Biol Chem. 1992;267:16816–16823. [PubMed] [Google Scholar]
- 8.Dell’Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5-bisphosphate. EMBO J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coghlan VM, et al. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 10.Klauck TM, et al. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 11.Oliveria SF, Dell’Acqua ML, Sather WA. AKAP79/150 anchoring of calcineurin controls neuronal L-type Ca2+ channel activity and nuclear signaling. Neuron. 2007;55:261–275. doi: 10.1016/j.neuron.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tunquist BJ, et al. Loss of AKAP150 perturbs distinct neuronal processes in mice. Proc Natl Acad Sci USA. 2008;105:12557–12562. doi: 10.1073/pnas.0805922105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshi N, et al. AKAP150 signaling complex promotes suppression of the M-current by muscarinic agonists. Nat Neurosci. 2003;6:564–571. doi: 10.1038/nn1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold MG, et al. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol Cell. 2006;24:383–395. doi: 10.1016/j.molcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Kinderman FS, et al. A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol Cell. 2006;24:397–408. doi: 10.1016/j.molcel.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heck AJ. Native mass spectrometry: A bridge between interactomics and structural biology. Nat Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 17.Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu Rev Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 18.Rinner O, et al. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoshi N, Langeberg LK, Gould CM, Newton AC, Scott JD. Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol Cell. 2010;37:541–550. doi: 10.1016/j.molcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dell’Acqua ML, Dodge KL, Tavalin SJ, Scott JD. Mapping the protein phosphatase-2B anchoring site on AKAP79. Binding and inhibition of phosphatase activity are mediated by residues 315–360. J Biol Chem. 2002;277:48796–48802. doi: 10.1074/jbc.M207833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Zhang L, Rao A, Harrison SC, Hogan PG. Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction. J Mol Biol. 2007;369:1296–1306. doi: 10.1016/j.jmb.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 22.Taverner T, et al. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc Chem Res. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharyya S, Biou V, Xu W, Schluter O, Malenka RC. A critical role for PSD-95/AKAP interactions in endocytosis of synaptic AMPA receptors. Nat Neurosci. 2009;12:172–181. doi: 10.1038/nn.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faux MC, Scott JD. Regulation of the AKAP79-protein kinase C interaction by Ca2+/Calmodulin. J Biol Chem. 1997;272:17038–17044. doi: 10.1074/jbc.272.27.17038. [DOI] [PubMed] [Google Scholar]
- 25.Perrino BA, Martin BA. Ca(2+)- and myristoylation-dependent association of calcineurin with phosphatidylserine. J Biochem. 2001;129:835–841. doi: 10.1093/oxfordjournals.jbchem.a002927. [DOI] [PubMed] [Google Scholar]
- 26.Colledge M, et al. Targeting of PKA to glutamate receptors through a MAGUK-AKAP complex. Neuron. 2000;27:107–119. doi: 10.1016/s0896-6273(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 27.Bauman AL, et al. Dynamic regulation of cAMP synthesis through anchored PKA-adenylyl cyclase V/VI complexes. Mol Cell. 2006;23:925–931. doi: 10.1016/j.molcel.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 29.Smith FD, et al. AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nat Cell Biol. 2010;12:1242–1249. doi: 10.1038/ncb2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gold MG, Smith FD, Scott JD, Barford D. AKAP18 contains a phosphoesterase domain that binds AMP. J Mol Biol. 2008;375:1329–1343. doi: 10.1016/j.jmb.2007.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 32.Mondragon A, et al. Overexpression and purification of human calcineurin alpha from Escherichia coli and assessment of catalytic functions of residues surrounding the binuclear metal center. Biochemistry. 1997;36:4934–4942. doi: 10.1021/bi9631935. [DOI] [PubMed] [Google Scholar]
- 33.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 34.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 35.Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.