Abstract
Primers and probes were developed for the element-specific detection of cry1A.105 and cry2Ab2 genes, based on their DNA sequence as present in GM maize MON89034. Cry genes are present in many genetically modified (GM) plants and they are important targets for developing GMO element-specific detection methods. Element-specific methods can be of use to screen for the presence of GMOs in food and feed supply chains. Moreover, a combination of GMO elements may indicate the potential presence of unapproved GMOs (UGMs). Primer-probe combinations were evaluated in terms of specificity, efficiency and limit of detection. Except for specificity, the complete experiment was performed in 9 PCR runs, on 9 different days and by testing 8 DNA concentrations. The results showed a high specificity and efficiency for cry1A.105 and cry2Ab2 detection. The limit of detection was between 0.05 and 0.01 ng DNA per PCR reaction for both assays. These data confirm the applicability of these new primer-probe combinations for element detection that can contribute to the screening for GM and UGM crops in food and feed samples.
Electronic supplementary material
The online version of this article (doi:10.1007/s00216-011-4875-9) contains supplementary material, which is available to authorized users.
Keywords: GMO, UGM, Real-time PCR, Cry genes
Introduction
In the European Union (EU) regulations have been implemented containing harmonized provisions for the risk assessment and authorization of genetically modified organisms (GMOs) as well as for traceability, labelling and post-marketing surveillance of the use of GMOs in the food and feed chain [1, 2]. At present (January 2011) a total of 31 genetically modified (GM) crop events in 6 different species are authorized in the EU including 17 for maize, 6 for cotton, 3 for oilseed rape, 3 for soya beans, 1 for potato and 1 for sugar beet [3]. Appropriate methods for detection and quantification of GMOs are required to control the compliance with legal provisions [4]. The European Union Reference Laboratory (EURL) provides official methods for the event-specific detection and quantification of material from all authorized GMOs [5] and event-specific or construct-specific methods for a number of unauthorised GMs (UGMs). UGMs can be GMOs that are approved in other countries but not within the EU or that have not been approved elsewhere in the world [6]. The EU threshold for UGMs is zero, which means that their presence is not allowed in food or feed materials [7].
The detection and identification of specific elements like promoters, coding sequences and terminators present in GM events can be very helpful to identify UGMs [8, 9]. If the presence of specific elements cannot be accounted for by the detected events in a particular sample, this is indicative for the presence of a UGM. Querci et al. [10] developed a real-time PCR based ready-to-use multi-target analytical system for simultaneous identification of 39 GM events by using event-specific, construct-specific and species-specific primers and probe combinations. This system allows identification of GM plants approved in EU and in the process of being approved. However, as this system only specifically identifies these GMO events, it is not suitable for the detection of UGMs. For this the so-called GMO matrix approach is required. This approach allows to describe and to identify each GMO by the presence of the respective target sequences in comparison with the screening results obtained for the analyzed sample. This matrix basically consists of a table in which the rows represent the content of genetic elements of the respective GMO, while the columns define the targeted sequence elements analyzed in the matrix [10]. Up to date GMO databases and a network of collaborating laboratories are required for the addition of further targets which are frequently present in GM plants [11, 12]. With respect to this development for new approaches for GMO detection, including UGMs, it is important to continuously expand and validate detection methods for GM events and elements, including promoters, coding sequences and terminators.
Cry genes are the most commonly applied genes present in GM events to achieve insect resistance. There are more than 200 cry toxins listed by the Bt delta-endotoxin nomenclature committee [13]. Their origin is from the soil bacterium Bacillus thuringiensis (Bt), derived GM elements may result from codon optimization or even domain swapping. Cry toxins expressed in GM plants show insecticidal activity against one or more insect pests. They have a defined spectrum of activity generally restricted to a few species of insects or their larvae. For this reason, it is common to find more than one cry gene in the same GM plant to render the crops resistant to a broader range of insects. These toxins are classified according to their amino acid sequence homology. There are three domains and five conserved sequence blocks common to a large majority of these proteins [13, 14]. Different cry proteins can show 19–90% of identity among them [15]. PCR amplification using gene-specific primers has been performed for the detection of known and novel cry genes [16–21].
Cry1A.105 and cry2Ab2 are present in maize MON89034 (MON-89Ø34-3) and confer resistance to Lepidopteran pests. They provide control of Ostrinia species (e.g. European corn borer, Asian corn borer), and Diatraea species (e.g. South-western corn borer, sugarcane borer). GM maize MON89034 also provides a high level control of fall armyworm (Spodoptera frugiperda) and improves protection from damage caused by corn earworm (Helicoverpa zea) [22]. Cry1A.105 and cry2Ab2 are also present in other related stacked GM maize varieties that include MON89034: MON89034 × MON88017, MON89034 × NK603 [23, 24] and MON89034 × TC1507 × MON88017 × DAS-59122-7 [25].
The cry1A.105 gene codes for a protein consisting of 3 domains that are identical with other cry proteins used in GM plants. Cry1A.105 shows 93.6%, 90%, and 76.7% of amino acid sequence homology respectively with the cry1Ac, cry1Ab and cry1F proteins from B. thuringiensis. The amino acid sequences of domains I and II are identical with the respective domains from cry1Ab and cry1Ac proteins. Domain III is almost identical to cry1F, while the C-terminal domain is identical to cry1Ac [26].
The cry2Ab protein present in MON89034 is also present in GM cotton MON15985 and 3 stacked varieties of this cotton: LLcotton25 × MON15985, MON15985 × MON1445, MON15985 × MON88913 [27].
Molecular characterization of GM maize MON89034 demonstrated that the DNA inserted into the genome is present at a single locus and contains one functional copy of the cry1A.105 and the cry2Ab2 expression cassettes [22]. Current detection of these genes is based on validated methods for event-specific detection of GM maize MON89034 and GM cotton MON15985 [28–30]. It is important to also develop primers for element-specific detection of these cry targets. This will enable the use of these elements in a matrix-based approach for the detection of UGMs, as their presence, in the absence of the MON89034 and MON15985 events, may identify GM varieties that are not yet approved for the European market. The specificity and sensitivity of the methods need to be tested by the use of reference materials [11].
On this basis, the present study shows the design of real-time PCR assays for specific detection of the cry1A.105 and cry2Ab2 genes, based on their DNA sequence present in GM maize MON89034. These assays were evaluated by real-time PCR in terms of specificity, limit of detection and efficiency.
Materials and methods
Sample material and DNA isolation
Certified Reference Materials (CRMs), certified as being non-GM maize containing less than 2.0 g/kg of MON810, MON863, NK603, MON88017 and MON89034, and samples certified to contain a percentage of a defined GM maize containing a cry-related sequence, were selected in order to determine the specificity of cry1A.105 and cry2Ab2 primers: ERM-BF412f (SYN-BTØ11-1 or Bt11, 4.89%), ERM-BF411f (SYN-EV176-9 or Bt176, 5%), ERM-BF413f (MON-ØØ81Ø-6 or MON810, 5%), ERM-BF418d (DAS-Ø15Ø7-1 or TC1507, 9.86%), ERM-BF416d (MON-ØØ863-5 or MON863, 9.85%), ERM-BF424d (DAS-59122-7 or DAS59122, 9.87%), ERM-BF423d (SYN-IR6Ø4-5 or MIR604, 9.85%), ERM-BF422d (DAS-21Ø23-5 × DAS-24236-5 or 281-24-236 × 3006-210-23, 10%) (European Reference Materials, ERM, Geel, Belgium) and AOCS 0906-E (MON-89Ø34-3 or MON89034, >99.42%), AOCS 0406-D (MON88Ø17-3 or MON88017 >99.05%), AOCS 0406-A (non-transgenic maize, <0.2%), AOCS 0804-C (MON-ØØ531-6 or MON531 >97.39%), AOCS 0804-D (MON-15985-7 or MON15985 >98.45%) (American Oil Chemists’ Society, AOCS; Illinois, USA). The DNeasy Plant Mini Kit (Qiagen) was used for DNA isolation according to the manufacture’s protocol, only modified by including a lysis treatment with CTAB buffer (20 g/L CTAB; 1.4 M NaCl; 0.1% Tris; 20 mM Na2EDTA, pH 8.0) instead of AP1 buffer supplied in the kit. Also 20 μL of proteinase K (20 mg/mL) was added to each sample during the incubation at 60 ° C for 15 min. DNA concentrations were determined on a Thermo Scientific NanoDrop™ spectrophotometer with measurements at UV region from 220 to 350 nm.
Primer design
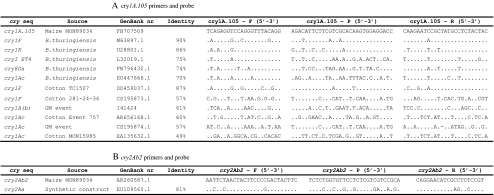
Primers and probes were designed based on the sequences of the cry1A.105 and cry2Ab2 genes that are present in MON89034, GenBank accession: FB707509 and AR260587.1, respectively [31]. Sequence analysis was performed at the GenBank database by using the Blastn tool (Basic Local Alignment Search Tools Nucleotide) of the National Center for Biotechnology Information (NCBI). The alignments among similar sequences were performed by using the software ClustalW [32] with other sequences and cry genes that are present in GM events. Beacon Designer 7.0 was used to obtain the primers and probes (Premier Biosoft, California, USA). The software settings were adjusted for the following parameters as follows: melting temperature (Tm) of 59 ± 1 ° C, length from 18 to 30 bp and amplicon from 75 to 150 bp. The probes were designed with 18 to 30 bp, Tm 10 ± 2 ° C higher than primer Tm and they were labelled with 6-carboxyfluorescein (FAM) and TAMRA-quenched oligonucleotide. Primers and probes are described in Table 1. The endogenous hmg (high mobility group) maize gene was amplified by primers ZM1-F, ZM1-R and probe ZM to confirm the quality of the isolated DNA [28]. The performance of the hmg PCR using DNA isolated with the DNA isolation method described in this article has been in-house validated as the endogenous reference gene of several event-specific methods [43].
Table 1.
Primers and probes for cry genes and hmg gene detection. F = forward primer, R = reverse primer, P = probe
| Primer | Sequence (orientation 5′- 3′) | Amplicon size | Reference |
|---|---|---|---|
| cry1A.105 - F1 | TCAGAGGTCCAGGGTTTACAGG | 133 bp | this work |
| cry1A.105 - R1 | GTAGTAGAGGCATAGCGGATTCTTG | ||
| cry1A.105 - P | AGACATTCTTCGTCGCACAAGTGGAGGACC | ||
| cry2Ab2 - F | AATTCTAACTACTTCCCCGACTACTTC | 121 bp | this work |
| cry2Ab2 - R | ACGGAGAGGCGATGTTCCTG | ||
| cry2Ab2 - P | TCTCTGGTGTTCCTCTCGTCGTCCGCA | ||
| ZM1 – F (hmg) | TTGGACTAGAAATCTCGTGCTGA | 79 bp | [27] |
| ZM1 – R (hmg) | GCTACATAGGGAGCCTTGTCCT | ||
| probe ZM (hmg) | CAATCCACACAAACGCACGCGTA |
Real-time PCR
PCR amplification was performed in a 96-well microtiter plate in a total volume of 25 μL containing 12.5 μL Diagenode Mastermix 2× (real-time PCR Mastermix, Diagenode, Liege, Belgium), 300 nM of each primer, 150 nM probe, 5.6 μL milliQ water and 5.0 μL of DNA. Samples were amplified on iQ5 and MyiQ™ (Bio-Rad Laboratories, California, USA) according to the following protocol: 2 min at 50 °C, 10 min at 95 °C, 45 cycles of: 15 s at 95 °C and 1 min at 60 °C. All samples were analyzed in duplicate. The real-time PCR conditions were the same for all PCR reactions.
The specificity of cry1A.105 and cry2Ab2 primers/probe was verified by testing all CRMs listed in Table 3. The duplicate test was performed by using a final concentration of 50 ng DNA per PCR reaction.
Table 3.
List of Certified Reference Materials used to assess the specificity of the cry1A.105 and cry2Ab2 primers and the results obtained after duplicate testing
| Sample | Reference material | % GMO | Estimated copy number in 50 ng genomic DNA | cry element | cry1A.105 detection | cry2Ab2 detection |
|---|---|---|---|---|---|---|
| maize | MON89034 | 100% | 18349 | cry1A.105, cry2Ab2 | + | + |
| Bt11 | 4.89% | 897 | cry1Ab | - | - | |
| Bt176 | 5% | 917 | cry1Ab | - | - | |
| MON810 | 5% | 917 | cry1Ab | - | - | |
| TC1507 | 9.86% | 1809 | cry1F | - | - | |
| MON863 | 9.85% | 1807 | cry3Bb1 | - | - | |
| DAS59122 | 9.87% | 1811 | cry34Ab1, cry35Ab1 | - | - | |
| MIR604 | 9.85% | 1807 | cry3A | - | - | |
| MON88017 | >99.05% | >18174 | cry3Bb1 | - | - | |
| Non-transgenic maize* | <0.2%* | <37 | none | - | - | |
| cotton | 281-24-236x3006-210-23 | 10% | 2146 | cry1Ac, cry1F | - | - |
| MON531 | >97.39% | >20899 | cry1Ac | - | - | |
| MON15985 | >98.45% | >21127 | cry2Ab2 | - | + |
*Non-transgenic maize AOCS 0406-A contained <0.2% of MON810, MON863, NK603, MON88017 and MON89034 (http://www.aocs.org/files/TechnicalPDF/0406A.pdf)
The real-time PCR analysis was evaluated for each primer/probe pair by the serial dilution of CRM MON89034 100% (AOCS 0906-E) with water to final concentrations of 150, 30, 6, 1.20, 0.60, 0.25, 0.05 and 0.01 ng DNA per PCR reaction. The copy numbers in the calibration curve samples are obtained by dividing the amount of sample DNA (picograms) by the published average 1C value for the Zea mays genome corresponding to 2.725 pg [29, 33]. Therefore, the estimated copy numbers in the calibration curves were calculated to be 55,046; 11,009; 2,202; 440; 220; 88; 18 and 4 copies of haploid genome, respectively. Concentrations from 0.01 to 150 ng DNA per PCR were tested in 9 different PCR runs in duplicate in a total period of 39 days of experiment. The lowest concentrations, 0.01 and 0.05 ng DNA per PCR, were tested an additional 6 times in 8 of the PCR runs. Outliers were identified and excluded of the analysis based on quartile distribution of Ct-values [34]. All real-time PCR runs were evaluated separately by using the automatic settings for each run. Two observations for the 0.05 ng DNA per PCR were used for the evaluation of the efficiency and the correlation coefficient (R2).
The 95% confidence interval (CI) was calculated based on the difference between the upper and lower 95% confidence limits with the standard formula: upper/lower 95% confidence limit = sample mean ± 1.96 (standard error of the sample mean).
Results
DNA quality
The DNA isolated from 100 mg of GM maize MON89034 (AOCS0906 - E) and non-GM maize (AOCS 0406 - A) showed a final concentration range of 30 ng/μL to 54 ng/μL and an absorbance ratio A260/280 nm of 1.83 to 1.92. The DNA quality of all CRMs from Table 1 was also checked by spectrophotometer and PCR. Expected amplification products for GM maize MON89034 were obtained by using 8 different DNA concentrations from 0.01 to 150 ng DNA per PCR reaction with hmg primers and probe. Amplification of hmg was observed for all maize samples as expected. The Ct-values for hmg detection ranged from 22.82 to 37.25 for the DNA concentration range tested.
Primer and probe design
Primers and probes were designed based on the sequences of the cry1A.105 and cry2Ab2 genes present in GM maize MON89034 [31]. Blastn search and ClustalW alignment showed that several regions of the cry1A.105 sequence between positions 5,542 and 5,748 are completely identical with GM events that contain the cry1Ac gene, for instance: brinjal (GenBank DM460255.1); soybean (GenBank HB386795.1) and cotton (GenBank EA731070.1). Therefore, the most dissimilar region with other GM sequences was selected for primer design in order to avoid cross-reactivity with non-target sequences. The resulting amplicons of 133 bp for cry1A.105 and 121 bp for cry2Ab2 were also analysed by Blastn analysis against the patent sequence databases and nucleotide sequence collection at the NCBI website [35]. For the patent database, both the cry1A.105 and cry2Ab2 amplicons returned 100 blast hits from a total of 26 and 16 different patents, respectively. All hits found showed high sequence coverage between 93% and 100%. For the nucleotide sequence collection, cry1A.105 and cry2Ab2 amplicons were also submitted to a Blastn analysis. For the hits returned after the cry1A.105 amplicon query, 18 hits showed a high coverage of at least 96%, all from different cry genes of B. thuringiensis. Those with at least 70% sequence identity were aligned with the cry1A.105 sequence. For the cry2Ab2 amplicon, 3 of 20 blast hits were correspondent to cry genes and only 1 showed coverage of more than 90% and was aligned with the cry2Ab2 sequence. The remaining sequences showed coverage of 41% or less and were not analysed further. Both amplicons were aligned using ClustalW with sequences that showed sequence coverage between 90% and 98% and with an identity of at least 70%. Cry genes present in GM events were also included in the alignment. Table 2 shows the identity of cry1A.105 and cry2Ab2 primers and probe sequences with the selected sequences.
Table 2.
Comparison of primers and probe sequences with related cry sequences from B. thuringiensis and GM events. Only nucleotides that differed from cry1A.105 (A) or cry2Ab2 (B) are indicated. Identity was calculated based only on the primers and probe sequences. F = forward primer, R = reverse primer, P = probe
Specificity
Thirteen CRMs were tested with both the cry1A.105 and the cry2Ab2 method. A cry1A.105 element was present in 1 of these and a cry2Ab2 element in 2. The positive control maize MON89034 produced the expected amplification signal with Ct mean ± SD equal to 24.64 ± 0.06 for cry1A.105 and 24.98 ± 0.09 for cry2Ab2 detection. GM cotton MON15985 generated the expected amplicon with cry2Ab2 primers and probe and it showed a Ct mean ± SD equal to 25.58 ± 0.10. Neither of the two methods showed amplification for any of the non-target samples tested (Table 3).
Limit of detection
To estimate the limit of detection (LOD), GM maize MON89034 was analyzed from a pool of 3 independent DNA isolations. Two different DNA concentrations, 0.01 and 0.05 ng DNA per PCR reaction were analysed in a total of 8 replicates coming on 8 different days of PCR analysis. At 0.01 ng DNA, cry1A.105 and cry2Ab2 elements were detected in respectively 77% and 69% of the samples tested. The amount of 0.05 ng showed 100% of detection for cry1A.105 and 94% of detection for cry2Ab2 (Table 4). Non GM maize was also tested in a total of 18 times in 9 PCR runs, showing a positive signal 5 times (28%, Ct mean 38.72) for cry1A.105 detection and 4 times (22%, Ct mean 37.31) for cry2Ab2 detection. No amplification was observed for the 4 extraction controls analyzed or the 16 water controls.
Table 4.
Limit of detection for cry1A.105 and cry2Ab2 in a total of 64 samples tested in 8 independent PCR runs for 2 different concentrations (0.05 and 0.01 ng DNA/PCR)
| ng/PCR | Estimated Copy number per PCR reaction | Ct mean | positive/total | % positive |
|---|---|---|---|---|
| cry1A.105 | ||||
| 0.05 | 18 | 35.43 | 64/64 | 100 |
| 0.01 | 4 | 37.85 | 49/64 | 77 |
| cry2Ab2 | ||||
| 0.05 | 18 | 35.36 | 60/64 | 94 |
| 0.01 | 4 | 36.96 | 44/64 | 69 |
Efficiency
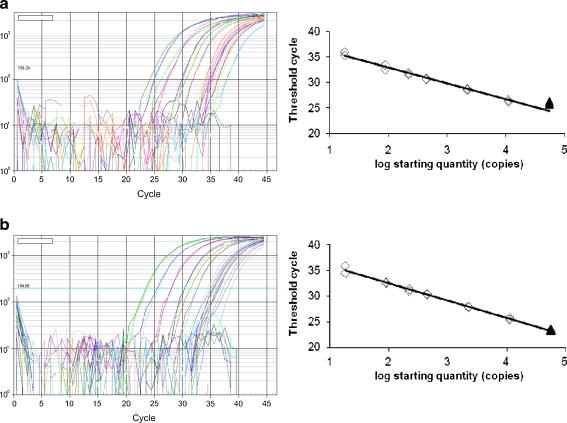
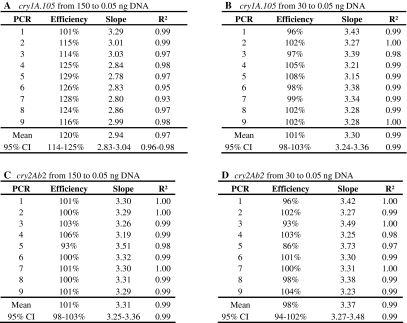
The efficiency and linear response were evaluated for both PCR methods in 9 separate experiments. Visual inspection of the calibration lines showed a clear linear trend for all tests, when Ct values were plotted against the log value of the DNA amount. The only exception was the 150 ng sample for the cry1A.105 PCR that was above the regression line in all cases. Figure 1 shows the amplification plots and standard curve of the 8th PCR run that showed results close to the average of all PCRs for each assay. For generation of the regression lines, the full range from 0.05 to 150 ng DNA per reaction was used as well as a smaller range, omitting the 150 ng samples. The main characteristics of the PCR regression lines are shown in Table 5. The concentration and the Ct-values obtained for each PCR run are shown in the Electronic Supplementary Material (Table S1).
Fig. 1.
Amplification plots and linear regression of 8th PCR run for GM maize MON89034. a cry1A.105 detection. b cry2Ab2 detection
Table 5.
Parameters of PCR standard curve for cry1A.105 and cry2Ab2 detection based on 9 PCR runs with samples tested in duplicate for 150, 30, 6, 1.20, 0.60, 0.25 and 0.05 ng DNA per PCR reaction
Discussion
Primer design and analysis of specificity
The cry1A.105 amplicon showed a high identity with cry1F, cry1Ac and cry1Ab nucleotide sequences from B. thuringiensis. This was anticipated since the cry1A.105 protein has high identity with cry1F, cry1Ac and cry1Ab proteins [13, 26]. There are five conserved regions common to the majority of cry proteins [14]. Moreover, several cry sequences are nearly identical representing slight variants of the same gene [15, 19]. The cry2Ab2 amplicon also showed a high identity of 80% with a cry2Aa synthetic gene (GenBank EU109585.1), which was applied in GM chickpea that is in development [16]. Therefore, it is very important to perform a thorough analysis of homology in the development phase of the PCR. Also in the testing phase it is very important to include as many related cry-containing samples as possible to rule out unwanted cross-reactions. The primer/probe combination designed to detect the cry1A.105 gene showed the expected result for amplification of this gene in GM maize MON89034. Both the cry1A.105 and the cry2Ab2 primers and probe successfully detected the target gene in GM maize MON89034. The cry2Ab gene in GM cotton MON15985 was also detected although the primers and probe were not specifically designed for this variety due to the undisclosed MON15985 sequence [27]. Given the results, it is very likely that the nucleotide sequence of the 121 bp cry2Ab2 amplicon analyzed in this work share similarities between both GM events. Both primer and probe pairs also proved to be very specific since no cross-reactivity was observed with any of the non-target samples tested. We tested all twelve available certified reference materials containing one or more cry sequences. Although 100% material was not available, the estimated number of copies tested was at least 897, which should be enough for a substantial cross-reaction to show up during the PCR analysis. Of course, the emergence of new varieties means that specificity testing for GM elements will be an ongoing process. The performance of the hmg PCR using DNA isolated with the DNA isolation method described in this article was already in-house validated as the endogenous reference gene of several event-specific methods [43].
Limit of detection
The LOD, corresponding to the lowest amount in a sample that can be reliably detected, was experimentally determined to be between 0.01 and 0.05 ng DNA per PCR reaction. Theoretically, this corresponds to 4–18 copies of the haploid genome of GM maize MON89034. It is also equivalent to 0.02–0.10% of MON89034 in case of 50 ng DNA input per PCR reaction, or 0.01–0.05% of MON89034 in case of 100 ng DNA input. According to the ENGL Method Performance Requirements [36] analytical methods should detect the presence of an analyte at 1/20th of the threshold relevant for legislative requirements at least 95% of the time. The threshold for labelling GM products in EU is 0.9% of GMO which corresponds to an LOD value of less than 0.045% of GM in terms of ENGL criteria [36]. For 50 ng DNA input, the 0.045% is in between the upper and lower estimations for the LOD, so it cannot be guaranteed that 50 ng input DNA would be sufficient to meet the ENGL criteria for either method. For 100 ng input of DNA in a PCR reaction, this percentage is just below the estimated upper level of the LOD. It can be concluded that both methods meet the ENGL Method Performance Requirements for LOD when 100 ng maize DNA is used in the PCR reaction.
The non-GM maize AOCS 0406-A showed amplification for both methods in a limited number of reactions. Ct-values were similar to the Ct-values found for the lowest DNA concentration tested. However, the total percentage of positive reactions was approximately 3 times lower than that of the lowest DNA concentration. According to the analysis certificate of the non-GM maize AOCS 0406-A material used in this study, it contained less than 0.2% of MON810, MON863, NK603, MON88017 and MON89034 [36]. The certificate showed results for MON810, MON863, NK603 and MON88017 testing. MON810 was identified in 13 out of 15 samples while NK603 was identified in 1 out of 15 samples. The percentages were in the range of 0.05–0.2%, for presence of MON810 and NK603. MON863 and MON88017 were not detected in any of the 15 samples. The certificate showed no data on testing for MON89034 [37]. Also other non-GM CRMs have sometimes been reported to contain low levels of GM material [38–41]. In our experiments none of the 4 extraction controls and none of the 16 MQ controls showed any amplification for either PCR method. It is therefore unlikely that the unexpected positive results were due to a contamination in our laboratory. A more likely explanation would be the unintended low level presence in the CRM of 0% MON89034, or another cry1A.105 and cry2Ab containing GM.
Efficiency and quantitative response
The cry2Ab2 method performed in a linear quantitative manner in the full range tested, between 0.05 and 150 ng DNA per PCR reaction. In 9 experiments, the average slope of the trend line ranged from −3.19 to −3.51 corresponding to 106–93% of efficiency. The R² average value for cry2Ab2 detection ranged from 0.98 to 1.00. According to Table 5c and Fig. 1, results for cry2Ab2 are in agreement with ENGL requirements that specify an efficiency range between 90 and 110% and an R² higher than or equal to 0.98, in order for a PCR method to be used for quantification [36].
For the cry1A.105 detection, 150 ng DNA input seemed not to fit a linear regression (Table 5 and Fig. 1). In fact, for all repetitions, the 150 ng samples showed a higher Ct value than would be expected on the basis of a linear regression for the other samples. The average Ct-value for the amount of 150 ng DNA was 25.72 ± 0.73 and 25.96 ± 0.29 for 30 ng DNA, which can be expressed as a delta Ct of 0.24 ± 0.44 (available as Electronic Supplementary Material, Table S1). In contrast, the expected delta Ct for a fivefold dilution is 2.32, assuming a PCR efficiency of 100%. Also, the R² overall average value for cry1A.105 decreased from 0.99 to 0.97 when the 150 ng DNA was included. Finally, the overall efficiency dropped from a theoretically impossible 120% to a very acceptable 101% when 150 ng was excluded from the analysis. All these observations indicated that the cry1A.105 PCR was sensitive to an inhibitory factor that may have been present in the DNA isolation that was used for these experiments. An option to overcome this possible limitation would have been the design of new primers and probe. However, this might have compromised the specificity, given the high homology between the cry1A.105 element and other cry elements. For cry2Ab2 detection, inhibition at 150 ng DNA per PCR reaction was not observed and the R² and efficiency did not change when this concentration was omitted from the analysis. Data showed that the designed PCR methods performed in a linear quantitative manner between 0.05 and 30 ng DNA per PCR reaction for cry1A.105 and between 0.05 and 150 ng DNA per PCR reaction for cry2Ab2.
Conclusions
New primers and probes specific for cry1A.105 and cry2Ab2 detection were developed. They showed a high specificity for target detection without cross-reactivity with other non-target GMOs tested. Also the limit of detection and quantitative response make both real time PCR methods adequate for the screening and detection of GMOs and GMO-derived products. These methods can be included in further interlaboratory studies to contribute to the development of strategies for GMO detection. To use these element-specific methods for quantification relative to the endogenous maize gene hmg it is necessary to perform an in-house validation including relative repeatability and reproducibility standard deviation and trueness. The parameters should comply with the ENGL Method Performance Requirements.
The detection of cry1A.105 and cry2Ab2 genes can be used to detect approved GMO- and UGM-derived products, which will aid the efforts to maintain GMO regulations within the European Union as well as worldwide.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 335 kb)
Acknowledgments
AZD was supported by a doctoral fellowship from CAPES (Coordination for the Improvement of Higher Education Personnel), Ministry of Education, Brazil, according the cooperation program CAPES-Wageningen (project 005/09).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Regulation (EC) No. 1829/2003 of the European Parliament and of the Council of 22 September 2003 on Genetically Modified Food and Feed. Off J Eur Comm L268:1–23
- 2.Regulation (EC) No. 1830/2003 of the European Parliament and of the Council of 22 September 2003 Concerning the Traceability and Labelling of Genetically Modified Organisms and the Traceability of Food and Feed Products Produced from Genetically Modified Organisms and Amending Directive 2001/18/EC. Off J Eur Comm L268:24–28
- 3.European Commission. Food and feed safety. Genetically modified food and feed. List of GMOs authorized in the EU. http://ec.europa.eu/food/dyna/gm_register/index_en.cfm. Accessed 27 Jan 2011
- 4.Marmiroli N, Maestri E, Gullì M, Malcevschi A, Peano C, Bordoni R, De Bellis G. Methods for detection of GMOs in food and feed. Anal Bioanal Chem. 2008;392:369–384. doi: 10.1007/s00216-008-2303-6. [DOI] [PubMed] [Google Scholar]
- 5.European Union Reference Laboratory for GM Food and Feed (2010) Status of dossiers. EURL-GMFF validation process. http://gmo-crl.jrc.ec.europa.eu/statusofdoss.htm. Accessed 26 June 2010
- 6.Holst-Jensen A. Testing for genetically modified organisms (GMOs): Past, present and future perspectives. Biotech Adv. 2009;27:1071–1082. doi: 10.1016/j.biotechadv.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 7.Davison J. GM plants: Science, politics and EC regulations. Plant Sci. 2010;178:94–98. doi: 10.1016/j.plantsci.2009.12.005. [DOI] [Google Scholar]
- 8.Prins TW, Van Dijk JP, Beenen HG, Van Hoef AMA, Voorhuijzen MM, Schoen CD, Aarts HJM, Kok EJ. Optimised padlock probe ligation and microarray detection of multiple (non-authorised) GMOs in a single reaction. BMC Genomics. 2008;9:584. doi: 10.1186/1471-2164-9-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruttink T, Demeyer R, Gulck E, Droogenbroeck B, Querci M, Taverniers I, Loose M. Molecular toolbox for the identification of unknown genetically modified organisms. Anal Bioanal Chem. 2010;396:2073–2089. doi: 10.1007/s00216-009-3287-6. [DOI] [PubMed] [Google Scholar]
- 10.Querci M, Foti N, Bogni A, Kluga L, Broll H, Van den Eede G. Real-time PCR-based ready-to-use multi-target analytical system for GMO detection. Food Anal Methods. 2009;2:325–336. doi: 10.1007/s12161-009-9093-0. [DOI] [Google Scholar]
- 11.Waiblinger HU, Grohmann L, Mankertz J, Engelbert D, Pietsch K. A practical approach to screen for authorized and unauthorised genetically modified plants. Anal Bioanal Chem. 2010;396:2065–2072. doi: 10.1007/s00216-009-3173-2. [DOI] [PubMed] [Google Scholar]
- 12.Querci M, Van den Bulcke M, Žel J, Van den Eede G, Broll H. New approaches in GMO detection. Anal Bioanal Chem. 2010;396:1991–2002. doi: 10.1007/s00216-009-3237-3. [DOI] [PubMed] [Google Scholar]
- 13.Crickmore N, Zeigler DR, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean DH. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62(3):807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Maagd RA, Bravo A, Crickmore N. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 2001;17:193–199. doi: 10.1016/S0168-9525(01)02237-5. [DOI] [PubMed] [Google Scholar]
- 15.Hofte H, Whiteley HR. Insecticidal Crystal Proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53(2):242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acharjee S, Sarmah BK, Kumar PA, Olsen K, Mahon R, Moar WJ, Moore A, Higgins TJV. Transgenic chickpeas (Cicer arietinum L.) expressing a sequence-modified cry2Aa gene. Plant Sci. 2010;178:333–339. doi: 10.1016/j.plantsci.2010.02.001. [DOI] [Google Scholar]
- 17.Singh CK, Ojha A, Bhatanagar RK, Kachru DN. Detection and characterization of recombinant DNA expressing vip3A-type insecticidal gene in GMOs - standard single, multiplex and construct-specific PCR assays. Anal Bioanal Chem. 2008;390:377–387. doi: 10.1007/s00216-007-1714-0. [DOI] [PubMed] [Google Scholar]
- 18.Porcar M, Juarez-Perez V. PCR-based identification of Bacillus thuringiensis pesticidal crystal genes. FEMS Microbiol Rev. 2003;26:419–432. doi: 10.1111/j.1574-6976.2003.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 19.Berón CM, Curatti L, Salerno GL. New Strategy for identification of novel cry-type genes from Bacillus thuringiensis strains. Appl Env Microbiol. 2005;71(2):761–765. doi: 10.1128/AEM.71.2.761-765.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davolos C, Guidelli-Thuler AM, Abreu IL, Sena JAD, Lemos MVF. Cry1 genes from Bacillus thuringiensis: specificity determination and implications for primer design. Biotechnol Lett. 2009;31:1891–1897. doi: 10.1007/s10529-009-0088-0. [DOI] [PubMed] [Google Scholar]
- 21.Tan F, Zhu J, Tang J, Tang X, Wang S, Zheng A, Li P. Cloning and characterization of two novel crystal protein genes, cry54Aa1 and cry30Fa1, from Bacillus thuringiensis strain BtMC28. Curr Microbiol. 2009;58:654–659. doi: 10.1007/s00284-009-9386-y. [DOI] [PubMed] [Google Scholar]
- 22.Anderson HM, Allen JR, Groat JR, Johnson SC, Kelly RA, Korte J, Rice JF (2008) Corn plant and seed corresponding to transgenic event MON89034 and methods for detection and use thereof. United States Patent Application 2008/0260932 A1
- 23.Taylor M, Hartnell G, Nemeth M, Lucas D, Davis D. Comparison of broiler performance when fed diets containing grain from second-generation insect-protected and glyphosate-tolerant, conventional control or commercial reference corn. Poult Sci. 2007;86:1972–1979. doi: 10.3382/ps.2007-00139. [DOI] [PubMed] [Google Scholar]
- 24.Taylor M, Lucas D, Nemeth M, Davis S, Hartnell G. Comparison of broiler performance and carcass parameters when fed diets containing combined trait insect-protected and glyphosate-tolerant corn (MON 89034 × NK603), control, or conventional reference corn. Poult Sci. 2007;86:1988–1994. doi: 10.3382/ps.2007-00139. [DOI] [PubMed] [Google Scholar]
- 25.United States Environmental Protection Agency (2009) Pesticide Fact Sheet of MON-89Ø34-3 x DAS- Ø15Ø7-1 x MON-88Ø17-3 x DAS-59122-7. http://cera-gmc.org/docs/decdocs/09-211-001.pdf. Accessed 10 Jan 2011
- 26.Canadian food Authority (2009) Decision Document DD2008-74: Determination of the Safety of Monsanto Canada Inc.’s Corn (Zea mays L.) Event MON 89034. http://cera-gmc.org/docs/decdocs/08-192-001.pdf. Accessed 10 Jan 2011
- 27.Corbisier P, Bhat S, Partis L, Xie VRD, Emslie KR. Absolute quantification of genetically modified MON810 maize (Zea mays L.) by digital polymerase chain reaction. Anal Bioanal Chem. 2010;396(6):2143–2150. doi: 10.1007/s00216-009-3200-3. [DOI] [PubMed] [Google Scholar]
- 28.Joint Research Centre (2008) Event-specific Method for the Quantification of Maize Line MON89034 using Real-time PCR. Protocol, 21 Oct., 2008 http://gmo-crl.jrc.ec.europa.eu/summaries/MON89034_validated_Method.pdf. Accessed 22 June 2010
- 29.Joint Research Centre (2008) Event-specific Method for the Quantification of Maize Line MON89034 using Real-time PCR. Validation report, 21 Oct., 2008. http://gmo-crl.jrc.ec.europa.eu/summaries/MON89034_validated_Method.pdf. Accessed 22 June 2010
- 30.Joint Research Centre (2008). Event-specific Method for the Quantification of Cotton Line MON15985 using Real-time PCR. Protocol, 19 June 2008. http://gmo-crl.jrc.ec.europa.eu/summaries/MON15985_validated_Method.pdf. Accessed 22 June 2010
- 31.GMO method detection database. http://gmdd.shgmo.org/index/search. Accessed 10 June 2010
- 32.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 33.Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. doi: 10.1007/BF02672069. [DOI] [Google Scholar]
- 34.Burns JM, Nixon GJ, Foy CA, Harris N. Standardisation of data from real-time quantitative PCR methods – evaluation of outliers and comparison of calibration curves. BMC Biotechnol. 2005;5:31. doi: 10.1186/1472-6750-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Center for Biotechnology Information (2010). http://www.ncbi.nlm.nih.gov/. Accessed 25 May 2010
- 36.European Network of Genetically Modified Organism Laboratories. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing, 13 Oct., 2008. http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf. Accessed 22 June 2010
- 37.Clapper G, Cantrill R (2009) Certification report – The certification of conventional and MON88017 Corn Certified Reference Materials AOCS 0406-A and AOCS 0406-D. Champaign, IL, USA. http://www.aocs.org/files/TechnicalPDF/0406AD%20report.pdf. Accessed 25 May 2010
- 38.Clapper G, Cantrill R (2007) Certification report - Certified Reference Materials AOCS 0906-A and AOCS 0906-B Report of the certification process for Conventional and MON89788 Soybean Seed Certified Reference Materials. AOCS, Urbana, IL, USA. http://www.aocs.org/files/TechnicalPDF/MON89788%20Final%20Report.pdf. Accessed 04 Oct 2010
- 39.Clapper G, Cantrill R (2009) Certification report - The certification of Conventional, Roundup Ready®, Bollgard®, and Bollgard II® Cottonseed Reference Materials AOCS 0804-A, AOCS 0804-B, AOCS 0804-C, AOCS 0804-D. Champaign, IL, USA. http://www.aocs.org/files/TechnicalPDF/0804ABCD%20report.pdf. Accessed 25 May 2010
- 40.Trapmann S, Catalani P, Conneely P, Corbisier P, Gancberg D, Hannes E, Le Guern L, Kramer GN, Prokisch J, Robouch P, Schimmel H, Zeleny R, Pauwels J (2002) The certification of reference materials of dry-mixed soya powder with different mass fractions of Roundup ready soya IRMM-410 S. European Commission, Joint Research Centre Institute for Reference Materials and Measurements, Geel, Belgium. http://www.erm-crm.org/ERM_products/search/reports/BF410a.pdf. Accessed 04 Oct 2010
- 41.Trapmann S, Conneely P, Contreras M, Corbisier P, Gancberg D, Hannes E, Gioria S, Muñoz-Pineiro A, Van Nyen M, Schimmel H, Szilagy S, Emons H (2005) Certification Report - The certification of reference materials of dry-mixed maize powder with different mass fractions of 1507 maize certified reference materials ERM®-BF418. European Commission, Joint Research Centre Institute for Reference Materials and Measurements, Geel, Belgium. http://www.erm-crm.org/ERM_products/search/reports/BF418a.pdf. Accessed 04 Oct 2010
- 42.Demeke T, Jenkins GR. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal Bioanal Chem. 2010;396:1977–1990. doi: 10.1007/s00216-009-3150-9. [DOI] [PubMed] [Google Scholar]
- 43.Scholtens IMJ, Kok EJ, Hougs L, Molenaar B, Thissen JTNM, Van der Voet H. Increased efficacy for in-house validation of real-time PCR GMO detection methods. Anal Bioanal Chem. 2010;396:2213–2227. doi: 10.1007/s00216-009-3315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 335 kb)