Abstract
Atherosclerosis is an inflammatory disorder of the vasculature that is orchestrated by the action of cytokines. Macrophages play a prominent role in all stages of this disease, including foam cell formation, production of reactive oxygen species, modulation of the inflammatory response and the regulation of the stability of atherosclerotic plaques. The role of the matrix metalloproteinase family in the control of plaque stability is well established. A disintegrin and metalloproteinase with thrombospondin motif (ADAMTS) family has been implicated in several diseases and the expression of ADAMTS-4 in macrophages of atherosclerotic lesions has suggested a potential role for this protease in atherosclerosis. However, the action of cytokines on the expression of ADAMTS-4 in macrophages is poorly understood. We have investigated here the effect of transforming growth factor-β (TGF-β) on ADAMTS-4 expression in macrophages along with the regulatory mechanisms underlying its actions. Consistent with the anti-atherogenic role of TGF-β, this cytokine decreased the expression of ADAMTS-4 mRNA and protein in human macrophages. Transient transfection assays showed that the −100 to +10 promoter region contained the minimal TGF-β response elements. Small-interfering RNA-mediated knockdown revealed a critical role for Smads, p38 mitogen-activated protein kinase and c-Jun in the action of TGF-β on ADAMTS-4 mRNA expression. These studies show for the first time that TGF-β inhibits the expression of ADAMTS-4 in human macrophages and identifies the signalling pathways underlying this response. The inhibition of macrophage ADAMTS-4 expression is likely to contribute to the anti-atherogenic, plaque stabilisation action of TGF-β.
Keywords: A disintegrin and metalloproteinase with thrombospondin motifs 4 family, Atherosclerosis, Inflammation, Transforming growth factor-β, Macrophages
1. Introduction
A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) proteases are non-membrane bound enzymes that are able to interact with and degrade components of the extracellular matrix (ECM), such as pro-collagen and hyalectans (Salter et al., 2010; Jones and Riley, 2005; Porter et al., 2005). Of the ADAMTS proteases identified to date, ADAMTS-1 and ADAMTS-4 are the most abundant and consequently have been the subject of the majority of research on these enzymes (Salter et al., 2010; Jones and Riley, 2005). There has been much focus on the ability of ADAMTS proteases to cleave aggrecan, a proteoglycan present in articular cartilage (Salter et al., 2010; Naito et al., 2007; Tang, 2001). Cleavage of aggrecan in human cartilage contributes to the pathophysiology of osteoarthritis and rheumatoid arthritis and a role for ADAMTS-4 in this process has been defined (Salter et al., 2010; Naito et al., 2007; Tang, 2001).
A possible role for the ADAMTS proteases as contributors to inflammation and atherosclerosis has only recently come to light (Salter et al., 2010). Atherosclerosis, the primary cause of coronary heart disease, is an inflammatory disorder orchestrated by the action of cytokines. The latter stages of this disease are associated with the migration and the proliferation of vascular smooth muscle cells (VSMCs), which leads to the production of the extracellular matrix and the formation of a fibrous cap in the atherosclerotic plaque (Bui et al., 2009). The balance between the synthesis and the degradation of the ECM by matrix metalloproteinases (MMPs) and other proteases determines the susceptibility of the plaque to rupture and subsequent thrombosis and myocardial infarction (Li and Glass, 2002; Lusis et al., 2004).
The ADAMTS proteases were first associated with inflammation when it was shown that administration of lipopolysaccharide (LPS) in vivo could enhance ADAMTS-1 expression in the heart and in the kidneys (Kuno et al., 1997). ADAMTS-1, -4 and -8 have been found to be present within atherosclerotic lesions and mice overexpressing ADAMTS-1 crossed with ApoE-deficient mice, an animal model for atherosclerosis, showed increased thickening of the arterial intima (Wight, 2005; Jonsson-Rylander et al., 2005; Wågsäter et al., 2008). ADAMTS-4 is predominantly associated with macrophages of atherosclerotic lesions and its expression is increased following both monocyte–macrophage differentiation and during development of atherosclerosis in a mouse model system (Wågsäter et al., 2008). The ability of the ADAMTS proteases to cleave proteoglycans is likely to be central to any proposed role in atherosclerosis. Both ADAMTS-1 and -4 are able to cleave versican, a proteoglycan similar in structure to aggrecan but predominantly expressed in the vasculature (Sandy et al., 2001). Versican expression is upregulated in vascular disease, accumulates in atherosclerotic plaques (Worley et al., 2003) and can be cleaved by ADAMTS-1 and ADAMTS-4 at specific Glu-Ala bonds (Sandy et al., 2001). It has also been shown that in the human aorta, versican fragments can be generated by ADAMTS-1 and ADAMTS-4 digestion of intact human versican (Sandy et al., 2001).
Very few studies have investigated the regulation of ADAMTS protease expression in macrophages and further studies are necessary especially as ADAMTS-4 has been found to colocalize with macrophages of atherosclerotic lesions (Wågsäter et al., 2008). To our knowledge only one study has been carried out that investigated the expression of ADAMTS proteases in response to cytokines in human macrophages and this demonstrated that ADAMTS-4 expression was up-regulated by the pro-inflammatory cytokines interferon-γ (IFN-γ) and tumour necrosis factor-α (Wågsäter et al., 2008).
TGF-β is a major anti-atherogenic cytokine and inhibition of its actions, using neutralising antibodies or expression of a dominant negative receptor, has been found to accelerate the development of atherosclerosis in mouse models of this disease (Singh and Ramji, 2006a). The cytokine inhibits foam cell formation and an inverse relationship has been identified between circulating levels of TGF-β and the development of atherosclerosis (Singh and Ramji, 2006a). In addition, regions in the aorta with low TGF-β expression have a high probability of developing atherosclerosis (Singh and Ramji, 2006a). TGF-β is also known to inhibit the expression of MMPs, which decrease plaque stability via thinning of the fibrous plaque through cleavage of collagens, elastins and other proteoglycans in the ECM (Galis and Khatri, 2002; Singh and Ramji, 2006a; Newby, 2007). In light of such a potent anti-atherogenic role of TGF-β, we examined its action on ADAMTS-4 expression in human macrophages with a view to further investigating the molecular mechanisms underlying such regulation. We show here that TGF-β down-regulates ADAMTS-4 expression at the level of mRNA, protein and promoter activity. In addition, our studies demonstrate a critical role for Smads, p38 mitogen-activated protein kinase (MAPK) and c-Jun in the regulation of ADAMTS-4 by TGF-β.
2. Materials and methods
2.1. Reagents
The human monocytic leukaemia THP-1 cell line and the human hepatoma Hep3B cell line were obtained from the European Collection of Animal Cell Cultures (ECACC). Human recombinant TGF-β1 was from Peprotech, validated Smad-2, c-Jun and extracellular signal-regulated kinase (ERK) 1/2 small interfering RNA (siRNA) was from Qiagen and validated Smad-3 and p38 MAPK siRNA was from Invitrogen. Antibodies were from Affinity Bioreagents (ADAMTS-4), Cell Signaling Technology (Smad2/3, p38MAPK, ERK1/2), Santa Cruz Biotechnology (c-Jun) and Sigma (β-actin). Superfect™ transfection reagent was from Qiagen and INTERFERin™ from Polyplus Transfection.
2.2. Cell culture
The cell lines were maintained in either DMEM (Hep3B) or RPMI-1640 (THP-1) supplemented with 10% (v/v) heat-inactivated fetal calf serum, 100 U/ml penicillin and 100 μg/ml Streptomycin. The cultures were maintained in a humidified atmosphere at 37 °C containing 5% (v/v) CO2. THP-1 monocytes were differentiated into macrophages using 0.16 μg/ml phorbol 12-myristate 13-acetate (PMA). For human monocyte-derived macrophages (HMDMs), Ficoll–Hypaque purification was used to isolate the cells from buffy coats (McLaren et al., 2010a,b; Li et al., 2010). Blood layered over Lymphoprep™ (Nycomed Pharmaceuticals) in Accuspin™ tubes (Sigma) was centrifuged and platelets removed from the mononuclear cell interface by washing several times with PBS, containing 0.4% (v/v) tri-sodium citrate. Monocytes were plated out in RPMI-1640 supplemented as described above, except containing 5% (v/v) fetal calf serum, and left to differentiate into macrophages for 7 days.
2.3. Transfection of siRNA
THP-1 monocytes were transfected with 7.5 nM siRNA using INTERFERin™ essentially as described by the manufacturer (PolyPlus Transfection). The cells were then incubated for 24 h before differentiation into macrophages using PMA as described above and subsequent treatment with TGF-β (30 ng/ml). Gene silencing was measured 48 h after transfection by Western blot analysis.
2.4. Real-time quantitative PCR (RT-qPCR)
RT-qPCR was carried out using primers against ADAMTS-4 or the ribosomal protein L13A (RPL13A) or the glyceraldehyde 3-dehydrogenase (GAPDH) control genes and the SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma) on a DNA Engine Opticon 2® real-time PCR detection system (MJ Research). The sequences of the primers were 5′-GGGATAGTGACCACATTGTT-3′ and 5′-AGGCACTGGGCTACTACTAT-3′ for ADAMTS-4; 5′-CCTGGAGGAGAAGAGGAAAGAGA-3′ and 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′ for RPL13A; and 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′ for GAPDH. For each transcript a standard curve was constructed using a recombinant pGEM-T plasmid containing the PCR product generated for each specific set of primers. A melting curve was constructed to verify single product amplification and the comparative Ct method was used for analysis and normalisation to the control transcript levels.
2.5. Transient transfection assays
Hep3B and U937 cells were transfected using Superfect™. Transfection of U937 cells was carried out as previously described (Hughes et al., 2002; Irvine et al., 2005). For Hep3B, the cells were subcultured at a ratio of 1:6 into 12-well plates 24 h prior to transfection and medium replaced with fresh DMEM before transfection. The DNA:Superfect™ mix was prepared according to the manufacturer's instructions (Qiagen) in 50 μl medium containing no antibiotics or serum. The mixture was incubated at room temperature for 10 min and then diluted with DMEM medium before being added to the cells. The cells were then left for 30 min and then treated with vehicle or TGF-β (30 ng/ml) for 24 h. The cells were then harvested using 1× passive lysis buffer (Promega) and the luciferase activity measured using a commercially available kit (Promega). The luciferase activity was normalised to total protein levels as determined using the micro BCA protein assay kit according to the manufacturer's instructions (Pierce).
2.6. Western blot analysis
Samples were prepared in Laemmli buffer and size-fractionated under reducing conditions using 10% SDS-polyacrylamide gels and transferred to a PVDF membrane (Millipore) by Western blotting (Ali et al., 2010). Incubation with primary and secondary antibodies and chemiluminescent detection was carried out as described previously (Ali et al., 2010). Samples were subjected to electrophoresis alongside comparative molecular weight markers (GE Healthcare) to determine the size of the protein product. Membranes were developed using chemiluminescence detection reagents (GE Healthcare) and Kodak XAR sensitive film (Sigma).
2.7. Statistical analysis of data
Statistical comparisons of data were carried out using Student's t test with P < 0.05 considered statistically significant. Semi-quantitative, densitometric analysis of Western blot signals was performed using the GeneTools software (GRI).
3. Results
3.1. TGF-β inhibits ADAMTS-4 expression at the mRNA and protein level in THP-1 macrophages and HMDMs
The action of TGF-β on the expression of ADAMTS-4 in human macrophages has not been determined and was therefore investigated. The THP-1 monocytic cell line, which can be differentiated into macrophages using PMA, is used extensively to investigate macrophage gene expression in relation to atherosclerosis because of demonstrated conservation of responses with primary macrophage cultures, in particular gene regulation by cytokines (Auwerx, 1991; McLaren et al., 2010a,b; Li et al., 2010).
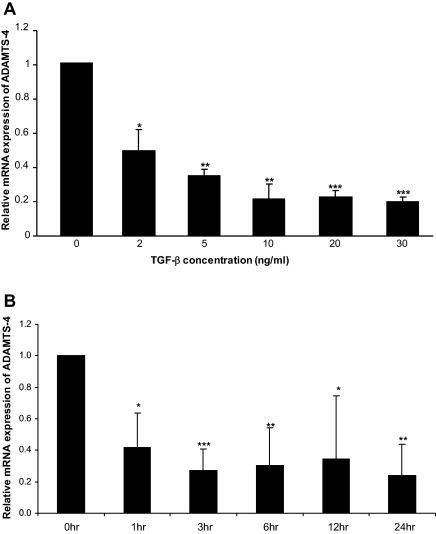
Our previous studies on the TGF-β-mediated activation of apolipoprotein E expression showed maximal response at 24 h with 30 ng/ml of the cytokine (Singh and Ramji, 2006b). We therefore carried out dose–response RT-qPCR experiments on ADAMTS-4 using the 24 h incubation period. As shown in Fig. 1A, a marked inhibition of ADAMTS-4 mRNA expression was observed with 2 ng/ml TGF-β with maximal suppression using 30 ng/ml of this cytokine. Further time course experiments using 30 ng/ml of the cytokine showed that ADAMTS-4 mRNA expression was down-regulated by TGF-β within 1 h of treatment and this response was sustained over the 24 h time period. Subsequent experiments were carried out using 30 ng/ml of TGF-β for an incubation period of 24 h, unless otherwise stated.
Fig. 1.
TGF-β inhibits ADAMTS-4 mRNA expression in THP-1 macrophages. THP-1 macrophages were treated for 24 h with the indicated concentration of TGF-β (A) or with 30 ng/ml of this cytokine for the time points shown (B). Total RNA was isolated and subjected to RT-qPCR. The Ct method was used for analysis and the values for ADAMTS-4 were normalised to the control gene GAPDH (A) or RPL13A (B). The data shows the expression relative to the 0 ng/ml (A) or 0 h (B) (arbitrarily assigned as 1) (mean ± SD from three independent experiments). Asterisks indicate significant down-regulation of ADAMTS-4 mRNA expression (*P < 0.05, **P < 0.01, ***P < 0.001).
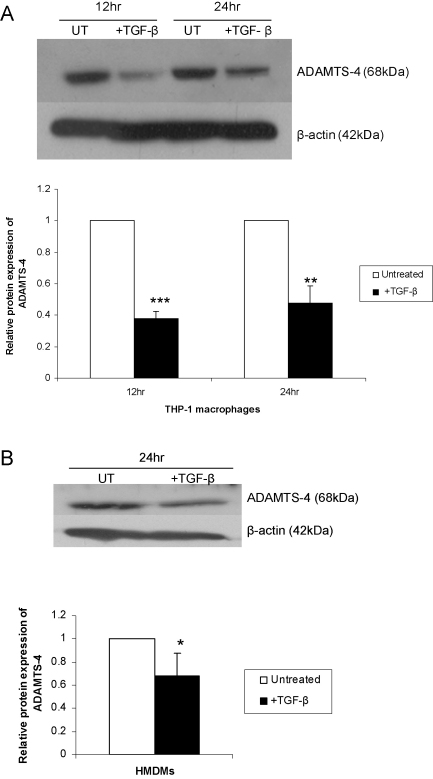
To determine whether this down-regulation was accompanied by a corresponding change in the level of protein, the expression of ADAMTS-4 was investigated using Western blot analysis. Preliminary experiments showed that the expression of ADAMTS-4 protein was decreased with delayed kinetics compared to RNA (data not shown) so longer incubation periods were used for Western blot analysis. Fig. 2A shows that ADAMTS-4 protein expression was significantly down-regulated in THP-1 macrophages by TGF-β treatment. In the light of these results and to confirm that the TGF-β-mediated decrease in ADAMTS-4 expression was not peculiar to the THP-1 cell line, we decided to carry out representative experiments in primary cultures of HMDMs. For this, the action of TGF-β on ADAMTS-4 protein expression was determined. Fig. 2B shows that protein expression of ADAMTS-4 was down-regulated by TGF-β in these primary cultures.
Fig. 2.
TGF-β inhibits ADAMTS-4 protein expression in macrophages. THP-1 macrophages (A) or HMDMs (B) were either left untreated (UT) or incubated with 30 ng/ml TGF-β for the time points indicated. An equal amount of the protein extract was subjected to Western blot analysis using antisera against ADAMTS-4 and the control β-actin, as shown. The relative ADAMTS-4 expression normalised to β-actin, as determined by densitometric analysis, from three independent experiments is shown (mean ± SD) with the values from untreated cells arbitrarily assigned as 1 (*P < 0.05; **P < 0.01; ***P < 0.001).
3.2. TGF-β inhibits ADAMTS-4 promoter activity
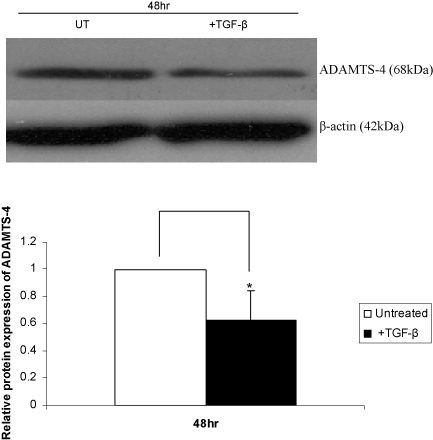
In order to investigate whether the action of TGF-β was mediated, at least in part, at the transcriptional level through the regulation of promoter activity, the effect of this cytokine on ADAMTS-4 promoter activity in transfected cells was determined. Because of difficulties in transfecting the THP-1 cell line with exogenous DNA, most experiments were carried out using the human hepatoma Hep3B cell line. These cells are used extensively to investigate the regulation of gene expression by cytokines in relation to inflammation (acute-phase response) with demonstrated conservation of responses to primary cultures (Foka et al., 2003, 2009 and references therein). Western blot analysis showed that TGF-β also inhibits ADAMTS-4 expression in these cells (Fig. 3), thereby showing that the response was also conserved in hepatocytes. The 48 h time point was chosen for experiments in these cells as it corresponded to maximal decrease in ADAMTS-4 expression as identified by preliminary time course analysis (data not shown).
Fig. 3.
TGF-β inhibits ADAMTS-4 protein expression in Hep3B cells. The cells were either left untreated (UT) or incubated with TGF-β for 48 h. Equal amounts of protein were subjected to Western blot analysis using antisera against ADAMTS-4 and the control β-actin, as indicated. The relative ADAMTS-4 expression normalised to β-actin, as determined by densitometric analysis, from three independent experiments is shown (mean ± SD) with the value from untreated cells arbitrarily assigned as 1 (*P < 0.05).
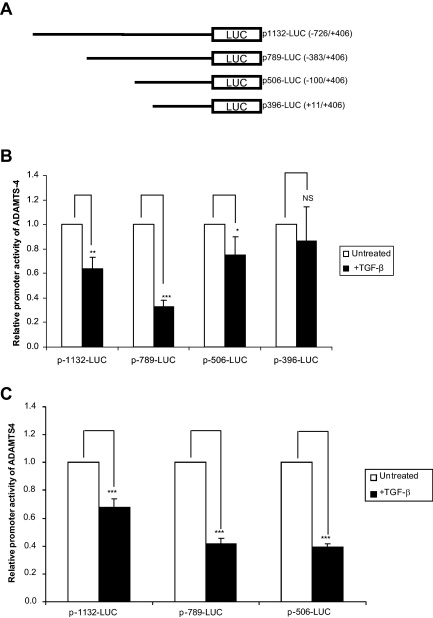
We employed four promoter deletion constructs linked to the luciferase gene: −726/+406 (p1132-LUC); −383/+406 (p789-LUC); −100/+406 (p506-LUC); and +11/+406 (p396-LUC) (Fig. 4A) (Mizui et al., 2000). A TGF-β-mediated decrease in promoter activity was obtained with the p1132-LUC, p789-LUC and p-506-LUC constructs but not with the p-396-LUC construct (Fig. 4B). This suggests that the minimal TGF-β response elements reside between the −100 and the +10 region. Similar results were obtained when representative experiments were carried out using human U937 macrophages (Fig. 4C) [the luciferase reporter gene activity from the +11/+406 (p396-LUC) construct in these cells was negligible and was therefore not included for further analysis].
Fig. 4.
TGF-β inhibits ADAMTS-4 promoter activity. (A) Schematic representation of the different ADAMTS-4 promoter-luciferase DNA constructs used. Hep3B cells (B) and U937 monocytes (C) were transfected with the ADAMTS-4 promoter-luciferase DNA constructs using Superfect™. The cells were either left untreated or treated overnight with 30 ng/ml TGF-β (1 μM PMA was also included for U937 only to initiate differentiation) and cell lysates were measured for luciferase activity. Data shown is the ratio of luciferase activity:protein concentration expressed as the fold change in activity relative to the equivalent untreated sample (arbitrarily assigned as 1) and is representative of three independent experiments (NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001).
3.3. The action of TGF-β on ADAMTS-4 expression requires Smads, p38 MAPK and c-Jun
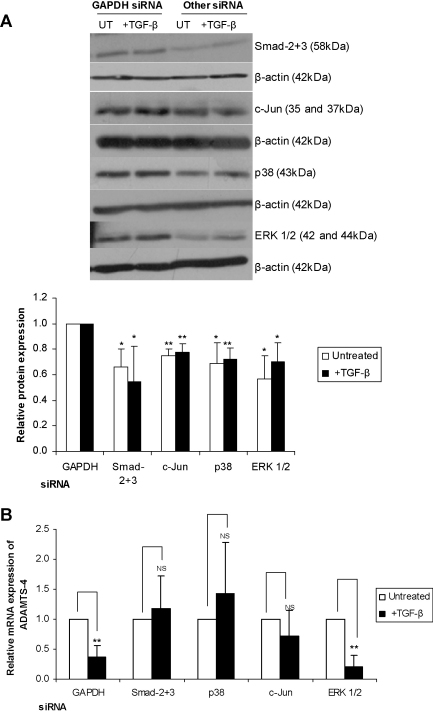
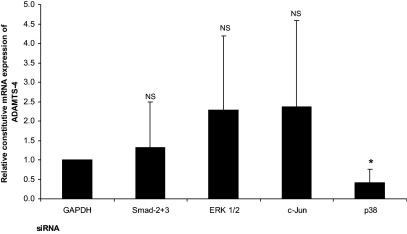
TGF-β mediates many of its actions through the classical Smad signalling pathway with the three MAPK pathways [ERK, c-Jun N-terminal kinase (JNK) and p38 MAPK] required in the regulation of certain genes (Ten Dijke and Hill, 2004). Most of these studies have been carried out in relation to the TGF-β-mediated induction of gene expression and very little is currently understood in relation to the inhibition of gene expression. We therefore analysed whether the Smad or MAPK pathways had any role in the regulation of ADAMTS-4 expression by this cytokine. This was investigated using siRNA-mediated RNA interference assays and RT-qPCR analysis. As in a number of previous studies, GAPDH siRNA was used as a control (Ali et al., 2010) and this had no effect on the TGF-β-mediated inhibition of ADAMTS-4 expression (Fig. 5). Knockdown of Smad-2 plus 3, ERK 1/2, c-Jun, a key downstream target of the JNK pathway, and p38 MAPK was achieved using commercially validated siRNA sequences and relative expression of each protein following siRNA-mediated knockdown was confirmed in THP-1 macrophages by Western blot analysis (Fig. 5A). The TGF-β-mediated inhibition of ADAMTS-4 expression was attenuated by knockdown of Smad-2 plus -3 (Fig. 5B). In addition, the TGF-β-mediated inhibition of ADAMTS-4 expression was attenuated by knockdown of c-Jun and p38 MAPK but not ERK1/2 (Fig. 5B). Because the expression of ADAMTS-4 is induced during monocyte–macrophage differentiation (Wågsäter et al., 2008), the effect of these knockdown on the constitutive expression of ADAMTS-4 in macrophages, driven by the differentiation process, was also determined. This was inhibited by knockdown of only p38 MAPK but not Smad-2 plus -3, ERK1/2 or c-Jun (Fig. 6).
Fig. 5.
The effect of siRNA-mediated knockdown of key components of the TGF-β signalling pathway on the inhibition of ADAMTS-4 expression by this cytokine. THP-1 monocytes were transfected with validated siRNA against Smad-2 and -3, ERK 1/2, c-Jun or p38 MAPK as shown. Cells were incubated with siRNA for 24 h before differentiation for 24 h with PMA and subsequently being either left untreated (UT) or stimulated with 30 ng/ml TGF-β for 24 h. Total RNA and protein was then purified. (A) siRNA-mediated knockdown was verified by Western blot analysis using antisera against total Smad-2 and -3, ERK 1/2, c-Jun, p38 MAPK (p38) and β-actin, as indicated. The image shown is representative of three independent experiments. The histogram shows relative expression (mean ± SD) of each protein normalised to β-actin, as determined by densitometric analysis, with the values from GAPDH siRNA-transfected cells being arbitrarily assigned as 1 (*P < 0.05, **P < 0.01). (B) Total RNA was subjected to RT-qPCR and the ADAMTS-4 expression was normalised to the control gene RPL13A and presented as the fold change relative to the untreated sample for each siRNA, which was arbitrarily assigned as 1. The data shown is representative of three independent experiments (NS, not significant, **P < 0.01).
Fig. 6.
The effect of siRNA-mediated knockdown on the constitutive expression of ADAMTS-4 in THP-1 macrophages. The experiments were carried out as Fig. 5 and the relative constitutive mRNA expression of ADAMTS-4 (i.e. in the absence of TGF-β), normalised to the control gene RPL13A and presented as a fold change relative to the GAPDH-transfected sample (assigned as 1) from three independent experiments, is shown (NS, not significant, *P < 0.05).
4. Discussion
Several members of the ADAMTS family have been linked with inflammation but their role in this and in pathophysiological inflammatory states including atherosclerosis is only beginning to be researched (Salter et al., 2010). Although the exact role of the ADAMTS proteases in atherosclerosis remains unclear it is thought that the ability of ADAMTS-1 and -4 to cleave versican, the primary proteoglycan component of the vasculature, is likely to be central to any hypothesised role in the disease (Salter et al., 2010; Worley et al., 2003). The production and secretion of proteases by smooth muscle cells and by macrophages is a key regulatory action during atherosclerosis (Bui et al., 2009). The vulnerability of the atherosclerotic plaque is determined by the balance between the synthesis and the degradation of the ECM. This degradation is achieved through the action of MMPs and other proteases such as the ADAMTS proteases, which cleave the collagens and proteoglycans of the ECM (Galis and Khatri, 2002; Newby, 2007).
We demonstrate for the first time that ADAMTS-4 mRNA and protein expression is inhibited by the anti-atherogenic cytokine TGF-β in THP-1 macrophages and HMDMs (Figs. 1 and 2). The response is also conserved in hepatocytes (Fig. 3). This novel finding is consistent with the work of Wågsäter et al. (2008) who found that ADAMTS-4 mRNA expression can be up-regulated by the pro-inflammatory cytokines IFN-γ and TNF-α in THP-1 macrophages. In addition to this, TGF-β has been demonstrated to inhibit the expression of a number of MMPs, including MMP-1 (White et al., 2000), MMP-3 (Kerr et al., 1990) and MMP-9 (Ogawa et al., 2004). In our studies, down-regulation of ADAMTS-4 mRNA expression by TGF-β was observed within 1 h and continued up to 24 h of treatment with this cytokine (Fig. 1).
In addition to characterising the response of ADAMTS-4 to TGF-β, we also identify the regulatory region of the ADAMTS-4 promoter involved in this response. We show here that ADAMTS-4 promoter activity was suppressed by TGF-β treatment (Fig. 4). Transient transfection of ADAMTS-4 promoter deletion constructs demonstrates an involvement of the −100/+10 region in the negative regulation of the ADAMTS-4 promoter by TGF-β (Fig. 4). Of the ADAMTS proteases, only the promoters of ADAMTS-4 and ADAMTS-5 have been characterised (Mizui et al., 2000; Thirunavukkarasu et al., 2006, 2007). Mizui et al. (2000) have identified the −383 to +10 sequence of the ADAMTS-4 promoter as the region required for full promoter activity. In addition, analysis of the promoter activity of various deletion constructs suggested that the region between −726 and −384 was likely to contain silencer elements (Mizui et al., 2000).
We also show that the Smad pathway is required in the action of TGF-β on ADAMTS-4 expression. The Smads are the classical transducers of the TGF-β signal (Ross and Hill, 2008) and Smad-2, -3 and -4 are expressed in macrophages and foam cells of atherosclerotic lesions (Kalinina et al., 2004). Following siRNA-mediated knockdown of Smad-2 plus -3, the TGF-β-mediated inhibition of ADAMTS-4 mRNA expression was no longer significant (Fig. 5). Smads have been implicated in the repression of a number of genes by TGF-β including monocyte chemoattractant protein-1, involved in the recruitment of monocytes in inflammatory vascular disease states (Feinberg et al., 2004), and the scavenger receptor CD36, which has been shown to be regulated by a reduction in peroxisome proliferator-activated receptor-γ activity mediated by TGF-β through Smad-3 and the activator protein-1 (AP-1) family (Fu et al., 2003). In addition, our data suggest an involvement of MAPKs as the TGF-β-mediated inhibition of ADAMTS-4 was no longer significant following siRNA-mediated knockdown of p38 MAPK and c-Jun (a key downstream target for JNK pathway) (Fig. 5). In contrast, the ERK pathway was not involved (Fig. 5). TGF-β is able to activate the MAPK pathways (Ten Dijke and Hill, 2004) but whether this acts independently or in conjunction with the classical Smad pathway is unclear. Smads have been shown to interact with the c-Jun and c-fos proteins, components of the AP-1 transcription factor complex (Verrecchia and Mauviel, 2002), and the TGF-β-mediated inhibition of MMP-1 expression has been shown to be dependent on the interaction between Smads and the AP-1 site present within the MMP-1 promoter (Hall et al., 2003).
Several cytokines are present in atherosclerotic lesions and the action of combinations of cytokines on gene expression and cellular changes is likely to be more profound than that produced by a single cytokine. For example, our previous studies have shown that there is a synergism between IFN-γ and TNF-α in the regulation of macrophage lipoprotein lipase gene expression (Tengku-Muhammad et al., 1998). On the other hand, IFN-γ antagonises the action of TGF-β (Ulloa et al., 1999). Future studies should therefore investigate the action of combinations of cytokines on ADAMTS-4 expression.
In conclusion, we have shown that ADAMTS-4 is negatively regulated by TGF-β in human macrophages and that this regulation requires Smads, p38 MAPK and c-Jun. We have identified the minimal regulatory promoter region involved in this response. These findings are likely to be important for atherosclerosis given the potent anti-atherogenic action of TGF-β, as demonstrated by numerous studies in vitro and in vivo, and the demonstrated crucial role of proteases in the regulation of plaque stability. It is therefore essential that the role of ADAMTS-4 in the regulation of atherosclerosis in mouse model systems is determined.
Acknowledgements
We thank the British Heart Foundation for financial support (Grant PG/08/073/25520) and Dr. Yoshiharu Mizui of Eisai Co. Ltd., Japan for providing the ADAMTS-4 promoter constructs. Rebecca Salter and Tim Ashlin were recipients of BBSRC Studentships.
References
- Ali S., Singh N.N., Yildirim H., Ramji D.P. Requirement for nuclear factor kappa B signalling in the interleukin-1-induced expression of the CCAAT/enhancer binding protein-δ gene in hepatocytes. Int J Biochem Cell Biol. 2010;42:113–119. doi: 10.1016/j.biocel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J. The human leukemia cell line, THP-1: a multifaceted model for the study of monocyte–macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. [DOI] [PubMed] [Google Scholar]
- Bui Q.T., Prempeh M., Wilensky R.L. Atherosclerotic plaque development. Int J Biochem Cell Biol. 2009;41:2109–2113. doi: 10.1016/j.biocel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Feinberg M.W., Shimizu K., Lebedeva M., Haspel R., Takayama K., Chen Z. Essential role for Smad3 in regulating MCP-1 expression and vascular inflammation. Circ Res. 2004;94:601–608. doi: 10.1161/01.RES.0000119170.70818.4F. [DOI] [PubMed] [Google Scholar]
- Foka P., Irvine S.A., Kockar F., Ramji D.P. Interleukin-6 represses the transcription of the CCAAT/enhancer binding protein-α gene in hepatoma cells by inhibiting its ability to autoactivate the proximal promoter region. Nucleic Acids Res. 2003;31:6722–6732. doi: 10.1093/nar/gkg861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foka P., Singh N.N., Salter R.C., Ramji D.P. The tumour necrosis factor-α-mediated suppression of the CCAAT/enhancer binding protein-α gene transcription in hepatocytes involves inhibition of autoregulation. Int J Biochem Cell Biol. 2009;41:1189–1197. doi: 10.1016/j.biocel.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Fu M., Zhang J., Lin Y., Zhu X., Zhao L., Ahmad M. Early stimulation and late inhibition of peroxisome proliferator-activated receptor γ (PPARγ) gene expression by transforming growth factor-β in human aortic smooth muscle cells: role of early growth-response factor-1 (Egr-1), activator protein 1 (AP1) and Smads. Biochem J. 2003;370:1019–1025. doi: 10.1042/BJ20021503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galis Z.S., Khatri J.J. Matrix metalloproteinases in vascular remodelling and atherogenesis: the good, the bad and the ugly. Circ Res. 2002;90:251–262. [PubMed] [Google Scholar]
- Hall M.C., Young D.A., Waters J.G., Rowan A.D., Chantry A., Edwards D.R. The comparative role of activator protein 1 and Smad factors in the regulation of Timp-1 and MMP-1 gene expression by transforming growth factor-β1. J Biol Chem. 2003;278:10304–10313. doi: 10.1074/jbc.M212334200. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Tengku-Muhammad T.S., Irvine S.A., Ramji D.P. A novel role of Sp1 and Sp3 in the interferon-γ-mediated suppression of macrophage lipoprotein lipase gene transcription. J Biol Chem. 2002;277:11097–11106. doi: 10.1074/jbc.M106774200. [DOI] [PubMed] [Google Scholar]
- Irvine S.A., Foka P., Rogers S.A., Mead J.R., Ramji D.P. A critical role for the Sp1-binding sites in the transforming growth factor-β-mediated inhibition of lipoprotein lipase gene expression in macrophages. Nucleic Acids Res. 2005;33:1423–1434. doi: 10.1093/nar/gki280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.C., Riley G.P. ADAMTS proteinases: a multi-domain, multi-functional family with roles in extracellular matrix turnover and arthritis. Arthritis Res Ther. 2005;7:160–169. doi: 10.1186/ar1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson-Rylander A.C., Nilsson T., Fritsche-Danielson R., Hammarstrom A., Behrendt M., Andersson J.O. Role of ADAMTS-1 in atherosclerosis: remodelling of carotid artery, immunohistochemistry, and proteolysis of versican. Arterioscler Thromb Vasc Biol. 2005;25:180–185. doi: 10.1161/01.ATV.0000150045.27127.37. [DOI] [PubMed] [Google Scholar]
- Kalinina N., Agrotis A., Antropova Y., Ilyinskaya O., Smirnov V., Tararak E. Smad expression in human atherosclerotic lesions. Evidence for impaired TGF-β/Smad signalling in smooth muscle cells of fibrofatty lesions. Arterioscler Thromb Vasc Biol. 2004;24:1391–1396. doi: 10.1161/01.ATV.0000133605.89421.79. [DOI] [PubMed] [Google Scholar]
- Kerr L.D., Miller D.B., Matrisian L.M. TGF-β1 inhibition of transin/stromelysin gene expression is mediated through a Fos binding sequence. Cell. 1990;61:267–278. doi: 10.1016/0092-8674(90)90807-q. [DOI] [PubMed] [Google Scholar]
- Kuno K., Kanada N., Nakashima E., Fujiki F., Ichimura F., Matsushima K. Molecular cloning of a gene encoding a new type of metalloproteinase–disintegrin family protein with thrombospondin motifs as an inflammation-associated gene. J Biol Chem. 1997;272:556–562. doi: 10.1074/jbc.272.1.556. [DOI] [PubMed] [Google Scholar]
- Li A.C., Glass C.K. The macrophage foam cell as a target for therapeutic intervention. Nat Med. 2002;8:1235–1242. doi: 10.1038/nm1102-1235. [DOI] [PubMed] [Google Scholar]
- Li N., McLaren J.E., Michael D.R., Clement M., Fielding C.A., Ramji D.P. ERK is integral to the IFN-γ-mediated activation of STAT1, the expression of key genes implicated in atherosclerosis, and the uptake of modified lipoproteins by human macrophages. J Immunol. 2010;185:3041–3048. doi: 10.4049/jimmunol.1000993. [DOI] [PubMed] [Google Scholar]
- Lusis A.J., Mar R., Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- McLaren J.E., Calder C.J., McSharry B.P., Sexton K., Salter R.C., Singh N.N. The TNF-like protein 1A-death receptor 3 pathway promotes macrophage foam cell formation in vitro. J Immunol. 2010;184:5827–5834. doi: 10.4049/jimmunol.0903782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren J.E., Michael D.R., Salter R.C., Ashlin T.G., Calder C.J., Miller A.M. IL-33 reduces macrophage foam cell formation. J Immunol. 2010;185:1222–1229. doi: 10.4049/jimmunol.1000520. [DOI] [PubMed] [Google Scholar]
- Mizui Y., Yamazaki K., Kuboi Y., Sagane K., Tanaka I. Characterization of the 5′-flanking region of human aggrecanase-1 (ADAMTS4) gene. Mol Biol Rep. 2000;27:167–173. doi: 10.1023/a:1007253930568. [DOI] [PubMed] [Google Scholar]
- Naito S., Shiomi T., Okada A., Kimura T., Chijiiwa M., Fujita Y. Expression of ADAMTS-4 (aggrecanase-1) in human osteoarthritic cartilage. Pathol Int. 2007;57:703–711. doi: 10.1111/j.1440-1827.2007.02167.x. [DOI] [PubMed] [Google Scholar]
- Newby A.C. Metalloproteinases and vulnerable atherosclerotic plaques. Trends Cardiovasc Med. 2007;17:253–258. doi: 10.1016/j.tcm.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K., Chen F., Kuang C., Chen Y. Suppression of matrix metalloproteinase-9 transcription by transforming growth factor-β is mediated by a nuclear factor-kappaB site. Biochem J. 2004;381:413–422. doi: 10.1042/BJ20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S., Clark I.M., Keveorkian L., Edwards D.R. The ADAMTS metalloproteinases. Biochem J. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S., Hill C.S. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Salter R.C., Ashlin T.G., Kwan A.P., Ramji D.P. ADAMTS proteases: key roles in atherosclerosis. J Mol Med. 2010;88:1203–1211. doi: 10.1007/s00109-010-0654-x. [DOI] [PubMed] [Google Scholar]
- Sandy J.D., Westling J., Kenagy R.D., Iruela-Arispe M.L., Verscharen C., Rodriguez-Mazaneque J.C. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J Biol Chem. 2001;276:13372–13378. doi: 10.1074/jbc.M009737200. [DOI] [PubMed] [Google Scholar]
- Singh N.N., Ramji D.P. The role of transforming growth factor-β in atherosclerosis. Cytokine Growth Factor Rev. 2006;17:487–499. doi: 10.1016/j.cytogfr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Singh N.N., Ramji D.P. Transforming growth factor-β-induced expression of the apolipoprotein E requires c-Jun N-terminal kinase, p38 kinase, and casein kinase 2. Arterioscler Thromb Vasc Biol. 2006;26:1323–1329. doi: 10.1161/01.ATV.0000220383.19192.55. [DOI] [PubMed] [Google Scholar]
- Tang B.L. ADAMTS: a novel family of extracellular matrix proteases. Int J Biochem Cell Biol. 2001;33:33–44. doi: 10.1016/s1357-2725(00)00061-3. [DOI] [PubMed] [Google Scholar]
- Ten Dijke P., Hill C.S. New insights into TGF-β-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Tengku-Muhammad T.S., Cryer A., Ramji D.P. Synergism between interferon γ and tumour necrosis factor α in the regulation of lipoprotein lipase in the macrophage J774.2 cell line. Cytokine. 1998;10:38–48. doi: 10.1006/cyto.1997.0254. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K., Pei Y., Moore T.L., Wang H., Yu X.P., Geiser A.G. Regulation of human ADAMTS-4 promoter by transcription factors and cytokines. Biochem Biophys Res Commun. 2006;345:197–204. doi: 10.1016/j.bbrc.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K., Pei Y., Wei T. Characterization of the human ADAMTS-5 (aggrecanase-2) gene promoter. Mol Biol Rep. 2007;34:225–231. doi: 10.1007/s11033-006-9037-3. [DOI] [PubMed] [Google Scholar]
- Ulloa L., Doody J., Massagué J. Inhibition of transforming growth factor-β/SMAD signalling by the interferon γ/STAT pathway. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- Verrecchia F., Mauviel A. Transforming growth factor-β signalling through the Smad pathway: role of extracellular matrix gene expression and regulation. J Invest Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
- Wågsäter D., Bjork H., Zhu C., Bjorkegren J., Valen G., Hamsten A. ADAMTS-4 and -8 are inflammatory regulated enzymes expressed in macrophage-rich areas of human atherosclerotic plaques. Atherosclerosis. 2008;196:514–522. doi: 10.1016/j.atherosclerosis.2007.05.018. [DOI] [PubMed] [Google Scholar]
- White L.A., Mitchell T.I., Brinckerhoff C.E. Transforming growth factor β inhibitory element in the rabbit matrix metalloproteinase-1 (collagenase-1) gene functions as a repressor of constitutive transcription. Biochim Biophys Acta. 2000;1490:259–268. doi: 10.1016/s0167-4781(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Wight T.N. The ADAMTS proteases, extracellular matrix and vascular disease. Walking the sleeping giant(s)! Arterioscler Thromb Vasc Biol. 2005;25:12–14. doi: 10.1161/01.ATV.0000150043.43083.aa. [DOI] [PubMed] [Google Scholar]
- Worley J.R., Baugh M.D., Hughes D.A., Edwards D.R., Hogan A., Sampson M.J. Metalloproteinase expression in PMA-stimulated THP-1 cells. Effects of peroxisome proliferator-activated receptor-gamma (PPARγ) agonists and 9-cis-retinoic acid. J Biol Chem. 2003;278:51340–51346. doi: 10.1074/jbc.M310865200. [DOI] [PubMed] [Google Scholar]