Abstract
Intratumor heterogeneity—heterogeneity of cancer cells within a single tumor—is considered one of the most problematic factors of treatment. Genetic heterogeneity, such as in somatic mutations and chromosome aberrations, is a common characteristic of human solid tumors and is probably the basis of biological heterogeneity. Using mutations in APC, TP53 and KRAS as markers to identify distinct colorectal cancer subpopulations, we analyzed a total of 42 primary colorectal cancer tissues and six paired liver metastases with multipoint microsampling, which enabled analysis of mutation patterns and allelic imbalances with a resolution of 0.01 mm2 (about 200 cells). There was usually more than one subpopulation in each primary tumor. Only two of 15 (13.3%) cases with three gene mutations and eight of 27 (29.6%) cases with two gene mutations had a single subpopulation. Cells with mutations in all of the examined genes usually constituted the major population. Multipoint microsampling of six primary and metastatic tumor pairs revealed that the majority of discrepancies in mutation patterns found with the bulk tissue analysis were due to loss of subpopulations in the metastatic tissues. In addition, multipoint microsampling uncovered substantial changes in subpopulations that were not detected with bulk tissue analysis. Specifically, the proportion of KRAS mutation-negative subpopulations increased in the metastatic tumors of four cases. Because KRAS mutation status is linked to cetuximab/panitumumab efficacy, subpopulation dynamics could lead to differences in response to cetuximab/panitumumab in primary versus metastatic tumors.
Electronic supplementary material
The online version of this article (doi:10.1007/s10585-011-9381-0) contains supplementary material, which is available to authorized users.
Keywords: Intratumor heterogeneity, Molecular target drug, Colorectal cancer, Somatic mutation
Introduction
Intratumor heterogeneity, which is the heterogeneity of cancer cells within a single tumor, is considered one of the most problematic factors of treatment. During anti-cancer therapy, the initial regression of the tumor eventually leads to the outgrowth of drug-resistant cells. Resistant cells likely exist in a heterogeneous primary cancer cell population rather than evolve from it. A recently emerging topic related to heterogeneity is the cancer stem cell (CSC) hypothesis [1], which assumes that only a fraction of cancer cells have the ability to initiate tumor formation. In addition, the CSC hypothesis claims that currently available drugs are not effective because they target the total cancer cell population.
In addition to the heterogeneity of biological features presented by CSC, genetic heterogeneity, such as in somatic mutations and chromosome aberrations, is a common characteristic of human solid tumors and is probably the basis of biological heterogeneity. A number of studies have described genetic heterogeneity [2–11]. Although these studies have demonstrated that cancer cell subpopulations with different mutation patterns are present in most solid tumors, there were several technical limitations. In particular, these studies did not use laser microdissection, which is now a standard technique, and they had a relatively high threshold of detection, which may have hampered the detection of rare clones.
Genetic heterogeneity within primary tumors is a single aspect of tumor heterogeneity. Although tumors develop at one site, some cancer cells leave the primary tumor and develop metastases at distant sites. Because metastases account for the majority of cancer-related deaths, understanding the underlying mechanism of their development is extremely important. Analysis of genetic heterogeneity, such as in somatic mutations, may help clarify the issue. Although intratumor heterogeneity has been intensely studied, the question of heterogeneity within metastases remains unexplored [12].
Colorectal cancer is one of the most common and well-studied cancer types. Because somatic mutations in the APC, KRAS and TP53 genes have been established as “drivers” for colorectal carcinogenesis [13], we chose these genes as markers to identify cancer cell subpopulations. We analyzed a total of 42 primary colorectal cancer tissues and 6 paired metastatic tissues with multipoint microsampling [11, 14], which is our proprietary technical approach for analyzing intratumor genetic heterogeneity. Multipoint microsampling enables analysis of mutation patterns and allelic imbalance with a resolution of 0.01 mm2 (about 200 cells). We examined the prevalence of intratumor genetic heterogeneity in primary tumors and the dynamics of cancer cell subpopulations in primary and metastatic tumors.
Materials and methods
Samples
In a previous study, a total of 86 bulk primary colorectal cancer tissues from our tumor tissue bank were examined for mutations in the APC, KRAS and TP53 genes. DNA was extracted from frozen bulk tumor tissues using the QIAamp DNA Micro Kit (Qiagen, Hilden, Germany), and the coding regions of APC, KRAS and TP53 were examined for mutations using High-Resolution Melting on a LightScanner (Idaho Technology, Salt Lake City, UT, USA). Samples with aberrant melting curves were analyzed using direct sequencing with the BigDye Terminator Cycle Sequencing Kit (ver. 3.1, Applied Biosystems, Carlsbad, CA, USA) on an ABI PRISM 3730 (Applied Biosystems). Cases with a single mutation, in which cells with no mutation were the only heterogeneous subpopulation, were excluded because such a subpopulation was difficult to distinguish from normal epithelial cells. Forty-two primary colorectal cancer tissues were included in the present study: 15 tumor tissues with mutations in all three genes (i.e., APC, KRAS and TP53 genes) and 27 tumors with only two mutated genes. The details of the mutations in these tumors are listed in Table S1. In addition, 6 liver metastatic tissues from the cases with mutations in all three genes were included in the study. The present study was approved by the Ethical Committee of the Osaka Medical Center for Cancer and Cardiovascular Diseases, and informed consent was obtained from all patients.
Multipoint microsampling
For the analysis of intratumor heterogeneity, 40-μm-thick sections from frozen cancer tissues were prepared on a Leica CM1900 cryostat (Leica Microsystems, Wetzlar, Germany) and stained with Mayer’s hematoxylin (Wako, Osaka, Japan). Forty to fifty small areas (100 × 100 μm) containing only tumor cells were microdissected from each sample. Microdissection was performed using the Leica AS LMD system (Leica Microsystems). The sampling was randomized, but we avoided repeated sampling from the same region. Genomic DNA was extracted using the prepGEM Kit (ZyGEM, Hamilton, New Zealand), according to the manufacturer’s protocol, and a 20-μl DNA mixture was prepared from each sample.
PCR amplification and SNaPshot assay
For each case, the DNA fragments that were found to contain mutations in bulk tissue analysis were simultaneously amplified using multiplex PCR on a GeneAmp PCR System 9700 (Applied Biosystems). The PCR was performed in a 10-μl reaction volume including 5 μl DNA (approximately 250 pg), 1× PCR buffer (Applied Biosystems), 2 mM MgCl2, 200 μM of each dNTP, 0.2 μM of each primer (Table S2) and 1 U AmpliTaqGold polymerase (Applied Biosystems). The cycling conditions consisted of an initial denaturation step at 94°C for 5 min, 40 cycles of denaturing at 94°C for 30 s, annealing at 54–56°C for 30 s, extension at 72°C for 40 s and a final synthesis at 72°C for 5 min.
The mutation status was quantitatively determined using the SNaPshot assay. Each primer was designed to bind to a complementary template immediately adjacent to the mutation site. The reaction was carried out in the presence of fluorescently labeled ddNTPs, and DNA polymerase was used to extend the primer by one nucleotide (adding a single ddNTP to its 3′ end). The following fluorescent dyes were used for the dideoxynucleotides: A, dR6G; C, dTAMRA; G, dR110; and T, dROX.
The PCR fragments were prepared for primer extension by incubating a mixture containing 7.5 μl PCR product, 0.5 U shrimp alkaline phosphatase (SAP) (TaKaRa, Otsu, Japan) and 1 U exonuclease I (TaKaRa) in a final volume of 10 μl at 37°C for 40 min, which was followed by inactivation of the enzymes at 80°C for 20 min. Primer extension was carried out in a 5-μl reaction, which contained 2 μl treated PCR product, 2.5 μl ABI Prism SNaPshot Multiplex Ready Reaction Mix (Applied Biosystems) and 0.5 μl extension primer mix (0.2 μM of each primer). The primer sequences are shown in Table S2. The cycling conditions, which were carried out according to the manufacturer’s protocol, included 25 cycles of denaturation at 96°C for 10 s, annealing at 50°C for 5 s and extension at 60°C for 30 s. To remove the unincorporated ddNTPs, 5 μl of the SNaPshot products was incubated for 40 min at 37°C with 0.5 U SAP (TaKaRa) in a final volume of 6 μl, and the enzyme was inactivated as described above. A 1-μl aliquot of the treated SNaPshot reaction was denatured in 9 μl distilled water in the presence of GeneScan 120 LIZ Size Standard (Applied Biosystems) for 5 min at 95°C and analyzed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Fragment analysis was performed with Gene Mapper Software v4.0 (Applied Biosystems).
Data analysis
The mutation status of each area was determined by calculating the mutant allele ratio (ΔM) for each gene using the following equation: ΔM = M/(M + N), where M is the mutant allele peak height and N is the normal peak height. The reproducibility of amplification and the SNaPshot assay was confirmed in our previous experiments [14]. In most cases of mutant allele loss, the corresponding peak height (M) was zero. However, there were several areas with residual peaks where we set the threshold for ΔM at 0.05.
Results
Intratumor heterogeneity of primary colorectal cancers
In a previous study, we established a method called multipoint microsampling to characterize genetic heterogeneity in colorectal cancer using laser microdissection [11, 14]. This method has two characteristics: (1) a sample size small enough to keep contamination from other cell populations to a minimum, but large enough to ensure unbiased amplification, and (2) a high enough sample number (~50) to obtain reproducible results from minor populations. To detect mutations, we used the SNaPshot assay, which is a quantitative primer extension assay that yields ratios of mutated and normal alleles.
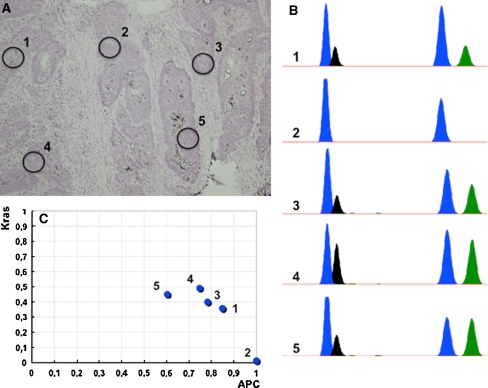
All 42 samples were subjected to multipoint microsampling. Figure 1 shows a microscopic image of a tumor tissue section from case 12 (Fig. 1a), an electropherogram of the SNaPshot assay (Fig. 1b) and a graphical presentation of the mutant allele ratios (ΔM) of APC and KRAS (Fig. 1c). Although all five regions in Fig. 1a contained cells with mutations in APC, not all of the regions carried mutations in KRAS; there was only one peak (ddG) for the KRAS allele in region 2 (Fig. 1b, c).
Fig. 1.
An example of intratumor heterogeneity. a A microscopic view of a colorectal cancer tissue section with black circles indicating the microdissected areas (1–5). b Electropherograms of the SNaPshot assay. The first two peaks represent the mutation status of APC (C>G), and the second two peaks represent the mutation status of KRAS (G>A). The blue peak is a fragment amplified with ddG, the black peak is a fragment amplified with ddC and the green peak is a fragment amplified with ddA. c Graphic representation of the SNaPshot results. The mutant allele ratio (ΔM) of APC is plotted on the x-axis, and the ΔM of KRAS is plotted on the y-axis
The mutation statuses of all samples taken from primary tumor tissues are summarized in Table 1. Representative results of areas from primary colorectal cancer tissues plotted in three dimensions were created using the ΔM values of APC, KRAS and TP53, and 2-D plots were made for the samples with two mutated genes. These results are shown in Fig. S1.
Table 1.
Summary of the genotypes in the 42 primary tumors
| Case | Clinical stage | Samples/tumor | Clone | Genotype | Areas | % | AIHb-APC | AIHb-KRAS | AIHb-TP53 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| APC muta | KRAS muta | TP53 muta | |||||||||
| 1 | IV | 42 | A | − | − | + | 3 | 7,1 | + | + | + |
| B | + | − | + | 1 | 2,4 | ||||||
| C | + | + | + | 38 | 90,5 | ||||||
| 3 | II | 48 | A | − | + | + | 2 | 4,2 | + | + | + |
| B | + | + | + | 46 | 95,8 | ||||||
| 11 | III | 41 | A | + | − | − | 7 | 17,1 | + | + | − |
| B | + | + | − | 4 | 9,8 | ||||||
| C | + | + | + | 30 | 73,2 | ||||||
| 19 | III | 45 | A | − | + | + | 2 | 4,4 | + | + | + |
| B | + | + | + | 43 | 95,6 | ||||||
| 24 | II | 49 | A | + | − | + | 2 | 4,1 | + | + | + |
| B | + | + | + | 47 | 95,9 | ||||||
| 33 | IV | 48 | A | + | − | − | 21 | 43,8 | − | + | + |
| B | + | + | − | 9 | 18,8 | ||||||
| C | + | + | + | 18 | 37,5 | ||||||
| 41 | III | 40 | A | − | − | + | 7 | 17,5 | + | + | + |
| B | − | + | + | 1 | 2,5 | ||||||
| C | + | + | + | 32 | 80 | ||||||
| 49c | IV | 40 | A | +/+ | + | + | 40 | 100 | + | + | − |
| 51 | IV | 42 | A | − | − | + | 1 | 2,4 | + | + | + |
| B | + | − | + | 2 | 4,8 | ||||||
| C | + | + | + | 39 | 92,9 | ||||||
| 65 | IV | 49 | A | + | − | − | 2 | 4,1 | + | + | + |
| B | + | − | + | 2 | 4,1 | ||||||
| C | + | + | + | 45 | 91,8 | ||||||
| 74 | I | 41 | A | + | + | + | 41 | 100 | + | − | + |
| 81 | IV | 42 | A | + | + | − | 4 | 9,5 | + | + | + |
| B | + | + | + | 38 | 90,5 | ||||||
| 82 | IV | 48 | A | + | − | + | 1 | 2,1 | + | + | + |
| B | − | − | + | 3 | 6,3 | ||||||
| C | − | + | + | 41 | 85,4 | ||||||
| D | + | + | + | 1 | 2,1 | ||||||
| 83 | IV | 45 | A | + | + | − | 4 | 8,9 | + | + | + |
| B | + | + | + | 41 | 91,1 | ||||||
| 85 | IV | 44 | A | + | − | + | 1 | 2,3 | + | + | + |
| B | − | + | − | 2 | 4,5 | ||||||
| C | − | − | + | 1 | 2,3 | ||||||
| D | − | + | + | 35 | 79,5 | ||||||
| E | + | + | + | 5 | 11,4 | ||||||
| 4 | III | 44 | A | + | + | − | 44 | 100 | + | + | ND |
| 6 | II | 40 | A | + | + | − | 40 | 100 | − | − | ND |
| 12 | III | 49 | A | + | − | − | 13 | 26,5 | + | + | ND |
| B | + | + | − | 36 | 73,5 | ||||||
| 13 | II | 45 | A | − | + | − | 1 | 2,2 | + | + | ND |
| B | + | + | − | 44 | 97,8 | ||||||
| 18 | III | 46 | A | + | − | − | 1 | 2,2 | + | + | ND |
| B | + | + | − | 45 | 97,8 | ||||||
| 21 | II | 50 | A | − | + | − | 2 | 4 | + | − | ND |
| B | + | + | − | 48 | 96 | ||||||
| 57 | III | 50 | A | + | + | − | 50 | 100 | − | − | ND |
| 8c | III | 50 | A | +/+ | − | − | 3 | 6 | + | ND | + |
| B | − | − | + | 9 | 18 | ||||||
| C | + | − | + | 8 | 16 | ||||||
| D | +/+ | − | + | 30 | 60 | ||||||
| 9 | IV | 50 | A | + | − | + | 50 | 100 | + | ND | − |
| 17 | III | 47 | A | + | − | − | 1 | 2,1 | + | ND | + |
| B | − | − | + | 3 | 6,4 | ||||||
| C | + | − | + | 43 | 91,5 | ||||||
| 20 | II | 42 | A | + | − | − | 4 | 9,5 | + | ND | + |
| B | − | − | + | 3 | 7,1 | ||||||
| C | + | − | + | 35 | 83,3 | ||||||
| 28 | II | 50 | A | + | − | + | 50 | 100 | − | ND | + |
| 36 | I | 50 | A | + | − | + | 50 | 100 | + | ND | + |
| 39 | III | 50 | A | − | − | + | 38 | 76 | − | ND | + |
| B | + | − | + | 12 | 24 | ||||||
| 43c | I | 50 | A | + | − | − | 2 | 4 | + | ND | + |
| B | + | − | + | 2 | 4 | ||||||
| C | +/+ | − | + | 46 | 92 | ||||||
| 46c | II | 50 | A | + | − | − | 5 | 10 | + | ND | + |
| B | + | − | +/+ | 45 | 90 | ||||||
| 48 | III | 41 | A | − | − | + | 2 | 4,9 | + | ND | − |
| B | + | − | + | 39 | 95,1 | ||||||
| 63c | II | 50 | A | +/+ | − | − | 1 | 2 | + | ND | + |
| B | +/+ | − | + | 49 | 98 | ||||||
| 66 | I | 50 | A | + | − | + | 50 | 100 | + | ND | + |
| 67 | III | 49 | A | − | − | + | 18 | 36,7 | + | ND | + |
| B | + | − | + | 31 | 63,3 | ||||||
| 70 | III | 44 | A | + | − | − | 2 | 4,5 | + | ND | + |
| B | + | − | + | 42 | 95,5 | ||||||
| 75 | III | 40 | A | + | − | − | 5 | 12,5 | + | ND | + |
| B | + | − | + | 35 | 87,5 | ||||||
| 76 | II | 40 | A | + | − | − | 2 | 5 | + | ND | + |
| B | − | − | + | 7 | 17,5 | ||||||
| C | + | − | + | 31 | 77,5 | ||||||
| 10 | II | 49 | A | − | − | + | 5 | 10,2 | ND | + | − |
| B | − | + | + | 44 | 89,8 | ||||||
| 26 | II | 42 | A | − | + | − | 4 | 9,5 | ND | + | + |
| B | − | − | + | 9 | 21,4 | ||||||
| C | − | + | + | 29 | 69 | ||||||
| 47 | III | 50 | A | − | + | + | 50 | 100 | ND | − | + |
| 64 | I | 46 | A | − | + | − | 23 | 50 | ND | + | + |
| B | − | + | + | 23 | 50 | ||||||
a mut mutations given in Table S1 for each tumor case
b AIH heterogeneity in allelic imbalance
cCases with two mutations in one gene (case 8, 43, 46, 49, 63). “+/+” means two mutations, “+” one mutation, “−” no mutation in the gene. For case 8 single mutation in APC is c.3139 G>T; for case 43-c.904 C>T
There was usually more than one subpopulation in each primary tumor. Indeed, only two of 15 (13.3%) cases with three gene mutations and eight of 27 (29.6%) cases with two gene mutations exhibited a single subpopulation. In most heterogeneous tumors, cells with all mutations constituted the major population (Table 1).
Because the SNaPshot assay measures relative levels of both alleles, we also revealed heterogeneity of allelic imbalance. Because the standard deviation of the SNaPshot assay was 0.08 [14], cases where the data ranges were over 0.32 (±2σ) were considered heterogeneous. For example, case 10 showed heterogeneity in the allele ratio of KRAS (ΔM = 0.0–0.53), but not of TP53 (ΔM = 0.94–1.0), whereas case 6 did not show heterogeneity for either of the two genes (ΔM = 0.46–0.61 for KRAS and 0.85–1.0 for APC) (Fig. S1). The status of the heterogeneity of allelic imbalance is shown in Table 1. The heterogeneity was probably due to a mixture of two or more cell subpopulations with different allelic ratios.
Comparison of intratumor genetic heterogeneity between primary and metastatic tumors
Among the 15 primary tumors with the three gene mutations, six cases had corresponding hepatic metastatic tumors. The details of mutations revealed by bulk tissue analysis are shown in Table 2. Except for two cases (82 and 85), metastatic tissues carried the same mutations as the corresponding primary tumors. We compared the intratumor heterogeneity of the three gene mutations between primary and metastatic tumors for each case. Table 3 summarizes the mutation status of subpopulations in the tissue pairs. The percentages of each subpopulation in a primary tumor and its paired metastasis are graphically presented for all six cases in Fig. 2.
Table 2.
Mutations found by bulk tissue analysis of six pairs of primary and metastatic tumors
| Case | Sample | APC mutation | KRAS mutation | TP53 mutation |
|---|---|---|---|---|
| 1 | T | c.4348 C>T R1450a | c.34 G>T G12C | c.818 G>A R273H |
| M | c.4348 C>T R1450a | c.34 G>T G12C | c.818 G>A R273H | |
| 51 | T | c.2626 C>T R876a | c.35 G>T G12V | c.818 G>A R273H |
| M | c.2626 C>T R876a | c.35 G>T G12V | c.818 G>A R273H | |
| 81 | T | c.1690C>T R564a | c.183 A>T Q61H | c.524 G>A R175H |
| M | c.1690C>T R564a | c.183 A>T Q61H | c.524 G>A R175H | |
| 82 | T | c.2626 C>T R876a | c.34 G>A G12S | c.755_63 del 9 bp |
| M | – | c.34 G>A G12S | c.755_63 del 9 bp | |
| 83 | T | c.4147 insA | c.38 G>A G13D | c.682 ins 8 bp |
| M | c.4147 insA | c.38 G>A G13D | c.682 ins 8 bp | |
| 85 | T | c.2626 C>T R876a | c.35 G>T G12V | c.733 G>A G245S |
| M | – | c.35 G>T G12V | c.733 G>A G245S |
aStop codon
Table 3.
Cell populations found in primary tumors and metastases of the patients
| Case | Clone | Genotype | Examined samples/tumor | Areas in primary tumorb | % | Examined samples/metastasis | Areas in metastasisb | % | ||
|---|---|---|---|---|---|---|---|---|---|---|
| APC muta | KRAS muta | TP53 muta | ||||||||
| 1 | A | – | – | c.818 G>A R273H | 42 | 3 | 7.1 | 50 | 0 | 0 |
| B | c.4348 C>T R1450c | – | c.818 G>A R273H | 1 | 2.4 | 36 | 72.0 | |||
| C | c.4348 C>T R1450c | c.34 G>T G12C | c.818 G>A R273H | 38 | 90.5 | 14 | 28.0 | |||
| 51 | A | – | – | c.818 G>A R273H | 42 | 1 | 2.4 | 50 | 0 | 0 |
| B | c.2626 C>T R876c | – | c.818 G>A R273H | 2 | 4.8 | 0 | 0 | |||
| C | c.2626 C>T R876c | c.35 G>T G12V | c.818 G>A R273H | 39 | 92.9 | 50 | 100.0 | |||
| 81 | A | c.1690C>T R564c | c.183 A>T Q61H | – | 42 | 4 | 9.5 | 50 | 7 | 14.0 |
| B | c.1690C>T R564c | c.183 A>T Q61H | c.524 G>A R175H | 38 | 90.5 | 43 | 86.0 | |||
| 82 | A | c.2626 C>T R876c | – | c.755_63 del 9 bp | 48 | 1 | 2.1 | 49 | 0 | 0 |
| B | – | – | c.755_63 del 9 bp | 3 | 6.3 | 25 | 51.0 | |||
| C | – | c.34 G>A G12S | c.755_63 del 9 bp | 41 | 85.4 | 24 | 49.0 | |||
| D | c.2626 C>T R876c | c.34 G>A G12S | c.755_63 del 9 bp | 3 | 6.3 | 0 | 0 | |||
| 83 | A | c.4147 insA | c.38 G>A G13D | – | 45 | 4 | 8.9 | 50 | 0 | 0 |
| B | c.4147 insA | c.38 G>A G13D | c.682 ins 8 bp | 41 | 91.1 | 50 | 100.0 | |||
| 85 | A | c.2626 C>T R876c | – | c.733 G>A G245S | 44 | 1 | 2.3 | 50 | 0 | 0 |
| B | – | c.35 G>T G12V | – | 2 | 4.5 | 0 | 0 | |||
| C | – | – | c.733 G>A G245S | 1 | 2.3 | 9 | 18.0 | |||
| D | – | c.35 G>T G12V | c.733 G>A G245S | 35 | 79.5 | 41 | 82.0 | |||
| E | c.2626 C>T R876c | c.35 G>T G12V | c.733 G>A G245S | 5 | 11.4 | 0 | 0 | |||
a mut mutation
bNumber of samples with a specific genotype
cStop codon
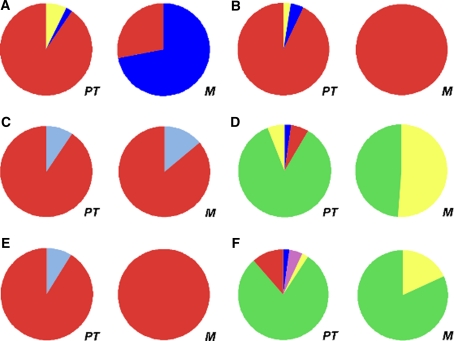
Fig. 2.
Genetic heterogeneity in six pairs of primary and metastatic tumor tissues. The percentages of areas with a particular mutation status in the primary tumor (PT) and its paired metastasis (M) are graphically presented for each case. The mutation patterns are: APCNKRASNTP53M, yellow; APCNKRASMTP53N, light purple; APCMKRASNTP53M, dark blue; APCNKRASMTP53M, green; APCMKRASMTP53N, light blue; and APCMKRASMTP53M, red (M, mutated gene; N, no mutation). a Case 1, b case 51, c case 81, d case 82, e case 83, and f case 85
In cases 82 and 85, there were discrepancies of mutation patterns; both cases lacked APC mutations in metastatic tumors (Table 2). Indeed, multipoint microsampling revealed that metastatic tumors lacked subpopulations with an APC mutation in both cases (i.e., subpopulations A and D in case 82 and subpopulations A and E in case 85) (Table 3 and Fig. 2d and f).
In cases 51 and 83, the major subpopulation of the primary tumor occupied all areas of the metastatic tumor (Fig. 2b and e, respectively). This result suggests that at least most cancer cells in the liver metastases were derived from the major subpopulations of the primary tumors in these two cases. In other cases, there was more than one subpopulation within the metastatic tumor tissue. In all cases, there were no de novo mutations, which suggests that cells from subpopulations in the primary tumors moved to the liver. In cases 1, 82 and 85, the proportions of KRAS mutation-negative subpopulations increased (Fig. 2a, d and f, respectively). Because both the primary and metastatic tumors in case 1 were mostly composed of a single major subpopulation, we performed array-CGH to exclude the possibility that the KRAS mutant allele would be lost by LOH after metastasizing. Notably, there was no LOH in the KRAS region (data not shown), which suggests that the mutation-negative subpopulation of the metastatic tumor was derived from the minor mutation-negative subpopulation of the primary tumor.
Discussion
Multipoint microsampling offers much higher resolution than techniques used in similar studies in the literature [2–11]. Consequently, we found intratumor heterogeneity in 87% of primary tumors with mutations in three genes (i.e., APC, KRAS and TP53) and in 70% of tumors with mutations in two of the three genes. We then compared mutation statuses between primary and metastatic tumors. The multipoint microsampling approach revealed that discrepancies of mutation patterns found by bulk tissue analysis were due to loss of subpopulations in the metastatic tissues. In addition, multipoint microsampling uncovered substantial changes in subpopulations that were not detected by bulk tissue analysis, such as the increase of KRAS mutation-negative cells in metastatic tumors.
A recent study of exome sequencing in pancreatic cancer reported a similar finding. Indeed, clonal populations that give rise to distant metastases were represented in one of the subpopulations taken from several parts of the primary tumors [15]. This study by Campbell et al. identified far more mutations than our study. In addition, the Campbell et al. study identified the original parental non-metastatic clones from which metastatic clones evolved. In the present study, we used multipoint microsampling, which has a markedly greater spatial resolution because the exome sequencing study was based on DNA prepared from more than 100 million cells. We found that the metastatic tumors included more than one subpopulation of the primary tumor in four cases (Fig. 2). In liver metastases, colorectal cancer cells move to the liver via the portal vein, which differs from ordinary distant metastases where cells move through the systemic circulation. The simultaneous migration of more than one subpopulation might be specific to liver metastases of colorectal cancer because of this anatomical characteristic.
Clinical trials with cetuximab or panitumumab revealed that patients with wild-type KRAS responded to therapy (up to 50%), whereas those with tumors exhibiting KRAS mutations had a low response rate (0–6%) [16–21]. Although the KRAS mutation status is predictive of cetuximab/panitumumab efficacy, there are cases in which the response did not match the prediction. Based on a hypothesis that the heterogeneity of KRAS mutations between primary and metastatic tumors may be responsible for this discrepancy, Italiano et al. analyzed 95 pairs of primary and metastatic tumors. Italiano et al. found that KRAS was mutated in primary tumors in six cases, but wild-type KRAS was found in metastatic tumors. In addition, eight cases showed that KRAS was mutated in metastatic tumors, but wild-type KRAS was found in primary tumors [22]. Moreover, Baldus et al. analyzed 20 pairs of a primary tumor and a distant metastasis, and found heterogeneity of the KRAS mutation status in two cases [23]. Heterogeneity between primary and lymph node metastases was more frequent (31% of cases) than that between primary and metastatic tumors. Both studies indicated that the discrepancy of the KRAS mutation status between a primary tumor and its distant metastasis was likely to be too rare to account for the discordance between the KRAS mutation status and cetuximab/panitumumab efficacy. The results from these two studies, however, were obtained with analysis of bulk tissues or macrodissection. The present study demonstrated that there were changes in proportions of subpopulations that could not be detected by bulk tissue analysis. Thus, the possibility remains that heterogeneity of the KRAS mutation status could be the cause of altered cetuximab/panitumumab efficacy. Our preliminary study on intratumor heterogeneity of EGFR mutations in lung cancer suggested that tumors that have cancer cells with wild-type EGFR exhibited an inferior response to gefitinib [11]. Because molecularly targeted drugs are becoming the mainstream of adjuvant therapy, intratumor heterogeneity of target genes might be an important factor requiring further intensive analysis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1 (Table_S1.xls). Genetic aberrations found in all 42 cases. (XLS 16 kb)
Table S2 (Table_S2.xls). Sequences of the primers used for PCR and the SNaPshot assay. (XLS 37 kb)
Figure S1 (Figure_S1.ppt). Graphic representation of genetic heterogeneity in eight of the primary colorectal cancer tissues. (PPT 1094 kb)
Acknowledgments
The authors thank Dr Kazuya Taniguchi for technical advice. This work was partly supported by the Knowledge Cluster Initiative (the Keihanna Science City area) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CSC
Cancer stem cell
- SAP
Shrimp alkaline phosphatase
References
- 1.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 2.Baisse B, Bouzourene H, Saraga EP, et al. Intratumor genetic heterogeneity in advanced human colorectal adenocarcinoma. Int J Cancer. 2001;93(3):346–352. doi: 10.1002/ijc.1343. [DOI] [PubMed] [Google Scholar]
- 3.Barnetson R, Jass J, Tse R, et al. Mutations associated with microsatellite unstable colorectal carcinomas exhibit widespread intratumoral heterogeneity. Genes Chromosomes Cancer. 2000;29(2):130–136. doi: 10.1002/1098-2264(200010)29:2<130::AID-GCC1023>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Giaretti W, Rapallo A, Sciutto A, et al. Intratumor heterogeneity of k-ras and p53 mutations among human colorectal adenomas containing early cancer. Anal Cell Pathol. 2000;21(2):49–57. doi: 10.1155/2000/747524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Garcia I, Sole RV, Costa J. Metapopulation dynamics and spatial heterogeneity in cancer. Proc Natl Acad Sci USA. 2002;99(20):13085–13089. doi: 10.1073/pnas.202139299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konishi N, Hiasa Y, Matsuda H, et al. Intratumor cellular heterogeneity and alterations in ras oncogene and p53 tumor suppressor gene in human prostate carcinoma. Am J Pathol. 1995;147(4):1112–1122. [PMC free article] [PubMed] [Google Scholar]
- 7.Losi L, Baisse B, Bouzourene H, et al. Evolution of intratumoral genetic heterogeneity during colorectal cancer progression. Carcinogenesis. 2005;26(5):916–922. doi: 10.1093/carcin/bgi044. [DOI] [PubMed] [Google Scholar]
- 8.Lyng H, Beigi M, Svendsrud DH, et al. Intratumor chromosomal heterogeneity in advanced carcinomas of the uterine cervix. Int J Cancer. 2004;111(3):358–366. doi: 10.1002/ijc.20258. [DOI] [PubMed] [Google Scholar]
- 9.Samowitz WS, Slattery ML. Regional reproducibility of microsatellite instability in sporadic colorectal cancer. Genes Chromosomes Cancer. 1999;26(2):106–114. doi: 10.1002/(SICI)1098-2264(199910)26:2<106::AID-GCC2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 10.Takeshima Y, Amatya VJ, Daimaru Y, et al. Heterogeneous genetic alterations in ovarian mucinous tumors: application and usefulness of laser capture microdissection. Hum Pathol. 2001;32(11):1203–1208. doi: 10.1053/hupa.2001.28956. [DOI] [PubMed] [Google Scholar]
- 11.Taniguchi K, Okami J, Kodama K, et al. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99(5):929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 14.Goranova TE, Ohue M, Kato K. Putative precursor cancer cells in human colorectal cancer tissue. Int J Clin Exp Pathol. 2009;2(2):154–162. [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Roock W, Piessevaux H, De Schutter J, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19(3):508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 17.Di Fiore F, Blanchard F, Charbonnier F, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96(8):1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht JR, Patnaik A, Berlin J, et al. Panitumumab monotherapy in patients with previously treated metastatic colorectal cancer. Cancer. 2007;110(5):980–988. doi: 10.1002/cncr.22915. [DOI] [PubMed] [Google Scholar]
- 19.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 20.Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 21.Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 22.Italiano A, Hostein I, Soubeyran I, et al. KRAS and BRAF mutational status in primary colorectal tumors and related metastatic sites: biological and clinical implications. Ann Surg Oncol. 2010;17(5):1429–1434. doi: 10.1245/s10434-009-0864-z. [DOI] [PubMed] [Google Scholar]
- 23.Baldus SE, Schaefer KL, Engers R, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16(3):790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (Table_S1.xls). Genetic aberrations found in all 42 cases. (XLS 16 kb)
Table S2 (Table_S2.xls). Sequences of the primers used for PCR and the SNaPshot assay. (XLS 37 kb)
Figure S1 (Figure_S1.ppt). Graphic representation of genetic heterogeneity in eight of the primary colorectal cancer tissues. (PPT 1094 kb)