Abstract
The proposed age of the striking biogeographic disjunction between the Arctic and southernmost South America varies from more than 65 million to a few thousand years, but no estimates based on explicit models and molecular data are available. Here we address the origin of bipolarity in crowberries (Empetrum), which are heath-forming dwarf shrubs with animal-dispersed fruits. We apply a fossil-calibrated relaxed molecular clock to model sequence evolution in two nuclear low-copy and two plastid DNA regions from 41 individual plants (420 clones for the nuclear regions) representing the entire geographic distribution of crowberries. The plastid region matK and four fossil calibration points were used to infer the ages of the crowberry stem and crown groups. All analyses resolved three major crowberry clades (A–C). Clade A contained sequences from the eastern Canadian pink-fruited crowberry (E. eamesii) as sister to clades B and C, which both contained sequences from the black-fruited northern hemisphere crowberry (E. nigrum). Clade B also contained a subclade with all sequences from the red-fruited southern hemisphere crowberry, which is often referred to as a distinct species, E. rubrum. Its closest relatives were consistently identified as black-fruited plants from northwestern North America. The median time to the most recent common ancestor for northern and southern hemisphere crowberries was estimated to 0.56–0.93 Ma, and 0.26–0.59 Ma for the southern plants only. We conclude that a single dispersal by a bird from northwestern North America to southernmost South America, taking place in the Mid-Pleistocene, is sufficient to explain the disjunction in crowberries.
Keywords: biogeography, Empetreae, Ericaceae, low-copy nuclear genes, molecular dating
Many terrestrial organisms have disjunct amphitropical distributions; that is, organisms belonging to the same or closely related species occur in the temperate regions of both hemispheres but are absent from the tropics. The most extreme amphitropical disjunctions are presented by the so-called bipolar organisms, typically occurring in the northernmost parts of the northern hemisphere as well as in southernmost South America. This element includes some 30 species of vascular plants (1, 2) and many bryophytes and lichen-forming fungi (3). The bipolar distribution pattern has intrigued biogeographers for a long time (1–5). Most studies have simply documented the geographic distribution of particular species and suggested possible explanations and timing for the origin of the disjunction. The classical hypotheses range from very old disruptions of continuous distributions across the tropics [e.g., the current populations are remnants from Mesozoic vicarious events (6) or Cretaceous or Paleogene ancestors (3)] to much more recent migration events [e.g., Late Pliocene or Pleistocene long-distance dispersals via air currents or migrating animals (1, 2) or north–south migration along Pleistocene transtropical highland bridges (4)].
For bipolar plants, there are still no studies presenting well-supported evolutionary hypotheses based on explicit models and molecular data to estimate the absolute age of their extreme disjunction. There are a few molecular studies of bipolar plant groups inferring “recent” origin of the disjunction simply from low levels of sequence divergence (7, 8; also see ref. 5). A few studies have addressed the origin and evolution of less extreme amphitropical disjunctions between North and South America based on molecular data and rigorous analytical methods, showing that these disjunctions largely result from Miocene to Holocene long-distance dispersals from north to south (e.g., 9, 10; also see ref. 5). In Euphrasia, the disjunction between Eurasian and southern hemisphere taxa (including South America, Australia, and New Zealand) was estimated to be 5–7 million y old, but the relationship between the northern hemisphere and South American taxa was not resolved (11).
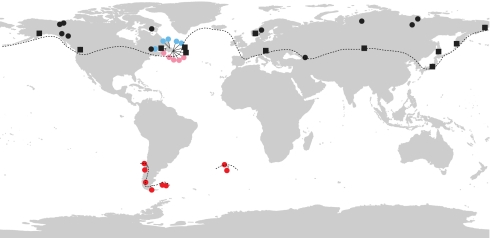
Crowberries (genus Empetrum L.) are evergreen dwarf shrubs and represent a classic example of extreme intercontinental disjunctions among land plants (e.g., 1–3, 6). The genus has a wide circumpolar/circumboreal distribution in the northern hemisphere, whereas it is limited to a small area in the southern hemisphere (Fig. 1). It is rare in northern Patagonia but more common in southern Patagonia and Tierra del Fuego, as well as in the Falkland Islands and the south Atlantic Tristan da Cunha and Gough Islands. In the northern hemisphere, there are diploid plants with black or pink fruits as well as tetraploid plants with black or purple fruits, whereas only diploid plants with red fruits occur in the southern hemisphere (12). Whereas the red-fruited plants in the southern hemisphere are usually referred to as a distinct species (E. rubrum Vahl ex Willd.), the taxonomy of northern hemisphere crowberries is very complex (e.g., 12). Here we follow ref. 13 and treat black-fruited di- and tetraploid plants as a single species, E. nigrum L., diploid eastern Canadian plants with pink fruits as E. eamesii Fernald & Wiegand, and tetraploid eastern North American plants with purple fruits as E. atropurpureum Fernald & Wiegand.
Fig. 1.
Total geographic range of crowberries (the genus Empetrum) and the origin of the accessions included in this study. Black squares and black dots indicate diploid and tetraploid E. nigrum, and blue, pink, and red dots indicate E. atropurpureum, E. eamesii, and E. rubrum, respectively. The dotted lines approximate the distribution of Empetrum, namely north of 40°N in the northern hemisphere and south of 36°S in South America and the Falkland, Tristan da Cunha, and Gough Islands.
Only two previous studies have focused on Empetrum using explicit phylogenetic analyses. Based on a parsimony analysis of morphological characters, a group of diploid plants with predominantly unisexual flowers and red fruits (the southern hemisphere “E. rubrum complex”) was hypothesized to be the progenitor of northern hemisphere diploid and tetraploid plants with black fruits and unisexual or hermaphroditic flowers, and the bipolar disjunction was suggested to result from breakup of a continuous transtropical distribution (12). In a limited parsimony analysis of combined nuclear ribosomal DNA (the internally transcribed spacer regions ITS 1 and ITS 2, and the 5.8S gene) and plastid matK sequences from five Empetrum accessions, a tetraploid North American E. nigrum (“E. hermaphroditum”) was resolved as sister to E. eamesii and a group formed by E. rubrum and a diploid European E. nigrum (7). Thus, in contrast to ref. 12, it was concluded that the modern bipolar distribution of crowberries results from long-distance dispersal from the north to the south. The disjunction was not explicitly dated, but based on low sequence divergence between northern and southern hemisphere plants, they inferred it to be recent.
Here we test the classical hypotheses on the age of the disjunction in bipolar plants by applying a fossil-calibrated relaxed molecular clock to infer the stem and crown group ages of crowberries, using the plastid-encoded matK region from 55 taxa representing the family Ericaceae. The inferred crown group age is used as a prior for the time to the most recent common ancestor (tMRCA) of plastid-encoded trnfMCAU-trnSUGA and trnSGCU-trnGUUC sequences and nuclear-encoded low-copy RPC2 and RPB2-I sequences in crowberries. We sampled plant material representing the entire geographic range (Fig. 1) as well as the morphological and ploidy-level variation in the genus to infer the relationship between northern and southern hemisphere crowberries and the timing of their disjunction.
Results
DNA Sequences.
Each of the plastid trnfMCAU-trnSUGA and trnSGCU-trnGUUC matrices consisted of 41 sequences from 41 accessions, and 997 and 1,443 aligned positions, respectively. For the nuclear RPC2 region, direct sequencing of the PCR products revealed high levels of within-individual sequence polymorphism in many diploids and tetraploids, probably representing a combination of single-locus heterozygosity and homeology resulting from whole-genome duplication in the tetraploids (SI Results). In total, 25 accessions contained two or more sequences and were therefore subjected to cloning. A total of 252 RPC2 clones were sequenced, and 57 consensus sequences were constructed (Table S1). The final RPC2 matrix consisted of 71 sequences from a total of 34 accessions and 925 aligned positions. For the nuclear RPB2-I region, 19 accessions showed one or more polymorphisms in direct sequenced PCR products. Accessions for which only a single polymorphism in a parsimony-informative character was detected were included in the analyses. Five accessions, including both diploids and tetraploids, were cloned and contributed two or more sequences to the matrix. A total of 168 RPB2-I clones were sequenced and 13 consensus sequences were constructed (Table S1). The final RPB2-I matrix consisted of 36 sequences from a total of 28 accessions and 2,511 aligned positions. GenBank accession numbers for the sequences produced by direct sequencing of PCR products as well as for consensus sequences produced from cloned PCR products are given in Table S1.
Phylogenetic Analysis of matK and Dating of Empetreae and Empetrum.
Comparing a GTR+Γ versus an SRD06 codon position model using Bayes factors showed that a GTR+Γ model provided a better fit (log10 Bayes factor > 30) to the Ericaceae matK data. The difference between the Yule and birth/death tree priors was of the same magnitude as the SE and thus considered not significant. All four analyses (SRD06 vs. GTR+Γ, Yule vs. birth/death tree prior) resulted in highly similar tree topologies and time estimates. The inferred phylogeny and all estimated node ages based on the combined results from the four separate analyses using a GTR+Γ substitution model and Yule tree prior are presented in Fig. S1.
The split between Empetrum and its sister genera Ceratiola and Corema was estimated to have occurred 23.6 Ma [95% highest posterior probability density (HPD) 10.1–38.8 Ma, median 22.4 Ma], and the Empetrum crown group age was estimated to 5.5 Ma (95% HPD 1.1–11.1 Ma, median 4.9 Ma) (Fig. S1).
Phylogenetic Analysis and Dating in Empetrum.
The plastid sequences were concatenated and, following model selection using MrAIC (14), analyzed under an HKY substitution model (15). The nuclear RPC2 and RPB2-I sequences were analyzed under an HKY and an HKY+I substitution model, respectively. The clade support for analyses run without data (i.e., analyzing only the priors) was not above 0.33 for any clade, thus indicating a strong phylogenetic signal in the data. The inferred ages of clades are summarized together with the posterior probabilities (PP) in Table 1.
Table 1.
Posterior probabilities, mean, and median time to the most recent common ancestor (million years), 95% HPD interval, and the effective sample size inferred for clades A–F in Fig. 2 from the trnfMCAU-trnSUGA and trnSGCU-trnGUUC plastid regions (Fig. S2) and the nuclear RPC2 and RPB2-I regions (Figs. S3 and S4), respectively
| Clade | Posterior probability | Mean | Median | 95% HPD lower | 95% HPD upper | Effective sample size |
| Plastid/RPC2/RPB2 | Plastid/RPC2/RPB2 | Plastid/RPC2/RPB2 | Plastid/RPC2/RPB2 | Plastid/RPC2/RPB2 | Plastid/RPC2/RPB2 | |
| A | 1.0/1.0/1.0 | 0.47/1.51/0.16 | 0.39/1.40/0.11 | 0.04/0.45/0.00 | 1.11/2.84/0.50 | 3,310/2,016/6,946 |
| B | 1.0/1.0/0.86 | 0.76/2.19/1.67 | 0.69/2.08/1.59 | 0.16/0.84/0.74 | 1.52/3.76/2.73 | 2,363/2,097/4,511 |
| C | 0.95/0.98/0.73 | 0.53/2.78/1.62 | 0.45/2.67/1.52 | 0.08/1.09/0.44 | 1.18/4.67/2.96 | 2,402/1,900/5,020 |
| D | 1.0/0.72/0.80 | 0.30/0.65/0.33 | 0.26/0.59/0.29 | 0.03/0.09/0.03 | 0.66/1.35/0.73 | 2,442/1,932/4,927 |
| E | 1.0/1.0/1.0 | 0.76/1.00/0.60 | 0.69/0.93/0.56 | 0.16/0.33/0.17 | 1.52/1.83/1.12 | 2,363/1,450/3,037 |
| F | 1.0/0.90/1.0 | 1.34/3.77/2.16 | 1.24/3.66/2.09 | 0.35/1.64/1.01 | 2.57/6.06/3.42 | 2,543/2,001/4,952 |
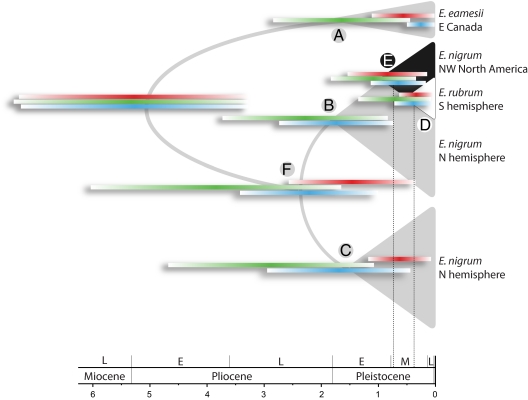
Three major clades (A–C) could be distinguished in the analyses of all three Empetrum datasets (Figs. S2–S4). A simplified tree displaying the relationships inferred in all three gene trees with the 95% HPD age intervals for each shared clade is presented in Fig. 2. The pink-fruited eastern Canadian E. eamesii was inferred as sister to all other crowberries in all three Empetrum datasets (clade A; Fig. 2 and Figs. S2–S4). The sequences from the northern hemisphere black-fruited di- and tetraploid E. nigrum were found both in clade B and clade C. The sequences from the southern hemisphere diploid E. rubrum were exclusively found in clade B, forming a subclade (D) that was well-supported by the plastid data and, to a lesser degree, also by the nuclear data (Table 1). The tMRCA (median) for the E. rubrum lineage (D) was estimated to 0.26 Ma from the plastid data, and to 0.59 and 0.29 Ma from RPC2 and RPB2-I, respectively (Table 1). Sequences from diploid E. nigrum from northwestern North America were consistently resolved as sister to E. rubrum with strong support (PP 1.0; Table 1 and Figs. S2–S4), forming subclade E within clade B (Fig. 2). No other diploids were included in subclade E, and with the exception of one sequence from a heterozygous diploid E. nigrum from Japan, no other diploids were included in the more inclusive clade B (Figs. S3 and S4). The tMRCA for the sequences belonging to subclade E was estimated to 0.69 Ma from the plastid data, and to 0.93 and 0.56 Ma from RPC2 and RPB2-I, respectively (Table 1). Clades B and C were resolved as sisters (clade F) in all analyses (Fig. 2). The plastid sequences from the purple-fruited tetraploid E. atropurpureum were resolved in clade C, but multiple sequences were identified in its nuclear regions. The analyses of the cloned RPC2 sequences resolved sequences from single individuals of this tetraploid in all three major clades (A–C), suggesting several allopolyploid origins.
Fig. 2.
Schematic phylogeny of E. eamesii, E. nigrum, and E. rubrum inferred from three DNA regions (Figs. S2–S4). Colored bars indicate the 95% HPD age interval for clades A–F inferred from the plastid trnfMCAU-trnSUGA and trnSGCU-trnGUUC regions (red), the nuclear region spanning exons 31–32 in RPC2 (green), and the nuclear region spanning exons 2–6 in RPB2-I (blue). See Table 1 for posterior probabilities and ages inferred from each DNA region for clades A–F. Note that the B and E clades coincide in the plastid tree (Fig. S2), and the red 95% HPD bar at clade E can also be placed at clade B. The timescale shows absolute age in million years. Holocene is not indicated. Dashed vertical lines indicate the average median time to the most recent common ancestor of the southern hemisphere E. rubrum (clade D) and its closest relative from the northern hemisphere, northwestern North American E. nigrum (clade E), estimated from all three DNA regions.
Discussion
Bipolar Disjunction in Crowberries Originated in the Middle Pleistocene.
Our study provides strong evidence for a Mid-Pleistocene origin of the bipolar disjunction in crowberries, and for northwestern North America as the source for colonization of the southern hemisphere. Although some sequences obtained from Canadian, Norwegian, Georgian, and Russian tetraploid E. nigrum as well as tetraploid Canadian E. atropurpureum are resolved as closely related to the diploid southern hemisphere E. rubrum (Figs. S3 and S4), the difference in ploidy level makes it unlikely that the tetraploids contributed to the diploid lineage. However, a close relationship between E. rubrum and northwestern North American diploid E. nigrum, excluding all other sampled diploids, is strongly supported in our analyses (subclade E; Figs. S2–S4). As northwestern North America was largely ice-free during the last glacial maximum (16), suitable habitats have likely been available throughout time, further supporting northwestern North America as the source for colonization of the southern hemisphere.
The median estimates of the tMRCA for extant diploid northern and southern hemisphere Empetrum (subclade E) ranged from 0.56 to 0.93 Ma and from 0.26 to 0.59 Ma for southern hemisphere plants only (subclade D; Fig. 2 and Table 1). This result is strikingly contradictory to earlier hypotheses of a many-million-year-old disjunction caused by disruption of a continuous distribution across the tropics in the Mesozoic (6) or in the Late Cretaceous or Paleogene (3, 12). Rather, our analysis corroborates the alternative hypotheses presented, for example, by refs. 1, 2, and 7 of a Mid-Pliocene or later age disjunction, probably via long-distance dispersal or stepwise migration along the Cordillera.
The arrival of Empetrum in the southern hemisphere before the end of the last glaciation is corroborated by pollen studies. Empetrum pollen has been recorded in a lake sediment core with an extrapolated age of >60,000 y from the Chilean Lake District [40°S (17)] and in some of the oldest pollen records (>41,000 y) from southern Patagonia and Tierra del Fuego (18). Furthermore, Empetrum is an important component in many high-latitude peat bog samples dated to 15,000–12,500 B.P. (18, 19), suggesting that Empetrum arrived in the southern hemisphere at the latest during the Late Pleistocene.
Today, crowberries occur in cold–temperate and moist areas with an oceanic climate, and it is therefore likely that they established in southern Patagonia and Tierra del Fuego during an interglacial period rather than during a glaciation, when the climate was more arid. Starting in the Late Miocene, southern Patagonia and Tierra del Fuego were heavily glaciated several times (20). The Patagonian Andes and the eastern piedmont areas between 36°S and 56°S were almost completely covered by a continuous ice sheet, the Great Patagonian Glaciation (GPG), with ice tongues reaching the Atlantic coast south of Río Gallegos (~51°S) toward the end of the Early Pleistocene (20, 21). The GPG was followed by four major glaciations (20) during the Middle and Late Pleistocene, the last having its maximum between 19,000 and 23,000 y ago (22). The upper age constraints for the end of the Danaglacial/post-GPG 1 (0.76 Ma) and the Gotiglacial/post-GPG 2 and 3 [0.065 Ma (20)] fit well with our estimated median ages for southern hemisphere Empetrum.
In records from the Falkland Islands, Empetrum pollen has been found in a sample dated >30,000 B.P. (23). Interestingly, although pollen records from Tristan da Cunha, Nightingale, and Gough Islands include Empetrum in samples as old as 11,000 B.P., pollen is lacking from older samples (36,000–43,700 B.P.) from Nightingale and Gough Islands (24–27). This may indicate a more recent arrival of Empetrum in the south Atlantic islands than in the Falkland Islands.
Although southern hemisphere pollen records indicate that our lower 95% HPD limit (0.03 Ma) is conservative, we find good correlation between our estimated divergence dates and the fossil record. The divergence between Empetrum and its sister genera Ceratiola and Corema was estimated to be 23.6 Ma (95% HPD, 10.1–38.8; median, 22.4 Ma) and the age of the Empetrum crown group was estimated to be 5.5 Ma (95% HPD, 1.1–11.1 Ma; median, 4.9 Ma; Table 1). The stem group age is consistent with the finding of a Middle Miocene Empetrum endocarp in a sand bed in Denmark (28) and seeds from Late Miocene sand beds in Germany (29). Applying a strict molecular clock (7) inferred the stem group age of Empetrum to be younger (14.8–22.8 and 11.2–16.5 Ma). The discrepancy between these estimates can be explained by several factors (reviewed in refs. 30–32). Whereas we used four fossils with parameterized age distributions reflecting the uncertainty of the maximum age of the clades (33, 34), ref. 7 used a single point estimate. In addition, ref. 7 used a Rhododendron fossil dated to 37 Ma to calibrate the divergence, compared with the 54.5 Ma-old seeds of Rhododendron newburyanum (35, 36) we used to constrain the minimum age for the Rhododendron clade. Finally, even though a more exact date is appealing, the use of a strict clock tends to result in a more precise—but not necessarily more accurate—estimate than relaxed clocks (37). Although a strict molecular clock could not be rejected by a likelihood ratio test in ref. 7, it performed much worse than an uncorrelated relaxed clock with our larger taxon sampling.
Bird-Mediated Long-Distance Dispersal from Northwestern North America.
The close relationship we have demonstrated between southern hemisphere E. rubrum and diploid plants from northwestern North America belonging to E. nigrum, as well as our age estimates of the origin of the bipolar disjunction, reject the earlier hypotheses invoking breakup of a very old, continuous transtropical distribution. However, did the crowberries reach southernmost South America via direct long-distance dispersal or via stepwise migration? Migration of cold-adapted plants from the north to the south along the American Cordillera during the Pleistocene climatic oscillations has already been proposed (4), and was recently invoked to explain bipolarity in Carex (38). However, without fossils or other evidence for occurrences in geographically intermediate areas, it is difficult to discern with certainty between direct long-distance dispersal and gradual migration. We find no data in the published fossil records indicating that crowberries ever occurred in the central parts of the American Cordillera, as would be expected if they migrated gradually from the north to the south. Rather, the monophyly we inferred for the southern hemisphere crowberry at the plastid loci, which are particularly prone to genetic drift, points to a strong bottleneck such as one following a single long-distance dispersal event. The lack of geographic structuring of the genetic variation across the entire circumarctic in the black-fruited crowberry (Figs. S2–S4) suggests that this species indeed has high capability for long-distance dispersal, as previously has been demonstrated for this and many other arctic plant species (17, 39, 40).
We therefore regard the long-distance dispersal hypothesis to be more parsimonious than one involving gradual migration, in particular because many birds are known to feed on fruits before migrating southward from their arctic/subarctic breeding grounds. For example, the whimbrel (Numenius phaeopus subsp. hudsonicus) and the American golden plover (Pluvialis dominica) have been reported to feed on Empetrum and Vaccinium before leaving their breeding grounds (41, 42). Their breeding ranges correspond well to the distribution of the Alaskan and other northwestern North American diploid E. nigrum populations inferred to be the closest extant relatives of the southern hemisphere E. rubrum. Notably, the whimbrel regularly winters in Patagonia, Tierra del Fuego, and the Falklands Islands (42), and whereas the American golden plover winters farther north in South America, it is also occasionally seen in Patagonia and Tierra del Fuego (41). A similar long-distance dispersal by birds was invoked to explain the New Amsterdam–Tristan da Cunha disjunction, separated by a distance of 8,000 km, in Phylica (Rhamnaceae) (43). Although current bird migratory patterns do not necessarily coincide with past migrations, these observations suggest that the bipolar disjunction in crowberries may have originated via bird-mediated long-distance dispersal. We therefore conclude that a single long-distance dispersal event from northwestern North America to southernmost South America, mediated by a Mid-Pleistocene bird, is sufficient to explain the extreme bipolar disjunction in today's crowberries and possibly also in other bipolar plant groups.
Materials and Methods
Plant material for DNA analysis and vouchers was collected in the field during 2002–2008 from southern South America and northern North America, Greenland, Europe, Russia, Japan, the Falkland Islands, and the Tristan da Cunha and Gough Islands group (Fig. 1 and Table S1). Leaf material was immediately dried in silica gel. The samples selected for this study represent the entire distribution and cover the morphological variation and the variation in ploidy level in the genus. The vouchers are deposited at the Botanical Museum (O), Natural History Museum, University of Oslo, and the DNA samples are deposited in the DNA bank at the National Centre for Biosystematics, Natural History Museum, University of Oslo.
The extraction of DNA from 41 accessions and the subsequent amplification of the plastid-encoded trnfMCAU-trnSUGA and trnSGCU-trnGUUC regions as well as the nuclear-encoded low-copy regions corresponding to exons 2–6 and intervening introns in RPB2-I and exons 31–32 and intervening introns in RPC2 of the second largest subunit of RNA polymerase II and III, respectively, were performed following standard procedures. Cloning was performed with the TOPO TA Cloning Kit (Invitrogen; see SI Materials and Methods for details). One or more consensus sequences from each of the cloned accessions were constructed as described (44). All sequences were manually aligned in Se-Al v. 2.0a.11 (45).
Phylogenetic Analyses and Dating.
Fifty-two plastid matK nucleotide sequences representing species of all eight subfamilies and 21 of the 24 tribes in Ericaceae (46) were downloaded from GenBank (Table S2). In addition, four new matK sequences representing the plastid diversity of Empetrum were included. The sequences were translated to amino acids and manually aligned using Se-Al v. 2.0a.11 (45).
We used four fossils to calibrate the phylogenetic tree. Flowers of Paleoenkianthus sayreville Nixon & Crepet, which are inferred to be the oldest known fossils (90 Myr) with ericaceous affinities (47), were used as a prior for the stem group age of Ericaceae. Leaves of Vaccinium creedensis Axelrod and Leucothoe nevadensis Axelrod dated to 26.5 Myr and 13–14 Myr (48, 49), respectively, were used as priors for the minimum age of Vaccinium and Leucothoe. Fossil seeds of Rhododendron newburyanum Collinson & Crane (35) dated to the Paleocene/Eocene transitional interval with a minimum age of 54.5 Myr (36) were used as a prior for the minimum age of Rhododendron.
Substitution model selection was done applying the Akaike information criterion as implemented in MrAIC v. 1.4.4 (14) combined with PHYML 3.0 (50), and resulted in a general time-reversible substitution model (51, 52) with rate heterogeneity across sites modeled using a discrete gamma model with four rate categories (53) as the best-fitting model (GTR+Γ). We also included the SRD06 codon position-based model (54) and tested it against the GTR+Γ model using Bayes factors as implemented in Tracer v. 1.5 (55, 56).
A relaxed molecular clock as implemented in BEAST v. 1.5.4 (37, 57) with uncorrelated lognormal distributed substitution rates for each branch was used to infer the phylogeny and estimate tMRCA of Empetrum. The tree was rooted with Enkianthus campanulatus (46) by constraining all other taxa to form a monophyletic group. The priors for tMRCA were set to lognormal distributions with logmean = 0, lognormal SD = 1.0, and offset set to 54.5 Myr, 26.5 Myr, and 13.5 Myr for Rhododendron, Vaccinium, and Leucothoe, respectively. The lognormal distribution for the priors thus fixed the minimum ages of the calibrated nodes but allowed the maximum ages to be sampled following a lognormal distribution with no hard limit (33). Because ref. 47 placed Paleoenkianthus sayreville near or within the family Ericaceae, the root age prior was set to a normal distribution with mean = 90 Myr and SD = 5.0, which approximated a distribution where 80.2 and 99.8 Myr formed the upper and lower limits of the 95% probability interval, respectively. Although the normal distribution puts no explicit hard boundaries on the maximum or minimum age of a node (33), an implicit minimum age of 54.8 Myr for the root of the tree was set by the lognormal prior on the Rhododendron tMRCA. Both a Yule speciation process and a birth/death speciation process were used as tree prior in separate analyses. The defaults in BEAUti v. 1.5.4 (57) were used for all other parameters. Four independent Markov chains were run for 20 million generations and the parameters were sampled every 2,000 generations. To test the influence of the priors on the posterior estimates, one additional chain was run for 100 million generations without data, sampling only the prior. The parameter estimates from each independent analysis were checked for stationarity and convergence using Tracer v. 1.5 (56), and joint estimates were produced using LogCombiner v. 1.5.4 (37, 57).
The phylogenetic relationships and the timing of lineage splits within Empetrum were coestimated independently from RPC2, RPB2-I, and the combined plastid dataset using BEAST v. 1.5.4 (37, 57). Substitution model selection was performed as described above for matK. Initial analyses using a relaxed molecular clock as implemented in BEAST v. 1.5.4 (37, 57) with uncorrelated lognormal distributed substitution rates for each branch showed that the marginal distribution of the SD of the rate variation included 0 and thus a strict molecular clock could not be rejected, and all subsequent analyses were consequently modeled using a strict molecular clock. The prior age of the Empetrum crown group was set to a normal distribution around the mean (5.5 Ma) estimated in the matK analysis and SD = 1.
The analyses were run with the prior number of “species” set to three (E. eamesii, E. nigrum, and E. rubrum) using the Species Tree Ancestral Reconstruction [i.e., *BEAST (58)] in BEAST v. 1.5.4. Each species was modeled under a separate coalescent prior (continuous population size and a constant root) and a strict molecular clock. Although *BEAST can be used to perform simultaneous analysis of multilocus data, the polyploid nature of many of our accessions violates the assumption of maximum two sequences per locus, and a simultaneous analysis of our three datasets was therefore inappropriate. Note that the separate coalescent priors do not imply monophyly for each species, only that the sequences assigned to each species are modeled under separate coalescent tree priors. The relationship between the species was simultaneously inferred under a Yule tree prior. Initial analyses showed that multiple nuclear sequences obtained from the allotetraploid E. atropurpureum were resolved with very strong support with either E. nigrum or E. eamesii and they were consequently assigned to the corresponding species. Markov chains were run as described for the matK analysis.
Supplementary Material
Acknowledgments
We thank Henry Hooghiemstra and Carlos Jaramillo for information on the lack of Empetrum fossils along the Cordillera, the National Centre for Biosystematics Journal Club and in particular Sanne Boessenkool for comments, and Reidar Elven for discussions on the taxonomy of Empetrum. All collectors (Table S1) are gratefully acknowledged. The work was funded by Grants 150322/720, 146515/420, and 170952/V40 from the Research Council of Norway (to C.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos.HM146928–HM147124 and HQ115639–HQ115642).
This article is a PNAS Direct Submission.
See Commentary on page 6341.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012249108/-/DCSupplemental.
References
- 1.Raven PH. Amphitropical relationships in the floras of North and South America. Q Rev Biol. 1963;38:151–177. [Google Scholar]
- 2.Thorne RF. Major disjunctions in the geographic ranges of seed plants. Q Rev Biol. 1972;47:365–410. [Google Scholar]
- 3.Du Rietz EG. Problems of bipolar plant distribution. Acta Phytogeogr Suec. 1940;13:215–282. [Google Scholar]
- 4.Darwin C. The Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: Penguin; 1859. 1st Ed; reprinted (1985) (Penguin Classics, Penguin Group, London) [PMC free article] [PubMed] [Google Scholar]
- 5.Wen J, Ickert-Bond SM. Evolution of the Madrean-Tethyan disjunctions and the North and South American amphitropical disjunctions in plants. J Syst Evol. 2009;47:331–348. [Google Scholar]
- 6.Humphries JC, Parenti LR. Cladistic Biogeography: Interpreting Patterns of Plant and Animal Distributions. Oxford: Clarendon; 1986. [Google Scholar]
- 7.Li JH, Alexander J, III, Ward T, Del Tredici P, Nicholson R. Phylogenetic relationships of Empetraceae inferred from sequences of chloroplast gene matK and nuclear ribosomal DNA ITS region. Mol Phylogenet Evol. 2002;25:306–315. doi: 10.1016/s1055-7903(02)00241-5. [DOI] [PubMed] [Google Scholar]
- 8.Wen J, Lowry PP, Walck JL, Yoo KO. Phylogenetic and biogeographic diversification in Osmorhiza (Apiaceae) Ann Mo Bot Gard. 2002;89:414–428. [Google Scholar]
- 9.Scherson RA, Vidal R, Sanderson MJ. Phylogeny, biogeography, and rates of diversification of New World Astragalus (Leguminosae) with an emphasis on South American radiations. Am J Bot. 2008;95:1030–1039. doi: 10.3732/ajb.0800017. [DOI] [PubMed] [Google Scholar]
- 10.Moore MJ, Jansen RK. Molecular evidence for the age, origin, and evolutionary history of the American desert plant genus Tiquilia (Boraginaceae) Mol Phylogenet Evol. 2006;39:668–687. doi: 10.1016/j.ympev.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 11.Gussarova G, Popp M, Vitek E, Brochmann C. Molecular phylogeny and biogeography of the bipolar Euphrasia (Orobanchaceae): Recent radiations in an old genus. Mol Phylogenet Evol. 2008;48:444–460. doi: 10.1016/j.ympev.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Anderberg AA. Phylogeny of the Empetraceae, with special emphasis on character evolution in the genus Empetrum. Syst Bot. 1994;19:35–46. [Google Scholar]
- 13.Murray DF, Mirré V, Elven R. (2009) in Flora of North America North of Mexico, ed Committee FoNAE (Oxford Univ Press, New York and Oxford), Vol 8.
- 14.Nylander JAA. 2004. MrAIC.pl ( http://www.abc.se/~nylander)
- 15.Hasegawa M, Kishino H, Yano TA. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 16.Abbott RJ, Brochmann C. History and evolution of the arctic flora: In the footsteps of Eric Hultén. Mol Ecol. 2003;12:299–313. doi: 10.1046/j.1365-294x.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- 17.Heusser CJ, Lowell TV, Heusser LE, Moreira A, Moreira S. Pollen sequence from the Chilean Lake District during the Llanquihue glaciation in marine oxygen isotope stages 4–2. J Quaternary Sci. 2000;15:115–125. [Google Scholar]
- 18.Markgraf V. Paleoenvironments and paleoclimates in Tierra del Fuego and southernmost Patagonia, South America. Palaeogeogr Palaeoclimatol Palaeoecol. 1993;102:53–68. [Google Scholar]
- 19.Heusser CJ, Rabassa J. Cold climatic episode of Younger Dryas age in Tierra-Del-Fuego. Nature. 1987;328:609–611. [Google Scholar]
- 20.Rabassa J, Coronato AM, Salemme M. Chronology of the Late Cenozoic Patagonian and their correlation with biostratigraphic glaciations units of the Pampean region (Argentina) J S Am Earth Sci. 2005;20:81–103. [Google Scholar]
- 21.Rabassa J, Coronato A. Glaciations in Patagonia and Tierra del Fuego during the Ensenadan Stage/Age (Early Pleistocene-earliest Middle Pleistocene) Quat Int. 2009;210:18–36. [Google Scholar]
- 22.Hulton NRJ, Purves RS, McCulloch RD, Sugden DE, Bentley MJ. The Last Glacial Maximum and deglaciation in southern South America. Quat Sci Rev. 2002;21:233–241. [Google Scholar]
- 23.Clark R, Huber UM, Wilson P. Late Pleistocene sediments and environmental change at Plaza Creek, Falkland Islands, South Atlantic. J Quaternary Sci. 1998;13:95–105. [Google Scholar]
- 24.Ljung K, Bjorck S, Hammarlund D, Barnekow L. Late Holocene multi-proxy records of environmental change on the South Atlantic island Tristan da Cunha. Palaeogeogr Palaeoclimatol Palaeoecol. 2006;241:539–560. [Google Scholar]
- 25.Ljung K, Bjorck S. Holocene climate and vegetation dynamics on Nightingale Island, South Atlantic—An apparent interglacial bipolar seesaw in action? Quat Sci Rev. 2007;26:3150–3166. [Google Scholar]
- 26.Wace NM, Dickson JH. Part II. Terrestrial botany of Tristan da Cunha islands. Philos Trans R Soc Lond B Biol Sci. 1965;249:273–360. [Google Scholar]
- 27.Bennett KD, Gribnitz KH, Kent LE. Pollen analysis of a Quaternary peat sequence on Gough Island, South-Atlantic. New Phytol. 1989;113:417–422. doi: 10.1111/j.1469-8137.1989.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 28.Friis EM. The Damgaard flora: A new Middle Miocene flora from Denmark. Bull Geol Soc Den. 1979;27:117–142. [Google Scholar]
- 29.Van der Burgh J. Miocene floras in the lower Rhenish Basin and their ecological interpretation. Rev Palaeobot Palynol. 1987;52:299–366. [Google Scholar]
- 30.Graur D, Martin W. Reading the entrails of chickens: Molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Gandolfo MA, Nixon KC, Crepet WL. Selection of fossils for calibration of molecular dating models. Ann Mo Bot Gard. 2008;95:34–42. [Google Scholar]
- 32.Donoghue PCJ, Benton MJ. Rocks and clocks: Calibrating the Tree of Life using fossils and molecules. Trends Ecol Evol. 2007;22:424–431. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Ho SYW. Calibrating molecular estimates of substitution rates and divergence times in birds. J Avian Biol. 2007;38:409–414. [Google Scholar]
- 34.Ho SYW, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- 35.Collinson M, Crane P. Rhododendron seeds from the Palaeocene of southern England. Bot J Linn Soc. 1978;76:195–205. [Google Scholar]
- 36.Collinson ME, Hooker JJ, Groecke DR. Cobham lignite bed and penecontemporaneous macrofloras of southern England: A record of vegetation and fire across the Paleocene-Eocene Thermal Maximum. Spec Pap Geol Soc Am. 2003;369:333–349. [Google Scholar]
- 37.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollan K, Heide OM, Lye KA, Heun M. Genetic variation, taxonomy and mountain-hopping of four bipolar Carex species (Cyperaceae) analysed by AFLP fingerprinting. Aust J Bot. 2006;54:305–313. [Google Scholar]
- 39.Alsos IG, et al. Frequent long-distance plant colonization in the changing Arctic. Science. 2007;316:1606–1609. doi: 10.1126/science.1139178. [DOI] [PubMed] [Google Scholar]
- 40.Brochmann C, Brysting AK. The Arctic—An evolutionary freezer? Plant Ecol Divers. 2008;1:181–195. [Google Scholar]
- 41.Johnson OW, Connors PG. In: The Birds of North America Online. Poole A, editor. Ithaca, NY: Cornell Lab of Ornithology; 1996. [Google Scholar]
- 42.Skeel MA, Mallory EP. In: The Birds of North America Online. Poole A, editor. Ithaca, NY: Cornell Lab of Ornithology; 1996. [Google Scholar]
- 43.Richardson JE, Fay MF, Cronk QCB, Chase MW. Species delimitation and the origin of populations in island representatives of Phylica (Rhamnaceae) Evolution. 2003;57:816–827. doi: 10.1111/j.0014-3820.2003.tb00293.x. [DOI] [PubMed] [Google Scholar]
- 44.Popp M, Erixon P, Eggens F, Oxelman B. Origin and evolution of a circumpolar polyploid species complex in Silene (Caryophyllaceae) inferred from low copy nuclear RNA polymerase introns, rDNA, and chloroplast DNA. Syst Bot. 2005;30:302–313. [Google Scholar]
- 45.Rambaut A. 1996. Se-Al: Sequence Alignment Editor ( http://tree.bio.ed.ac.uk/software/seal/)
- 46.Kron K, et al. Phylogenetic classification of Ericaceae: Molecular and morphological evidence. Bot Rev. 2002;68:335–423. [Google Scholar]
- 47.Nixon KC, Crepet WL. Late Cretaceous fossil flowers of Ericalean affinity. Am J Bot. 1993;80:616–623. [Google Scholar]
- 48.Axelrod DI. The Late Oligocene Creede Flora, Colorado. Berkeley, CA: University of California Press; 1987. [Google Scholar]
- 49.Axelrod DI. The Miocene Purple Mountain Flora of Western Nevada. Berkeley, CA: University of California Press; 1995. [Google Scholar]
- 50.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 51.Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- 52.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 1986;80:57–86. [Google Scholar]
- 53.Yang ZH. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 54.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- 55.Suchard MA, Kitchen CMR, Sinsheimer JS, Weiss RE. Hierarchical phylogenetic models for analyzing multipartite sequence data. Syst Biol. 2003;52:649–664. doi: 10.1080/10635150390238879. [DOI] [PubMed] [Google Scholar]
- 56.Rambaut A, Drummond AJ. 2007. Tracer ( http://beast.bio.ed.ac.uk/Tracer)
- 57.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.