Abstract
The SNF1 protein kinase of Saccharomyces cerevisiae is a member of the SNF1/AMP-activated protein kinase family, which is essential for metabolic control, energy homeostasis, and stress responses in eukaryotes. SNF1 is activated in response to glucose limitation by phosphorylation of Thr210 on the activation loop of the catalytic subunit Snf1. The SNF1 β-subunit contains a glycogen-binding domain that has been implicated in glucose inhibition of Snf1 Thr210 phosphorylation. To assess the role of glycogen, we examined Snf1 phosphorylation in strains with altered glycogen metabolism. A reg1Δ mutant, lacking Reg1-Glc7 protein phosphatase 1, exhibits elevated glycogen accumulation and phosphorylation of Snf1 during growth on high levels of glucose. Unexpectedly, mutations that abolished glycogen synthesis also restored Thr210 dephosphorylation in glucose-grown reg1Δ cells, indicating that elevated glycogen synthesis contributes to activation of SNF1 and that another phosphatase acts on Snf1. We present evidence that Sit4, a type 2A-like protein phosphatase, contributes to dephosphorylation of Snf1 Thr210. Finally, evidence that the effects of glycogen are not mediated by binding to the β-subunit raises the possibility that elevated glycogen synthesis alters glucose metabolism and thereby reduces glucose signaling to the SNF1 pathway.
Keywords: glucose regulation, signal transduction, yeast
The SNF1/AMP-activated protein kinase (AMPK) family is conserved from yeast to mammals and is essential for metabolic control and energy homeostasis (1, 2). The Saccharomyces cerevisiae SNF1 protein kinase is required for adaptation to glucose limitation and other stresses and for growth on carbon sources that are less preferred than glucose; it is named for the sucrose-nonfermenting phenotype of the snf1 mutant (2). SNF1 regulates transcription of a large set of genes (3) and the activity of metabolic enzymes involved in carbohydrate storage and fatty acid metabolism (4–6).
Members of the SNF1/AMPK family are heterotrimers composed of a catalytic α-subunit and regulatory β- and γ-subunits. The kinase is activated by phosphorylation of the conserved Thr residue in the activation loop of the catalytic subunit. In mammals, AMP binds to the γ-subunit (7, 8) and inhibits the dephosphorylation of the activation loop Thr (9). In S. cerevisiae, the γ-subunit of SNF1 has not been shown to bind nucleotide. Thr210 of the Snf1 catalytic subunit is phosphorylated in response to glucose limitation and other stresses (10, 11) by the Snf1-activating kinases Sak1, Tos3, and Elm1 (12–14). Reg1-Glc7 protein phosphatase 1 (PP1), comprising the regulatory subunit Reg1 (15) and the catalytic subunit Glc7, has been implicated in dephosphorylation of Thr210. Reg1 interacts physically with Snf1 (16, 17), and analysis of reg1Δ mutants indicated that Reg1 is required for maintaining Thr210 in the dephosphorylated state (10) and inhibiting SNF1 catalytic activity (18) during growth on high glucose. Studies suggested that dephosphorylation of Thr210 in response to addition of glucose is regulated at the level of access of Reg1-Glc7 to the activation loop (19).
S. cerevisiae encodes three alternate β-subunits, Sip1, Sip2, and Gal83, that affect interactions with substrates (20, 21) and confer specific subcellular localization of SNF1 (22). The β-subunits of SNF1 and AMPK contain a glycogen-binding domain (GBD) (23, 24) that is a member of the carbohydrate-binding module family (25). For mammalian AMPK, it has been reported that, in vitro, the binding of glycogen to the GBD inhibits the activity of AMPK and its phosphorylation by activating kinases (26). The GBD sequence is conserved in Gal83 and Sip2, except for substitution of a highly conserved aromatic residue with Leu, but is less conserved in Sip1 (23, 25). In vitro, Gal83 is able to bind glycogen, whereas Sip2 does so weakly (23). Glycogen is a major storage carbohydrate, and in glucose-grown cultures, glycogen is synthesized during late exponential phase dependent on SNF1 activity (4, 27) and is used after cells enter stationary phase (28). Deletion of the GBD from Gal83 relieved glucose inhibition of SNF1; in cells expressing Gal83ΔGBD as the only β-subunit, Snf1 Thr210 was highly phosphorylated during exponential growth on high levels of glucose (29). This phenotype most likely results from the absence of the GBD rather than loss of glycogen binding, because glycogen is low under these growth conditions, and cells with WT Gal83 but lacking glycogen synthase showed normal regulation of SNF1 (29). However, we considered the possibility that, under conditions when glycogen levels are high, the binding of glycogen could displace the GBD relative to the SNF1 heterotrimer, thereby relieving inhibition; according to this view, deletion of the GBD would mimic displacement.
To assess the role of glycogen in regulation of SNF1, we examined strains with altered glycogen metabolism, including the reg1Δ mutant, which exhibits elevated glycogen accumulation (15, 30, 31). Unexpectedly, we found that abolishing glycogen synthesis in the reg1Δ mutant restored dephosphorylation of Snf1 Thr210 during exponential growth on high levels of glucose. These findings implicate glycogen synthesis in regulation of SNF1 and indicate that Reg1-Glc7 phosphatase is not solely responsible for dephosphorylation of Thr210. We present evidence for a role of Sit4, a type 2A-like protein phosphatase, in the regulation of SNF1.
Results
Genetic Alterations of Glycogen Metabolism Do Not Affect Snf1 Thr210 Phosphorylation.
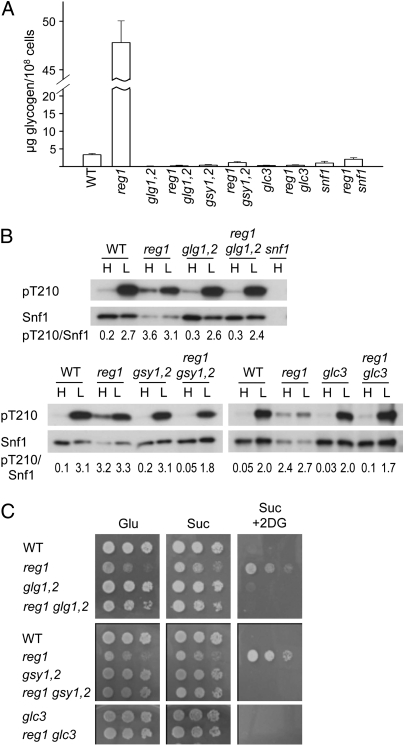
We first examined the regulation of Snf1 Thr210 phosphorylation in cells with elevated glycogen levels during exponential growth in glucose. Mutants lacking the degradative enzymes glycogen phosphorylase (Gph1) or glycogen debranching enzyme (Gdb1) and cells overexpressing glycogen synthase Gsy2 accumulated glycogen (Fig. 1 A and C). For assays of Thr210 phosphorylation, cells were grown in 2% glucose and collected by rapid filtration to preserve the phosphorylation state of Snf1, and an aliquot was subjected to glucose depletion by resuspension in 0.05% glucose for 10 min. Extracts were prepared, and phosphorylation was assayed by immunoblot analysis. In all cases, Thr210 was dephosphorylated in glucose-grown cells and became phosphorylated in response to glucose limitation (Fig. 1 B and D), indicating that the accumulation of glycogen to the levels achieved in these cells had no effect. Glycogen-deficient mutants lacking the self-glucosylating initiator proteins Glg1 and Glg2, the glycogen synthases Gsy1 and Gsy2, or the branching enzyme Glc3 also showed normal regulation of Thr210 phosphorylation (Fig. 1 A and B).
Fig. 1.
Regulation of Snf1 Thr210 phosphorylation in strains with altered glycogen metabolism. (A and B) Strains were WT and mutants, as indicated, in the W303 (Left) or BY4741 genetic background (Right); similar results were obtained for all mutants in both genetic backgrounds. (A) Glycogen content was determined. Values are averages of six determinations (bars show SD). (B) Cells were grown in SC plus 2% (H, high) glucose, and an aliquot of the culture was collected by rapid filtration. Another aliquot was collected, resuspended in SC plus 0.05% (L, low) glucose for 10 min, and collected. Extracts were prepared, and proteins (10 μg) were analyzed by immunoblotting with antiphospho-Thr-172-AMPK antibody to detect phosphorylation on Thr210 of Snf1 (pT210). Lower exposure did not reveal differences in phosphorylation in low glucose. Membranes were reprobed with antipolyhistidine antibody to detect Snf1. (C and D) W303-1A cells carrying vector pCM252 or pGSY2 expressing Gsy2-3xHA from the doxycycline-inducible tetO7 promoter (57) were grown overnight in selective SC plus 2% glucose, diluted in fresh media to an OD600 of 0.2 in the presence (+) or the absence (−) of 2 μg/mL doxycycline (dox), and grown for 6 h. Control cultures were treated with the drug vehicle (ethanol). Similar results were obtained with induction times up to 16 h. (C) Glycogen content was determined for induced cultures as above. (D) Aliquots were collected for analysis of phosphorylated Thr210 (pT210) and Snf1. Gsy2-3xHA was detected with 12CA5 antibody.
Abolishing Glycogen Synthesis Restores Dephosphorylation of Thr210 in the reg1Δ Mutant.
The reg1Δ mutant, lacking Reg1-Glc7 PP1, exhibits constitutive phosphorylation and activation of SNF1 (10, 18) and accumulates glycogen (15, 30, 31) (Fig. 2A). To explore effects of the associated glycogen accumulation on the defect in regulation of SNF1, we examined the reg1Δ mutant and several glycogen-deficient derivatives. During exponential growth on 2% glucose, the reg1Δ mutant exhibited highly elevated glycogen accumulation (Fig. 2A), with levels similar to those of WT cells in stationary phase (48 μg glycogen/108 cells), and phosphorylation of Thr210 was elevated relative to the amount of Snf1 protein (Fig. 2B). Surprisingly, reg1Δ glg1Δ glg2Δ, reg1Δ gsy1Δ gsy2Δ, and reg1Δ glc3Δ mutant cells exhibited normal glucose inhibition of Snf1 Thr210 phosphorylation (Fig. 2B), indicating that, in the absence of glycogen synthesis, Reg1-Glc7 is not required to maintain Snf1 Thr210 in the dephosphorylated state. Glycogen deficiency also suppressed other reg1Δ mutant phenotypes, including slow growth on glucose and resistance to the glucose analog 2-deoxyglucose, which inhibits SNF1 in glucose-limited cells (32) (Fig. 2C). These findings suggest that another phosphatase can dephosphorylate Thr210.
Fig. 2.
Mutations abolishing glycogen synthesis restore glucose inhibition of Snf1 in reg1Δ cells. Strains were W303 derivatives; the reg1 allele was reg1Δ::HIS3 or for studies of the effects of glc3, reg1Δ::URA3. (A) Glycogen content was determined as in Fig. 1A. (B) Immunoblot analysis of phosphorylated Thr210 and Snf1 as in Fig. 1B. Values indicate relative intensity of the bands corresponding to phosphorylated Snf1-Thr210 and total Snf1 protein (pT210/Snf1). (C) Cells were grown overnight in YEP plus 2% glucose and spotted with serial fivefold dilutions on solid YEP containing 2% glucose (Glu), 2% sucrose (Suc), or 2% sucrose plus 200 μg/mL 2-deoxy-D-glucose (Suc+2DG). Plates were incubated at 30 °C for 2 or 3 d (Suc+2DG) and photographed.
Reg1-Glc7 Contributes to Dephosphorylation of Activated Snf1 in Response to Increased Glucose Availability.
The addition of glucose to glucose-depleted cells results in the rapid dephosphorylation and inactivation of Snf1, thereby facilitating adaptation to this environmental change. To address the role of Reg1-Glc7 in this process, we grew cells on 2% glucose, shifted to 0.05% glucose for 10 min, and then restored glucose to 2% for 15 min. Replenishment of glucose resulted in rapid and complete dephosphorylation of Thr210 in WT and glc3Δ cells but not reg1Δ cells; partial dephosphorylation occurred in reg1Δ glc3Δ cells (Fig. 3A). We further examined cells growing exponentially in 2% glycerol plus 3% ethanol, the use of which requires SNF1 activity. Thr210 was phosphorylated in all cultures; WT and reg1Δ cells contained 26 and 30 μg glycogen/108 cells, respectively. When cells were shifted to 2% glucose for 15 min, Thr210 was dephosphorylated in WT and glc3Δ cells but remained highly phosphorylated in reg1Δ cells and partially phosphorylated in reg1Δ glc3Δ cells (Fig. 3B). Thus, regardless of the cell's capability for glycogen synthesis, Reg1-Glc7 clearly contributed to the rapid dephosphorylation of Snf1 in response to increased glucose availability. However, in reg1Δ glc3Δ cells, it was apparent that another phosphatase also contributes.
Fig. 3.
Reg1 is required for Snf1 dephosphorylation in response to glucose replenishment. Strains were as in Fig. 2. (A) Immunoblot analysis was as in Fig. 1B except that, after cells were resuspended in SC plus 0.05% glucose for 10 min, an aliquot was incubated in SC plus 2% glucose (+G) for 15 min and collected. Values indicate relative intensity of the bands corresponding to phosphorylated Snf1-Thr210 and total Snf1 protein (pT210/Snf1). (B) Cells were grown to midlog phase in SC plus 2% glycerol and 3% ethanol (GE); an aliquot was shifted to SC plus 2% glucose for 15 min (G). Cells were collected and analyzed as above.
Sit4 Phosphatase Has a Role in Dephosphorylation of Thr210.
We next assessed the involvement of other protein phosphatases in maintaining Snf1 Thr210 in the dephosphorylated state during growth in glucose. PP2A and PP2Cα dephosphorylate the corresponding Thr of mammalian AMPK in vitro (9, 33–35), Ppm1E and PP1-R6 have been implicated in mammalian cells (36, 37), and mammalian PP2A inactivates SNF1 in vitro (6). The PP2C family in S. cerevisiae includes seven members (Ptc1 to Ptc7); Ptc1 is the most closely related to Ppm1E. The seven single ptcΔ mutants and the triple ptc1Δ ptc2Δ ptc3Δ mutant (38) showed normal regulation of Thr210 phosphorylation during growth in glucose and in response to glucose depletion. PP2A comprises a catalytic subunit (Pph21 or Pph22), a regulatory subunit (Rts1 or Cdc55), and a scaffold subunit (Tpd3); we found no defect in pph21Δ pph22Δ, rts1Δ cdc55Δ, or tpd3Δ mutants.
We then examined mutants of the BY4741 background carrying deletions of additional genes encoding nonessential catalytic or regulatory subunits of protein phosphatases: bni4, bud14, fin1, gac1, gip1, gip2, glc8, mhp1, pig1, pig2, red1, ref2, reg2, shp1, sla1, ppz1, ppz2, sal6, pph3, psy2, psy4, ppg1, sit4, sap4, sap155, sap185, sap190, cnb1, ppt1, ptp1, ptp2, ptp3, mih1, ltp1, siw14, tep1, ymr1, dbf2, pps1, yvh1, msg5, and sdp1. The sit4Δ mutant was the only one of this set that showed elevated Thr210 phosphorylation during growth on 2% glucose (Fig. 4A).
Fig. 4.
Effects of sit4Δ on Snf1 regulation. Strains carried the indicated mutations introduced into strain CY4029 (W303-1A SSD1-v1); the SSD1-v1 allele is essential for viability of W303 sit4Δ mutants (41). (A) Cells were collected and analyzed as in Fig. 3A. (B) Glycogen content was determined as above. Similar results were obtained with the BY4741 sit4Δ mutant. (C) Sit4-TAP was expressed from the genomic locus of snf1Δ SIT4-TAP cells, and Snf1-3xHA was expressed from the native promoter on centromeric plasmid pYL225 or multicopy (2μ) plasmid pYL230, derivatives of vectors pRS316 (58) and pRS426 (59), respectively. Cells carrying pRS426 (V) served as control. Cells were grown to midlog phase in SC plus 2% glucose (H), and an aliquot was shifted to 0.05% glucose for 10 (L10) or 30 min (L30). Extracts were prepared, Sit4-TAP was isolated, and copurifying proteins were analyzed by immunoblotting, as described in Materials and Methods. TAP-purified proteins were recovered from 200 μg cell extract. Input was 5 μg cell extract.
Sit4 is a PP2A-like phosphatase that functions in the mitotic G1/S transition (39) and is involved in the target of rapamycin (TOR) pathway (40). Further analysis of the sit4Δ mutant showed that phosphorylation of Thr210 increased on glucose depletion, and subsequent glucose replenishment resulted in partial dephosphorylation (Fig. 4A). Sit4 associates with four noncatalytic subunits that are nonessential, Sap4, Sap155, Sap185, and Sap190 (41), and the cognate quadruple deletion mutant exhibited a defect in dephosphorylation of Thr210 similar to that of the sit4Δ mutant. Sit4 is involved in the control of glycogen metabolism (42, 43), and sit4Δ cells accumulated modest levels of glycogen during exponential growth in glucose (Fig. 4B). Analysis of sit4Δ glc3Δ cells showed that the absence of glycogen synthesis restored dephosphorylation of Thr210 in glucose-grown cells but did not abrogate the defect observed on glucose replenishment (Fig. 4A), which was also the case for reg1Δ glc3Δ cells. These findings indicate that Sit4, like Reg1-Glc7, contributes to the rapid dephosphorylation of Thr210 in response to increased glucose availability, and they raise the possibility that Reg1-Glc7 and Sit4 are together responsible for dephosphorylating Thr210 in glucose-grown cells. The reg1Δ sit4Δ and reg1Δ sit4Δ glc3Δ mutants are inviable.
Previous studies detected physical interaction of Reg1 with Snf1 (16, 17). To assess the association of Sit4 with Snf1, we expressed HA-tagged Snf1 from its native promoter on a centromeric or multicopy plasmid in a snf1Δ strain expressing tandem affinity purification (TAP)-tagged (44) Sit4 from the genomic SIT4-TAP locus. Extracts were prepared from cultures grown under different conditions, and Sit4-TAP was purified. The copurifying proteins were resolved by SDS/PAGE and analyzed by immunoblotting. Copurification of Snf1-HA with Sit4-TAP was barely detectable when Snf1-HA was expressed from the centromeric plasmid, but it was clearly evident when Snf1-HA was expressed at higher levels (Fig. 4C). This evidence for physical association is consistent with the idea that Sit4 directly dephosphorylates Thr210.
reg1Δ Affects Regulation of SNF1 Containing Different GBDs.
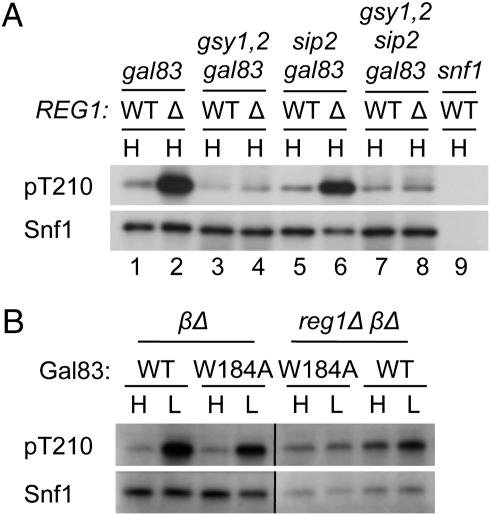
Having initiated this work because deletion of the GBD from Gal83 relieves glucose inhibition of SNF1, we returned to the possibility that, in reg1Δ cells, the binding of glycogen displaces the GBD relative to the SNF1 heterotrimer, thereby affecting Thr210 phosphorylation. The GBD of Gal83, the major β-subunit in glucose-grown cells (22, 45), bound glycogen in vitro, whereas the Sip2 GBD bound glycogen much less strongly, and the GBD is not well-conserved in Sip1 (23). To assess the role of the GBD, we examined the effects of reg1Δ in cells expressing only Sip1 and Sip2 (gal83Δ) or only Sip1 (gal83Δ sip2Δ). In both cases, reg1Δ caused phosphorylation of Thr210 in glucose-grown cells (Fig. 5A, compare lanes 1 and 2 and lanes 5 and 6), and introduction of gsy1Δ gsy2Δ restored dephosphorylation (Fig. 5A, lanes 4 and 8). The reg1Δ and gsy1Δ gsy2Δ mutations also affected 2-deoxyglucose resistance, as in Fig. 2C, regardless of the β-subunit present.
Fig. 5.
Regulation of Snf1 Thr210 phosphorylation in the presence of different β-subunits. Cells were grown in SC plus 2% glucose (H) or shifted to 0.05% glucose (L) and collected for immunoblot analysis of phosphorylated Thr210 and Snf1. (A) Strains were derivatives of W303 carrying REG1 (WT) or reg1Δ (Δ). (B) Gal83 (WT) and its derivative Gal83W184A were expressed as fusions to GFP from the native promoter on a centromeric plasmid (22) in gal83Δ sip1Δ sip2Δ (βΔ) or reg1Δ gal83Δ sip1Δ sip2Δ (reg1Δ βΔ) cells. All lanes are from the same blot.
We also examined the effect of reg1Δ in cells expressing Gal83W184A in which Ala is substituted for Trp184, which is highly conserved in carbohydrate-binding modules (25). Substitution of the analogous residue of the GBD of mammalian AMPK, Trp100, abolished binding to glycogen, and the crystal structure of AMPK GBD with bound β-cyclodextrin identified Trp100 as a key residue (24, 46). Previous studies showed that the GBD of Gal83W184A,R214Q did not bind glycogen in vitro (23). In glucose-grown reg1Δ gal83Δ sip1Δ sip2Δ (reg1Δ βΔ) cells, Thr210 phosphorylation was elevated to the same extent in cells expressing WT Gal83 or Gal83W184A (Fig. 5B). Together, these findings suggest that the effect of reg1Δ on Thr210 phosphorylation does not depend on glycogen binding to the GBD.
Discussion
Previous evidence that the GBD of the β-subunit Gal83 is required for glucose inhibition of SNF1 led us to assess the role of glycogen in regulation of SNF1. Analysis of the reg1Δ mutant, which exhibits both elevated glycogen accumulation and elevated Snf1 Thr210 phosphorylation during growth on high levels of glucose, gave an unexpected result. Mutations that abolished glycogen synthesis restored dephosphorylation of Snf1 Thr210, indicating that elevated glycogen synthesis has a role in the phosphorylation of Snf1 in glucose-grown reg1Δ cells and that Reg1-Glc7 PP1 cannot be solely responsible for dephosphorylation of Snf1.
We present evidence that the PP2A-related phosphatase Sit4 also contributes to the dephosphorylation of Snf1. The sit4Δ mutant similarly exhibited Thr210 phosphorylation and elevated glycogen accumulation during growth on high glucose, and abolishing glycogen synthesis similarly restored dephosphorylation. In glucose-depleted sit4Δ cells, the rapid dephosphorylation of activated Snf1 in response to glucose replenishment was impaired. We also show that Sit4, like Reg1, is physically associated with Snf1, consistent with a direct role in dephosphorylation. The sit4Δ reg1Δ and sit4Δ reg1Δ glc3Δ mutants are inviable, but we have found that introduction of snf1Δ restores viability. The simple model is that Reg1-Glc7 and Sit4 are together responsible for dephosphorylation of Thr210 (Fig. 6).
Fig. 6.
Model for the roles of Reg1-Glc7 and Sit4 in regulation of Snf1 phosphorylation during growth on high glucose. Reg1-Glc7 and Sit4 independently dephosphorylate Snf1 (arrows). The high glucose signal promotes dephosphorylation (arrow), possibly by promoting access of phosphatases to Snf1 Thr210 or by direct regulation of phosphatases. Reg1-Glc7 and Sit4 also inhibit (bars) glycogen synthesis; other mechanisms controlling glycogen synthesis are not shown. Elevated glycogen synthesis during growth on high glucose inhibits dephosphorylation of Snf1 Thr210; dashed lines indicate that possible mechanisms include direct inhibition of dephosphorylation and down-regulation of the glucose signal. In the absence of one of the phosphatases, increased glycogen synthesis inhibits dephosphorylation by the remaining phosphatase. If glycogen synthesis is abolished, the function of the remaining phosphatase is sufficient for dephosphorylation of Thr210.
It is possible that other phosphatases contribute to regulation of SNF1, perhaps under different growth conditions. Analysis of mutants lacking nonessential catalytic and regulatory subunits did not identify another phosphatase that plays a role in the inhibition of SNF1 during growth on high levels of glucose; however, such an activity could be encoded by multiple genes with overlapping functions or by an essential gene. Mass spectrometric analysis identified Snf1 as a kinase associated with the cell cycle phosphatase Cdc14 (47); in preliminary experiments, we did not detect phosphorylation of Snf1 Thr210 after growth of the temperature-sensitive cdc14-3 mutant (48) at restrictive temperature on high levels of glucose or after glucose depletion and replenishment.
Elevated glycogen synthesis is critical for the activation of SNF1 in reg1Δ and sit4Δ cells during growth on high glucose. However, elevated glycogen synthesis alone does not suffice, because overexpression of glycogen synthase in WT cells, containing both Reg1-Glc7 and Sit4, resulted in glycogen accumulation to a level twofold that of the sit4Δ mutant but did not cause Thr210 phosphorylation during growth on high glucose. We suggest that, in the absence of one of the phosphatases, increased glycogen synthesis inhibits dephosphorylation by the other phosphatase, whereas if glycogen synthesis is abolished, the function of the remaining phosphatase is sufficient for dephosphorylation (Fig. 6).
What is the role of glycogen in regulation of SNF1? Studies of SNF1 containing Sip1, Sip2, or Gal83W184A do not support the idea that glycogen binding regulates phosphorylation of SNF1 in vivo. We note that the SNF1 β-subunits carry a Leu substitution for a highly conserved aromatic residue in the putative glycogen-binding site (25, 46), raising the possibility that, in vivo, none of these subunits binds glycogen. Together with previous evidence (29, 49), the present findings suggest that the GBD contributes to glucose inhibition of SNF1 through its interaction with the rest of the SNF1 heterotrimer rather than through binding glycogen, although it remains possible that the GBD binds other carbohydrates. A direct effect of glycogen on the phosphatases has not been excluded.
We suggest that elevated glycogen synthesis in reg1Δ and sit4Δ cells promotes activation of SNF1 through its effects on glucose metabolism. Altered glucose metabolism may reduce glucose signaling to the SNF1 pathway. We propose that the absence of either Reg1-Glc7 or Sit4 phosphatase, in combination with decreased glucose signaling, impairs dephosphorylation of Snf1 Thr210 (Fig. 6).
Materials and Methods
Strains and Genetic Methods.
S. cerevisiae strains had the W303-1A (MATa ade2 can1 his3 leu2 trp1 ura3) or BY4741 (MATa his3 leu2 met15 ura3) genetic background. The alleles reg1Δ::HIS3 (16), reg1Δ::URA3 (15), snf1Δ::LEU2 (50), sip1Δ::kanMX6, sip2Δ::kanMX4, gal83Δ::TRP1 (23), snf1Δ::KanMX4, glg1Δ::KanMX4, gsy1Δ::KanMX4, gcl3Δ::KanMX4, gph1Δ::KanMX4, and gdb1Δ::KanMX4 (51) have been described. To construct gsy2Δ::LEU2 and glg2Δ::URA3, we replaced the KanMX4 marker in the cognate alleles (Open Biosystems) as described (52). To construct sit4Δ::nat1, we replaced the 0.7-kb XbaI-SmaI fragment of SIT4. The above alleles were introduced into the W303 background. Derivatives of BY4741 with KanMX4-marked deletions of genes affecting glycogen metabolism and genes encoding phosphatase subunits were obtained from Open Biosystems or constructed in this laboratory unless otherwise noted. We introduced snf1Δ::KanMX4 into strain BY4741 SIT4-TAP (Open Biosystems). Standard methods for genetic analysis and transformation were used. Yeast cultures were grown in rich yeast extract peptone (YEP) or synthetic complete (SC) medium lacking appropriate supplements to select for plasmids (53).
Preparation of Cell Extracts and Immunoblot Analysis of Snf1 Phosphorylation.
Cultures were grown to exponential phase (OD600 of 0.6–0.8) in SC medium containing 2% glucose. Cells (100 mL) were harvested by rapid filtration and immediately frozen in liquid nitrogen or resuspended in 0.05% glucose for 10 min and collected. For some experiments, after incubation in 0.05% glucose, an aliquot of cells was incubated in 2% glucose for 15 min and collected. Extracts were prepared by vortexing cells with glass beads 20 times for 10 s in 50 mM Tris·HCl, pH 7.5, 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM EDTA, 0.5% Triton X-100, 10% (vol/vol) glycerol, 1 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor mixture (Roche), and protein concentrations were determined by Bio-Rad assay. Proteins (10 μg) were separated on 7.5% SDS/PAGE and analyzed by immunoblotting using antiphospho-Thr-172-AMPK antibody (Cell Signaling Technologies). Membranes were incubated in 0.2 M glycine, pH 2, for 10 min and reprobed with antipolyhistidine antibody (Sigma) to detect Snf1, which contains polyhistidine. Antibodies were detected by enhanced chemiluminescence using ECLPlus (GE Healthcare). Intensity of the bands was quantified using ImageJ software from National Institutes of Health (54).
Purification of Sit4-TAP and Immunoblot Analysis.
Cells were collected, and extracts were prepared as above except that the extraction buffer contained 150 mM NaCl. Cell extract (1 mg) was incubated with 10 μL protein G Sepharose 4 Fast Flow (GE Healthcare) in 1 mL extraction buffer at 4 °C for 3 h. Beads were washed three times with 1 mL extraction buffer, and proteins were eluted by boiling in 50 μL 2× SDS/PAGE loading dye. Proteins were subjected to immunoblot analysis, as above, using 12CA5 antibody (Roche) to detect Snf1-3xHA and P1291 antibody (Sigma) to detect Sit4-TAP.
Determination of Glycogen Content.
Cultures were grown to exponential phase in SC plus 2% glucose, and cells (10 mL) were harvested by centrifugation. Assays of cellular glycogen content were performed as described (55) with minor modifications using 6.7 U/mL Aspergillus niger amyloglucosidase (catalog number A-7420; Sigma). The amount of glucose released was quantified using the glucose oxidase reaction (56).
Acknowledgments
We thank M. Momcilovic, Y. Liu, E. Herrero, J. Arino, M. J. Stark, K. T. Arndt, and A. Amon for gifts of plasmids and strains. We thank Y. Liu for valuable discussion and the reviewers for useful comments. M.C. is grateful to M. Johnston for his continuing interest and thoughtful suggestions. A.R. was a recipient of a Fulbright Fellowship. This work was supported by National Institutes of Health Grant GM34095 (to M.C.).
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2009.
The authors declare no conflict of interest.
References
- 1.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- 4.Hardy TA, Huang D, Roach PJ. Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:27907–27913. [PubMed] [Google Scholar]
- 5.Mitchelhill KI, et al. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 6.Woods A, et al. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- 7.Scott JW, et al. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao B, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 9.Suter M, et al. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J Biol Chem. 2006;281:32207–32216. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- 10.McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 11.Hong SP, Carlson M. Regulation of Snf1 protein kinase in response to environmental stress. J Biol Chem. 2007;282:16838–16845. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- 12.Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nath N, McCartney RR, Schmidt MC. Yeast Pak1 kinase associates with and activates Snf1. Mol Cell Biol. 2003;23:3909–3917. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland CM, et al. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- 15.Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanz P, Alms GR, Haystead TA, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong SP, Momcilovic M, Carlson M. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase α as Snf1-activating kinases in yeast. J Biol Chem. 2005;280:21804–21809. doi: 10.1074/jbc.M501887200. [DOI] [PubMed] [Google Scholar]
- 19.Rubenstein EM, et al. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated 2threonine 210 to the PP1 phosphatase. J Biol Chem. 2008;283:222–230. doi: 10.1074/jbc.M707957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincent O, Carlson M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 1999;18:6672–6681. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt MC, McCartney RR. β-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 2001;15:1104–1114. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiatrowski HA, et al. Mutations in the Gal83 glycogen-binding domain activate the Snf1/Gal83 kinase pathway by a glycogen-independent mechanism. Mol Cell Biol. 2004;24:352–361. doi: 10.1128/MCB.24.1.352-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polekhina G, et al. AMPK β subunit targets metabolic stress sensing to glycogen. Curr Biol. 2003;13:867–871. doi: 10.1016/s0960-9822(03)00292-6. [DOI] [PubMed] [Google Scholar]
- 25.Machovic M, Janecek S. The evolution of putative starch-binding domains. FEBS Lett. 2006;580:6349–6356. doi: 10.1016/j.febslet.2006.10.041. [DOI] [PubMed] [Google Scholar]
- 26.McBride A, Ghilagaber S, Nikolaev A, Hardie DG. The glycogen-binding domain on the AMPK β subunit allows the kinase to act as a glycogen sensor. Cell Metab. 2009;9:23–34. doi: 10.1016/j.cmet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson-Jaeger S, François J, Gaughran JP, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.François J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 29.Momcilovic M, Iram SH, Liu Y, Carlson M. Roles of the glycogen-binding domain and Snf4 in glucose inhibition of SNF1 protein kinase. J Biol Chem. 2008;283:19521–19529. doi: 10.1074/jbc.M803624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D, Chun KT, Goebl MG, Roach PJ. Genetic interactions between REG1/HEX2 and GLC7, the gene encoding the protein phosphatase type 1 catalytic subunit in Saccharomyces cerevisiae. Genetics. 1996;143:119–127. doi: 10.1093/genetics/143.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frederick DL, Tatchell K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol Cell Biol. 1996;16:2922–2931. doi: 10.1128/mcb.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedbacker K, Carlson M. Regulation of the nucleocytoplasmic distribution of Snf1-Gal83 protein kinase. Eukaryot Cell. 2006;5:1950–1956. doi: 10.1128/EC.00256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies SP, Helps NR, Cohen PT, Hardie DG. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C α and native bovine protein phosphatase-2AC. FEBS Lett. 1995;377:421–425. doi: 10.1016/0014-5793(95)01368-7. [DOI] [PubMed] [Google Scholar]
- 34.Wu Y, Song P, Xu J, Zhang M, Zou MH. Activation of protein phosphatase 2A by palmitate inhibits AMP-activated protein kinase. J Biol Chem. 2007;282:9777–9788. doi: 10.1074/jbc.M608310200. [DOI] [PubMed] [Google Scholar]
- 35.Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J. 2007;403:139–148. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Haro L, et al. The PP1-R6 protein phosphatase holoenzyme is involved in the glucose-induced dephosphorylation and inactivation of AMP-activated protein kinase, a key regulator of insulin secretion, in MIN6 β cells. FASEB J. 2010;24:5080–5091. doi: 10.1096/fj.10-166306. [DOI] [PubMed] [Google Scholar]
- 37.Voss M, et al. Ppm1E is an in cellulo AMP-activated protein kinase phosphatase. Cell Signal. 2011;23:114–124. doi: 10.1016/j.cellsig.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz A, González A, García-Salcedo R, Ramos J, Ariño J. Role of protein phosphatases 2C on tolerance to lithium toxicity in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2006;62:263–277. doi: 10.1111/j.1365-2958.2006.05370.x. [DOI] [PubMed] [Google Scholar]
- 39.Sutton A, Immanuel D, Arndt KT. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 41.Luke MM, et al. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jablonka W, Guzmán S, Ramírez J, Montero-Lomelí M. Deviation of carbohydrate metabolism by the SIT4 phosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta. 2006;1760:1281–1291. doi: 10.1016/j.bbagen.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 43.Posas F, Clotet J, Ariño J. Saccharomyces cerevisiae gene SIT4 is involved in the control of glycogen metabolism. FEBS Lett. 1991;279:341–345. doi: 10.1016/0014-5793(91)80183-4. [DOI] [PubMed] [Google Scholar]
- 44.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 45.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 46.Polekhina G, et al. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomson BN, et al. Regulation of Spo12 phosphorylation and its essential role in the FEAR network. Curr Biol. 2009;19:449–460. doi: 10.1016/j.cub.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449:492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- 50.Hubbard EJ, Yang XL, Carlson M. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics. 1992;130:71–80. doi: 10.1093/genetics/130.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 52.Voth WP, Jiang YW, Stillman DJ. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast. 2003;20:985–993. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]
- 53.Rose MD, Winston F, Hieter P. Methods in Yeast. Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 54.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 55.Parrou JL, François J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997;248:186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 56.Goldstein A, Lampen JO. β-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- 57.Bellí G, Garí E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]