Abstract
Some murine-endogenous retroviruses, making up ∼10 % of the mouse genome, are induced during the course of experimental sepsis in which lipopolysaccharide (LPS), a pathogenic component of Gram-negative bacteria, often plays a critical role. In this study, we investigated whether LPS stress induces the production of murine leukemia virus type-endogenous retrovirus (MuLV-ERV) virions in primary lymphoid cells. LPS treatment of cells (single-cell suspensions and sorted B- and T-cells) isolated from seven lymphoid organs of C57BL/6J mice resulted in a differential increase in the production of MuLV-ERV virions in most cells examined. Interestingly, among the 34 unique MuLV-ERV U3 sequences cloned from the viral genomic RNAs, the nuclear respiratory factor 1 (transcription factor) element was present only in the 20 U3 sequences that were derived from the LPS-induced MuLV-ERV U3 bands. Using the U3 sequences as a probe, 55 putative MuLV-ERV loci were mapped onto the C57BL/6J mouse genome and 15 of them retained full coding potential. Furthermore, one full-length recombinant MuLV-ERV originating from a locus on chromosome 13 was determined to be responsive to LPS stress. The findings from this study suggest that LPS stress differentially activates MuLV-ERV virion production in lymphoid organs in a cell type- and MuLV-ERV-specific manner. Further investigation is needed to define the role of MuLV-ERVs in the LPS signalling pathway(s) in general, as well as in the pathogenesis of sepsis.
INTRODUCTION
Infection of germ-line cells during the early stages of embryonic development by exogenous retroviruses allows for the permanent embedment of their proviral copies, called endogenous retroviruses (ERVs), into random loci of the germ-line genome (Boeke & Stoye, 1997). ERVs, which are passed to the offspring along with other genetic information in the germ-line genome, are reported to be present in all vertebrates examined so far and constitute ∼8 and ∼10 % of the human and mouse genomes, respectively (Griffiths, 2001; Herniou et al., 1998; Waterston et al., 2002).
Like all the other genetic elements in the genome, the ERVs' biological activities are primarily governed by the intracellular transcription environment, which is specific or non-specific for individual cells (Larsson & Andersson, 1998). Regulatory elements found in individual ERV promoters, which are located on the U3 region of the 5′ LTR, play a crucial role in controlling ERV expression within the transcription environment in which they reside (Corcoran et al., 1984; Selten et al., 1985; Trusko et al., 1989). The transcription environment is specifically and dynamically formulated for the short- and/or long-term in the individual cells, which are constantly subjected to various stressors (both proximal and distal) such as injury, infection and hormone change (Cho et al., 2008). Stressor-elicited activation of certain murine leukemia virus type-ERVs (MuLV-ERVs) may produce replication-competent virus particles whose genomes are either full length or defective (Kwon et al., 2009; Lee et al., 2007). These ‘activated’ virus particles have the potential to alter the response to stressors, such as sepsis or injury.
Lipopolysaccharide (LPS), which is found on the surface of Gram-negative bacteria, is one of the most extensively studied triggers of innate immune response (Alexander & Rietschel, 2001). Various cell types, such as fibroblasts, immature B-cells, B-cells, T-cells, monocytes and macrophages, are susceptible to LPS stress (Chaby & Girard, 1993; Morrison & Ulevitch, 1978). The binding of LPS to its membrane receptor complex, primarily composed of Toll-like receptor 4 and CD14, initiates a cascade of intracellular events associated with the production of pro-inflammatory cytokines and relevant pathogenic processes (O'Neill, 2003). In addition, the mitogenic properties of LPS are responsible for the initiation of proliferation and differentiation of B-cells into antibody-producing plasma cells in conjunction with V(D)J rearrangement of immunoglobulin genes (Yang et al., 2007).
A number of non-ecotropic MuLV-ERVs (i.e. xenotropic, polytropic and modified polytropic) have been identified and characterized in various mouse strains (Frankel et al., 1990; Jern et al., 2007; Kwon et al., 2009; Lee et al., 2007, 2008; Stoye & Coffin, 1987). It has been reported that certain endogenous xenotropic MuLVs are induced by LPS stimulation in the spleen cells of several strains of mice (Jongstra & Moroni, 1981; Kozak & Rowe, 1980; Krieg et al., 1988; Phillips et al., 1976). These findings provide evidence that LPS induces the expression of certain MuLV-ERVs in lymphoid tissues.
Recent studies conducted in our laboratory have demonstrated that stress signals elicited from burn injury or caecal ligation and puncture (CLP)-induced sepsis differentially altered the expression of MuLV-ERVs in a range of organs (Cho et al., 2008, 2009). It has been proposed that post-burn activation of Gram-negative bacterial activity in the gut contributes to the elevation of systemic LPS levels (Schwacha, 2003). In addition, LPS may play a critical role in the development of sepsis following CLP-induced polymicrobial peritonitis (Neviere et al., 1999). In this study, we tested the hypothesis that, among a range of stressors associated with the pathogenic processes of burn and/or sepsis, LPS induces expression of certain MuLV-ERVs in the cells of the lymphoid organs resulting in an increase in virion production.
RESULTS AND DISCUSSION
LPS-induced increase in the production of MuLV-ERV virions in a lymphoid organ- and cell type-specific manner
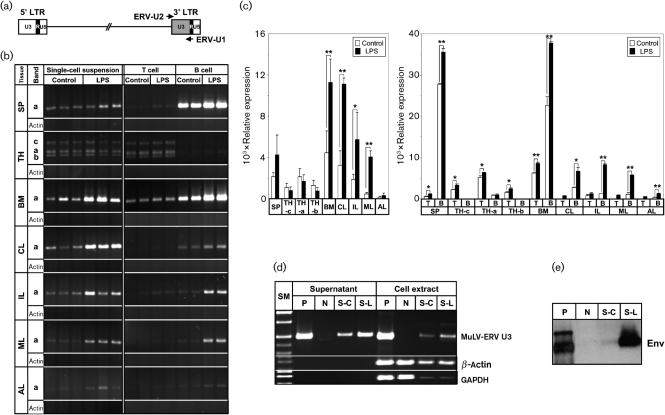
To investigate whether LPS stress induces MuLV-ERV virion production, primary cells derived from seven lymphoid tissues [spleen, thymus, bone marrow and four lymph nodes (axillary, cervical, inguinal and mesenteric)] of C57BL/6J mice were treated with LPS. Supernatants were collected from the LPS-treated and control primary single-cell suspensions and subjected to RT-PCR analysis for viral genomic RNA. A primer set for the MuLV-ERV U3 promoter regions within the 3′ LTR was used to examine changes in the levels of viral genomic RNAs (Fig. 1a). One distinct U3-region band (∼500 bp and labelled as ‘a’ in Fig. 1b) was amplified from viral genomic RNA from all lymphoid tissues; however, the thymus had two additional bands (∼450 bp, labelled as ‘b’ in Fig. 1b and ∼750 bp, labelled as ‘c’ in Fig. 1b). β-Actin was used as a negative control to confirm that there was no cellular mRNA contamination of the viral genomic RNAs isolated from the supernatants. A quantitative analysis of the amplification pattern of the ‘a’ band revealed that there was a significant increase (denoted by *, P<0.05 and **, P<0.01) in the level of MuLV-ERV genomic RNA in the culture supernatants of LPS-treated single-cell suspensions, which were derived from the bone marrow and three lymph nodes (cervical, inguinal and mesenteric) (Fig. 1c).
Fig. 1.
Differential induction of MuLV-ERV production in cells derived from various lymphoid organs in an organ- and cell type-specific manner. (a) Schematic representation of primer locations. A set of primers (ERV-U2 and ERV-U1) flanking the 3′ U3 region of a typical retroviral provirus are indicated by arrows. (b) RT-PCR analysis of changes in MuLV-ERV genomic RNA levels after LPS treatment in culture supernatants of primary cells derived from various lymphoid organs. Post-LPS treatment changes in the levels of MuLV-ERV genomic RNAs in the culture supernatants of primary cells [single-cell suspensions (left) and sorted B- and T-cells (right)] of seven lymphoid organs were analysed by RT-PCR. SP, Spleen; TH, thymus; BM, bone marrow; CL, cervical lymph node; IL, inguinal lymph node; ML, mesenteric lymph node; AL, axillary lymph node; a–c, different sizes of amplified U3 bands. (c) Quantitative presentation of the results (shown in b) from RT-PCR analysis of MuLV-ERV genomic RNA levels in the culture supernatants of primary cells (left graph, single-cell suspensions; right graph, sorted B- and T-cells). Density of the amplified U3 bands from each group were presented as mean±sd. Statistical significance is indicated by: *, P<0.05 and **, P<0.01. (d) RT-PCR analysis of MuLV-ERV genomic RNA from supernatants and mRNA from cell extracts of splenocyte cultures following LPS treatment. Using both β-actin and GAPDH as cellular mRNA controls, the presence of viral genomic RNA derived from the MuLV-ERV virions harvested from the culture supernatants was confirmed by RT-PCR. P, Positive control, SC-1/MuLV LP-BM5 cell; N, negative control, HEK293 cell-vehicle treatment; S-C, splenocyte-vehicle treatment; S-L, splenocyte-LPS treatment; SM, size marker. (e) Western blot analysis of MuLV-ERV Env protein in the culture supernatants of splenocytes following LPS treatment. Presence of MuLV-ERV virions in the culture supernatants of splenocytes following LPS treatment was confirmed by the detection of Env protein derived from MuLV-ERV virions. P, Positive control, SC-1/MuLV LP-BM5 cell; N, negative control, HEK293 cell-vehicle treatment; S-C, splenocyte-vehicle treatment; S-L, splenocyte-LPS treatment.
These findings suggest that LPS stress differentially induces production of MuLV-ERV virions, depending on the source of the lymphoid cells. It is probable that by-products of the well-studied LPS signalling pathway, such as nuclear factor κB and inflammatory cytokines, may participate in the activation of certain MuLV-ERVs (Lee et al., 2008; Verstrepen et al., 2008). Alternatively, transcription factors associated with unknown LPS signalling event(s) may play a role in the activation of these MuLV-ERVs, which in turn modulate the expression of certain molecules, including inflammatory mediators (e.g. interleukin-6 and cyclooxygenase-2). Furthermore, the activated MuLV-ERVs may enact their biological functions via their gene products (e.g. Env or Gag protein) and/or by the infection of host cells in conjunction with random genomic integration of their proviral copies (Dolei, 2005; Knerr et al., 2004; Mi et al., 2000).
To identify the specific cell type(s) in the tissues responsible for the increased production of MuLV-ERV virions in response to LPS treatment, the primary cells from the seven lymphoid organs were sorted into B- and T-cells. These sorted cells were also treated with LPS and the supernatants were collected and analysed for the presence of MuLV-ERV genomic RNAs by RT-PCR (Fig. 1b and c). Significant and differential increases (denoted by *, P<0.05 and **, P<0.01) in the levels of MuLV-ERV genomic RNA (‘a’ band) were observed in the supernatants of LPS-treated B-cells isolated from all lymphoid organs except for the thymus (Fig. 1b and c). Among the B-cell populations originating from four different lymph nodes, B-cells from the inguinal and mesenteric lymph nodes displayed a substantially greater response to LPS treatment, in regard to MuLV-ERV virion production, compared with the other two lymph nodes. Interestingly, the supernatants of the LPS-treated immature T-cells derived from the bone marrow had an evident increase in the level of MuLV-ERV genomic RNA compared with the no-treatment control.
The results from these experiments suggest that the B-cells residing in both central and peripheral immune organs are the main producers of MuLV-ERV virions in response to LPS stress, and the response is variable depending on the location of the lymphoid organs. Previous reports indicate that B-cells' response to LPS stress is variable depending on their locations, and leads to proliferation and differentiation into antibody-producing plasma cells (Kozak & Rowe, 1980; Phillips et al., 1976; Skalet et al., 2005; Yang et al., 2007). In addition, the animals' genetic background contributes to the B-cells' variable response to LPS stress, such as C57BL/6J (high LPS responder) versus BALB/c (low LPS responder) (Andersson et al., 1977; Wells et al., 2003). It may be interesting to examine whether the MuLV-ERV induction profile of B-cells derived from BALB/c mice in response to LPS stress is different from the profile observed in C57BL/6J mice in this study.
To confirm further that LPS stress contributes to the increase in production of MuLV-ERV virions, the levels of viral genomic RNA and mRNA were examined in culture supernatants and cell extracts of primary splenocytes, respectively, following LPS treatment (Fig. 1d). The presence of MuLV-ERV virions in the supernatants was shown by the detection of substantial levels of viral genomic RNAs by RT-PCR in the absence of evident amplification of β-actin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Both mRNA and genomic RNA levels of LPS-responsive MuLV-ERVs were increased in the LPS treatment group; however, no significant differences in the levels of β-actin and GAPDH in cell extracts were observed between control and LPS-treatment groups, confirming MuLV-ERV-specific induction. In addition, the production of MuLV-ERV virions by the LPS-treated splenocytes was substantiated by the detection of Env protein in the supernatants (Fig. 1e). Since we anticipated that a number of Env polypeptide variants (both known and unknown) exist in C57BL/6J mice, it is unclear which Env variants are represented in this blot.
Characteristics of U3 sequences derived from MuLV-ERV genomic RNAs
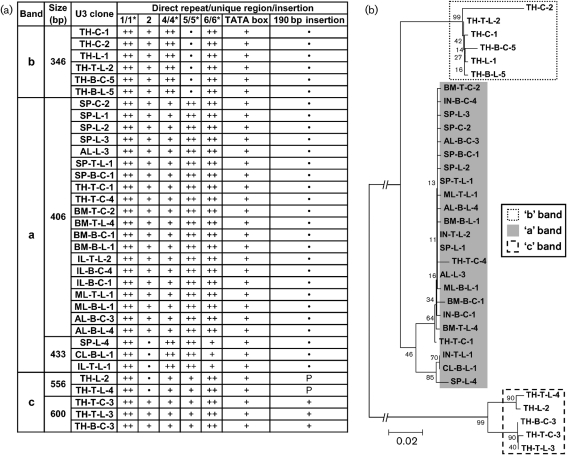
To examine the structure of the U3 regions of viral genomic RNA and to map the corresponding proviral loci on the genome, the amplified MuLV-ERV bands from the supernatants of single-cell suspensions and sorted B- and T-cells (both LPS treated and not treated) were sequenced. It should be noted that the sequence data represents the U3 regions of MuLV-ERV genomic RNAs near the 3′ end; however, the 3′ U3 region is presumed to be almost identical to the 5′ U3 region, which serves as a promoter in non-rearranged proviruses. The data obtained from this analysis revealed unique structural features among the different U3 size groups. From a total of 124 U3 sequences initially cloned from the three U3 bands (a, b and c), 34 unique sequences spanning five different sizes were identified. The majority (20 of 34 U3 sequences) were 406 bp in size (derived from the ‘a’ band), in which three direct repeats (1/1*, 5/5* and 6/6*) were present (Fig. 2a) (Tomonaga & Coffin, 1998, 1999). An insertion of 190 bp (complete or partial) and two direct repeats (1/1* and 6/6*) were found in all U3 sequences derived from the thymus ‘c’ band (556/600 bp in size). The U3 sequences of 346 bp in size, derived from the thymus ‘b’ band, had three direct repeats (1/1*, 4/4* and 6/6*) (Supplementary Fig. S1, available in JGV Online). Furthermore, phylogenetic analysis of the 34 unique U3 sequences revealed three main branches primarily clustered according to the size of the U3 sequences (Fig. 2b).
Fig. 2.
Structural features and phylogenetic relationship of the 34 unique U3 sequences isolated from MuLV-ERV genomic RNAs. (a) Sequence features of the 34 unique U3 sequences. Using an alignment protocol, sequence features [direct repeats (1/1*, 4/4*, 5/5* and 6/6*), unique sequences (2 and TATA box) and an insertion (190 bp insertion)] were determined for each U3 sequence (Tomonaga & Coffin, 1998, 1999). a, ‘a’ Band; b, ‘b’ band; c, ‘c’ band; ++, presence of direct repeat; +, presence of single unit; P, partial; •, absence. (b) Phylogenetic relationship of the 34 unique U3 sequences. The phylogenetic tree was established based on the multiple alignment data. The node values indicate the percentage probability for a specific branching. Bar, 0.02 nucleotide substitutions per site.
To examine the transcription potential of the 34 unique MuLV-ERV U3 sequences, the profile of putative transcription regulatory elements within each U3 sequence was surveyed (Supplementary Table S1, available in JGV Online). Among the 70 regulatory elements mapped, nine elements were shared by all 34 U3 sequences examined. Interestingly, one element specific for the binding of nuclear respiratory factor 1, (NRF1; Scarpulla, 1997) was only present in all 20 of the U3 sequences of 406 bp in size, which was one of the two size groups (406 and 433 bp) isolated from the ‘a’ band. The ‘a’ band was induced following LPS treatment in all lymphoid organs examined, except the thymus (Fig. 1b). The primary function of the NRF1 transcription factor is to regulate expression of nucleus-encoded subunits of the cytochrome c oxidase complex embedded in the mitochondrial membrane (Dhar et al., 2008; Evans & Scarpulla, 1990). NRF1 is also reported to bind to the promoter of hepatitis B virus X gene, which plays a role in the regulation of mitochondrial membrane potential (Tokusumi et al., 2004). Further investigation is needed to evaluate whether NRF1 and other transcription factors/co-factors play a specific role in the transcriptional activation of the MuLV-ERVs harbouring these unique U3 sequences in response to LPS stress.
In silico mapping of putative MuLV-ERVs using U3 sequences as a probe and characterization of their biological properties
In order to identify the MuLV-ERVs harbouring each unique U3 sequence, the National Center for Biotechnology Information (NCBI) mouse-genome database was surveyed using the 34 unique U3 sequences as data-mining probes. MuLV-ERVs with greater than 98 % identity, respective to the U3 probes, were mapped on various contiguous sequences of the database (Supplementary Table S2, available in JGV Online). A total of 56 MuLV-ERVs, ranging from 5312 to 9054 bp in proviral size, were identified and found to be distributed throughout the genome except for chromosome 17 (Supplementary Table S2). During chromosomal mapping of the 56 MuLV-ERVs using the Mouse blat search program (http://genome.ucsc.edu/), it was confirmed that 55 of them were present in the C57BL/6J mouse genome, while MuLV-ERV 34 was found within the SPRET/Ei mouse genome. Fifty-three of the 56 MuLV-ERVs have been reported in our previous studies (Kwon et al., 2009; Lee et al., 2007, 2008). In addition, 38 of the 56 MuLV-ERVs were found to be identical to ERVs reported previously (Supplementary Table S2) (Frankel et al., 1990; Jern et al., 2007). For each MuLV-ERV isolate, the complete proviral sequence plus flanking sequences (50 bp upstream and downstream) were subjected to the following analyses: ORF for coding potential, primer-binding site (PBS) for replication initiation, target site duplication (TSD) for recombination potential and LTR mutation rate for the estimated integration age (Supplementary Fig. S2, available in JGV Online).
In order to determine the coding potential of each MuLV-ERV isolate, the ORFs for gag, pol and env polypeptides were compared to the MuLV references (Supplementary Table S2) (Shinnick et al., 1981; Urisman et al., 2006). Fifteen of the 56 putative MuLV-ERVs retained intact coding sequences for all three polypeptides (gag, pol and env), while the rest had defective and/or partial ORFs for some or all of the polypeptides.
A stretch of 18 bp immediately downstream of the 5′ U5 region was examined to determine the PBS for each putative MuLV-ERV isolate by comparing it to the reference sequences (Harada et al., 1979; Nikbakht et al., 1985). Glutamine tRNA binding sites were identified in 53 of the MuLV-ERVs, while threonine tRNA-binding sites were present in MuLV-ERV 6, MuLV-ERV 40 and MuLV-ERV 46 (Supplementary Table S2) (Jern et al. 2007).
To determine whether there was a genetic rearrangement event(s) in the MuLV-ERV isolates since the initial integration into the genome, the TSDs flanking the 5′ and 3′ ends of each proviral sequence were surveyed (Supplementary Tables S2 and S3, available in JGV Online). TSDs are formed during the genomic integration event and any genomic rearrangements involving the proviral locus, primarily recombination via LTRs, replaces the TSDs with two different sequences (Sverdlov, 2000). Nine of the 56 MuLV-ERVs did not retain TSDs at their integration sites, suggesting that they are rearranged MuLV-ERVs, while the rest had TSDs of 4–12 bp. Alternatively, the absence of TSDs in the nine MuLV-ERVs may reflect random point mutations instead of rearrangement events. We were not able to confirm the recombination status of the nine MuLV-ERVs by an LTR phylogenetic analysis because of high sequence identity in each set of flanking LTR sequences (Supplementary Fig. S3, available in JGV Online). Interestingly, four of the nine MuLV-ERVs retained intact ORFs for all three polypeptides.
The integration age of MuLV-ERVs was estimated based on the mutation rate between the two flanking LTRs. Two MuLV-ERVs were determined to have estimated integration ages of 2.2 million years (Myr) (0.29 % mutation rate) and 2.1 Myr (0.27 % mutation rate). Both of them are defective proviruses, probably because of the accumulation of mutations in the coding sequences since their initial integration into the germ line. Since 45 of the 56 MuLV-ERVs had no mismatch between flanking LTRs (Supplementary Fig. S3), their estimated integration age was arbitrarily recorded as <1 Myr. On the other hand, the integration age of the rearranged MuLV-ERV isolates, as evidenced by the absence of TSDs, was not determined because the genetic rearrangement events may invalidate the calculation of the mutation rate of their LTR.
Identification of an LPS-responsive recombinant MuLV-ERV
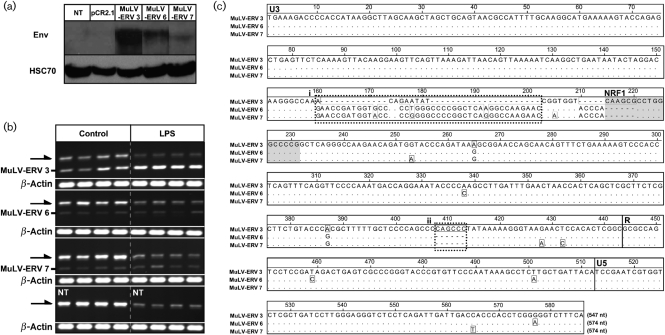
To further characterize MuLV-ERV LPS responsiveness, three MuLV-ERVs, which were isolated by using two U3 sequences (406 and 433 bp in size) as a probe (Supplementary Table S1), were cloned from C57BL/6J genomic DNA using a primer set specific for the sequences flanking each provirus. Among the three recombinant MuLV-ERVs, two [MuLV-ERV 3 (406 bp U3) and MuLV-ERV 7 (433 bp U3)] had intact coding sequences for all three polypeptides (gag, pol and env) and one [MuLV-ERV 6 (433 bp U3)] retained an intact coding sequences for pol and env only. Following transfection of the recombinant MuLV-ERVs into HEK293 cells, Western blot analysis using the cell extracts demonstrated varying levels of Env polypeptide expression among the recombinant MuLV-ERVs, with MuLV-ERV 3 having the greatest (Fig. 3a).
Fig. 3.
Identification of a LPS-responsive recombinant MuLV-ERV. (a) Confirmation of Env-protein expression from three recombinant MuLV-ERVs. Western blot analysis confirmed expression of Env protein in HEK293 cells transfected with the recombinant MuLV-ERVs (MuLV-ERV 3, 6 and 7). NT, No transfection; pCR2.1, transfection of pCR2.1 plasmid. (b) Effect of LPS treatment on the expression of recombinant MuLV-ERVs. RT-PCR analysis revealed that expression of the MuLV-ERV 3 recombinant, but not MuLV-ERV 6 and MuLV-ERV 7, increased in response to LPS treatment. Arrows indicate expression of MuLV-ERVs originating from RAW264.7 cells. NT, No transfection. (c) Alignment analysis of the LTR sequences of three recombinant MuLV-ERVs (MuLV-ERV 3, 6 and 7). In addition to the nuclear respiratory factor 1 (NRF1) site highlighted in grey, two sequence regions uniquely identified in the MuLV-ERV 3 LTR are indicated (i) and (ii).
To evaluate LPS responsiveness, the recombinant MuLV-ERVs were transfected into RAW264.7 alveolar macrophage cells and treated with LPS for 24 h. It has been reported that certain ecotropic and polytropic MuLVs are constitutively produced in this cell line (Hartley et al., 2008). RT-PCR analysis of the cell extracts revealed that LPS treatment resulted in a decrease in expression of MuLV-ERVs endogenous to RAW264.7 cells (Fig. 3b). In contrast, expression of the recombinant MuLV-ERV 3 was substantially augmented in the LPS treatment group compared with the vehicle control, while no evident changes were observed in the expression of the other two recombinants (MuLV-ERV 6 and MuLV-ERV 7). The LTR sequences of the three recombinant MuLV-ERVs were then subjected to alignment analysis to identify sequence features unique for the MuLV-ERV 3 LTR, especially in the U3 promoter region (Fig. 3c). In addition to the binding site for NRF1, as discussed above (Supplementary Table S1), two unique regions were identified in MuLV-ERV 3 compared with the LTRs of MuLV-ERV 6 and MuLV-ERV 7. The notion that these sequence features, including the NRF1 binding site, may contribute to the responsiveness of MuLV-ERV 3 to LPS treatment needs to be evaluated further. The results obtained from this experiment substantiate the finding that LPS stress increases the transcriptional activity of certain MuLV-ERVs, which reside in a specific genomic location and harbour unique U3-sequence feature(s).
Conclusions
LPS, a potent pathogenic molecule derived from Gram-negative bacteria, plays a crucial role during the onset and progression of sepsis. The mechanisms underlying sepsis pathogenesis have not been clearly defined yet. However, the involvement of LPS in inflammation has been investigated extensively, focusing on a set of signalling pathways. The findings from this study suggest that LPS stress can differentially activate MuLV-ERVs leading to virion production, primarily in B cells of a range of lymphoid organs. It may be important to investigate the roles of these MuLV-ERVs in LPS signalling events as well as in the pathogenesis of Gram-negative bacterial sepsis.
METHODS
Preparation of primary cells, sorting and LPS treatment.
Single-cell suspensions were prepared from the lymphoid organs [spleen, thymus, bone marrow and lymph nodes (axillary, cervical, inguinal and mesenteric)] of female C57BL/6J mice (11–13 weeks old; The Jackson Laboratory, Bar Harbor, ME, USA). The animal protocol was approved by the Animal Use and Care Administrative Advisory Committee of the University of California, Davis. The cells were suspended in culture medium [RPMI 1640 (Invitrogen) supplemented with 10 % FBS (J R Scientific) and 100 μg streptomycin ml−1 and 100 U penicillin G ml−1 (Invitrogen)] and adjusted to 8×105 cells ml−1. In a separate experiment, the lymphoid cell suspensions were subjected to sorting into B and T cells using Pan B (anti-B220) and Pan T (anti-Thy1.2) magnetic beads (Invitrogen), respectively, according to the manufacturer's protocol. Cells (single-cell suspensions and sorted) were incubated at 37 oC with 5 % CO2 for 24 h followed by LPS treatment [5 μg (ml culture medium)−1] RPMI 1640 served as a vehicle control for LPS treatment. SC-1/MuLV LP-BM5 cells (NIH AIDS Research and Reference Reagent Program, Germantown, MD, USA) and HEK293 cells served as positive and negative controls for MuLV-ERV virion production, respectively. SC-1/MuLV LP-BM5 and HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10 % FBS and 100 μg streptomycin ml−1 and 100 μg penicillin G ml−1. Cells and culture supernatants were harvested for the isolation of total RNA and viral genomic RNA, respectively.
Transfection and LPS treatment.
RAW264.7 alveolar macrophage cells were maintained in DMEM supplemented with 10 % FBS (Atlanta Biologicals), 100 units penicillin ml−1, 100 μg streptomycin ml−1 and 100 μg gentamicin ml−1. Twenty-four hours prior to transfection, cells (1×106 cells) in DMEM with 10 % FBS (Atlanta Biologicals), but without antibiotics, were seeded on six-well plates. Transient transfection of cells with each recombinant MuLV-ERV construct was performed using FuGENE HD (Roche Applied Science) according to the manufacturer's instructions. Subsequently, cells were incubated at 37 oC with 5 % CO2 for 24 h followed by a change of culture medium with or without LPS (5 μg ml−1). DMEM culture medium served as a vehicle control for LPS treatment. Cells were harvested for total RNA isolation followed by RT-PCR. To confirm the expression of Env protein from the individual recombinant MuLV-ERV constructs, HEK293 cells were maintained in DMEM as described above and transfection was performed using FuGENE 6 (Roche Applied Science) according to the manufacturer's protocol. At 2 days post-transfection, the cells were harvested for Western blot analysis. No transfection and transfection with pCR2.1 expression plasmid (Invitrogen) served as controls.
Isolation of virus particles.
Culture supernatants, which were harvested from the primary cells (single-cell suspensions as well as sorted B and T cells) and cell lines following LPS or vehicle treatment, were passed through a 0.45 μm filter (Corning) and centrifuged at 40 000 g for 1 h at 4 oC in a TLN100 or NVT65 rotor (Beckman Coulter) (Yee et al., 1994).
Extraction of RNAs (viral genomic RNA and cellular total RNA) and RT-PCR.
Viral genomic RNA from the virion preparations and total RNA from cells were isolated using a QIAamp Viral RNA mini kit (Qiagen) and RNeasy mini kit (Qiagen), respectively, according to the manufacturer's instructions. cDNA was synthesized from 1 μl of viral genomic RNA or 100 ng of total RNA using Sensiscript reverse transcriptase (Qiagen). A set of primers, ERV-U1 (5′-CGGGCGACTCAGTCTATCGG-3′) and ERV-U2 (5′-CAGTATCACCAACTCAAATC-3′), were adopted from a previous report to amplify the 3′-MuLV-ERV U3 region (Tomonaga & Coffin, 1999). The comparability between total RNA samples derived from cells, but not between genomic RNAs from virus particles, was determined using β-actin and/or GAPDH as an expression control. The density of the PCR products from viral genomic RNAs was determined using a Kodak Gel Logic 200 Imaging System (Carestream Health) and is presented as mean±sd. Statistical significance was determined by using Student's t-test.
Western blot.
Protein extracts were prepared from the virus particle preparations and transfected cells, and Western blotting was performed as described previously (Cho et al., 2000). Briefly, the membrane was blocked in 5 % non-fat dried milk powder, and goat antibody specific for gp69/71 of Rauscher MuLV (ViroMed Biosafety Laboratories) and anti-goat-HRP antibody (Jackson ImmunoResearch Laboratories) were used to detect MuLV-ERV Env protein. In addition, Western blot analysis for HSC70 protein in cell extracts was performed using the same antibody and protocol as described previously (Cho et al., 2004).
Cloning, sequencing and alignment.
The PCR amplified U3 bands were purified using a Qiaquick Gel Extraction kit (Qiagen) and cloned into pGEM-T Easy vector (Promega). Plasmid DNAs for sequencing were prepared using a Qiaprep Spin miniprep kit (Qiagen) and sequencing was performed at the Molecular Cloning Laboratory (South San Francisco, CA, USA). Initially, a total of 124 MuLV-ERV U3 clones were sequenced, and multiple-alignment analysis using Vector NTI (Invitrogen) identified 34 unique U3 sequences. Multiple alignments of the LTR sequences of three recombinant MuLV-ERVs were performed using the Lasergene program (version 8.0.2; dnastar).
Survey of U3 sequences for structural features and transcription regulatory elements.
Selective structural features (direct repeats, insertion and unique region) were surveyed in the U3 sequences by comparison to reference sequences (Tomonaga & Coffin, 1998, 1999). In addition, each unique U3 sequence, which serves as a promoter, was surveyed for a profile of transcription regulatory elements using MatInspector (Genomatix) with a core similarity of 0.90 (Quandt et al., 1995).
In silico mapping of putative MuLV-ERVs.
Using each unique MuLV-ERV U3 sequence as a probe, the corresponding putative proviral loci were initially identified by surveying the NCBI mouse-genome database followed by chromosomal mapping of the individual ERVs' loci using the Mouse blat search program at http://genome.ucsc.edu. Genomic U3 sequences sharing >98 % identity with a specific U3 probe were mapped for further in silico mapping of MuLV-ERVs. MuLV-ERV loci were identified based on the definition that they are ∼5 to ∼9 kb in size and flanked by LTRs. The coding potential of each MuLV-ERV was determined by examining the ORFs for gag, pol and env polypeptides using Vector NTI (Invitrogen). For truncated ORFs, the intactness (partial or defective) of each polypeptide gene depended on the presence of p12 for gag, RT (reverse transcriptase) for pol and SU (surface domain) for env. Within each polypeptide gene, if the specified coding sequence was intact, but the rest was defective, they were determined to be partial (P). If the specified sequence was not intact, it was classified as defective (−).
Analyses of PBS, recombination event and integration age.
Putative PBSs were determined by examining a stretch of 18 bp immediately downstream of the U5 region of the 5′ LTR using the conserved PBS sequences for tRNA as a reference (Harada et al., 1979; Nikbakht et al., 1985). The integration age was estimated using a formula of 0.13 % mutation rate between flanking LTRs=integration age of 1 Myr (Sverdlov, 2000; Tristem, 2000). The integration age was not determined for a single nucleotide difference between flanking LTRs in consideration of a potential error rate during reverse transcription, PCR, cloning and sequencing. A stretch of 4–12 bp flanking each MuLV-ERV was surveyed for TSDs, which indicates whether the provirus underwent any recombination event.
Construction of recombinant MuLV-ERVs.
Among the putative MuLV-ERVs identified by in silico mapping of the NCBI genome database, three proviral loci were selected for cloning to test for LPS stress response. The entire sequence of each target provirus was amplified from C57BL/6J genomic DNA using a pair of primers specifically designed with reference to its unique genomic locus and directly cloned into pCR2.1 expression plasmid (Invitrogen).
Supplementary Material
Acknowledgments
This study was supported by grants from Shriners of North America [no. 86800 to K. C., no. 84294 to D. N. K. (postdoctoral fellowship) and No. 84308 to Y. K. L. (postdoctoral fellowship)], and the National Institutes of Health (R01 GM071360 to K. C.).
Footnotes
Three supplementary figures and three supplementary tables are available with the online version of this paper.
References
- Alexander, C. & Rietschel, E. T. (2001). Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res 7, 167–202. [PubMed] [Google Scholar]
- Andersson, J., Coutinho, A. & Melchers, F. (1977). Frequencies of mitogen-reactive B cells in the mouse. I. Distribution in different lymphoid organs from different inbred strains of mice at different ages. J Exp Med 145, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D. & Stoye, J. P. (1997). Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In Retroviruses, pp. 343–435. Edited by Coffin, J. M., Hughes, S. H. & Varmus, H. E.. Cold Spring Harbor, NY. : Cold Spring Harbor Laboratory Press. [PubMed]
- Chaby, R. & Girard, R. (1993). Interaction of lipopolysaccharides with cells of immunological interest. Eur Cytokine Netw 4, 399–414. [PubMed] [Google Scholar]
- Cho, K., Zipkin, R. I., Adamson, L. K., McMurtry, A. L., Griffey, S. M. & Greenhalgh, D. G. (2000). Differential regulation of c-jun expression in liver and lung of mice after thermal injury. Shock 14, 182–186. [DOI] [PubMed] [Google Scholar]
- Cho, K., Pham, T. N., Crivello, S. D., Jeong, J., Green, T. L. & Greenhalgh, D. G. (2004). Involvement of CD14 and Toll-like receptor 4 in the acute phase response of serum amyloid A proteins and serum amyloid P component in the liver after burn injury. Shock 21, 144–150. [DOI] [PubMed] [Google Scholar]
- Cho, K., Lee, Y. K. & Greenhalgh, D. G. (2008). Endogenous retroviruses in systemic response to stress signals. Shock 30, 105–116. [DOI] [PubMed] [Google Scholar]
- Cho, K., Chiu, S., Lee, Y. K., Greenhalgh, D. & Nemzek, J. (2009). Experimental polymicrobial peritonitis-associated transcriptional regulation of murine endogenous retroviruses. Shock 32, 147–158. [DOI] [PubMed] [Google Scholar]
- Corcoran, L. M., Adams, J. M., Dunn, A. R. & Cory, S. (1984). Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell 37, 113–122. [DOI] [PubMed] [Google Scholar]
- Dhar, S. S., Ongwijitwat, S. & Wong-Riley, M. T. (2008). Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem 283, 3120–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolei, A. (2005). MSRV/HERV-W/syncytin and its linkage to multiple sclerosis: the usability and the hazard of a human endogenous retrovirus. J Neurovirol 11, 232–235. [DOI] [PubMed] [Google Scholar]
- Evans, M. J. & Scarpulla, R. C. (1990). NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev 4, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Frankel, W. N., Stoye, J. P., Taylor, B. A. & Coffin, J. M. (1990). A linkage map of endogenous murine leukemia proviruses. Genetics 124, 221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, D. J. (2001). Endogenous retroviruses in the human genome sequence. Genome Biol 2, reviews1017–reviews1017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, F., Peters, G. G. & Dahlberg, J. E. (1979). The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J Biol Chem 254, 10979–10985. [PubMed] [Google Scholar]
- Hartley, J. W., Evans, L. H., Green, K. Y., Naghashfar, Z., Macias, A. R., Zerfas, P. M. & Ward, J. M. (2008). Expression of infectious murine leukemia viruses by RAW264.7 cells, a potential complication for studies with a widely used mouse macrophage cell line. Retrovirology 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herniou, E., Martin, J., Miller, K., Cook, J., Wilkinson, M. & Tristem, M. (1998). Retroviral diversity and distribution in vertebrates. J Virol 72, 5955–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern, P., Stoye, J. P. & Coffin, J. M. (2007). Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet 3, 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra, J. & Moroni, C. (1981). Lipopolysaccharide induces retroviral antigen expression in 129/J mouse lymphocytes: evidence for assembly of a defective viral particle. J Virol 37, 1044–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knerr, I., Huppertz, B., Weigel, C., Dotsch, J., Wich, C., Schild, R. L., Beckmann, M. W. & Rascher, W. (2004). Endogenous retroviral syncytin: compilation of experimental research on syncytin and its possible role in normal and disturbed human placentogenesis. Mol Hum Reprod 10, 581–588. [DOI] [PubMed] [Google Scholar]
- Kozak, C. A. & Rowe, W. P. (1980). Genetic mapping of xenotropic murine leukemia virus-inducing loci in five mouse strains. J Exp Med 152, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg, A. M., Khan, A. S. & Steinberg, A. D. (1988). Multiple endogenous xenotropic and mink cell focus-forming murine leukemia virus-related transcripts are induced by polyclonal immune activators. J Virol 62, 3545–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, D. N., Greenhalgh, D. G. & Cho, K. (2009). Cloning and characterization of endogenous retroviruses associated with postinjury stress signals in lymphoid tissues. Shock 32, 80–88. [DOI] [PubMed] [Google Scholar]
- Larsson, E. & Andersson, G. (1998). Beneficial role of human endogenous retroviruses: facts and hypotheses. Scand J Immunol 48, 329–338. [DOI] [PubMed] [Google Scholar]
- Lee, Y. K., Chew, A., Fitzsimon, L., Thomas, R., Greenhalgh, D. & Cho, K. (2007). Genome-wide changes in expression profile of murine endogenous retroviruses (MuERVs) in distant organs after burn injury. BMC Genomics 8, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. K., Chew, A., Phan, H., Greenhalgh, D. G. & Cho, K. (2008). Genome-wide expression profiles of endogenous retroviruses in lymphoid tissues and their biological properties. Virology 373, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, S., Lee, X., Li, X., Veldman, G. M., Finnerty, H., Racie, L., LaVallie, E., Tang, X. Y., Edouard, P. & other authors (2000). Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789. [DOI] [PubMed] [Google Scholar]
- Morrison, D. C. & Ulevitch, R. J. (1978). The effects of bacterial endotoxins on host mediation systems. A review. Am J Pathol 93, 526–618. [PMC free article] [PubMed] [Google Scholar]
- Neviere, R. R., Cepinskas, G., Madorin, W. S., Hoque, N., Karmazyn, M., Sibbald, W. J. & Kvietys, P. R. (1999). LPS pretreatment ameliorates peritonitis-induced myocardial inflammation and dysfunction: role of myocytes. Am J Physiol 277, H885–H892. [DOI] [PubMed] [Google Scholar]
- Nikbakht, K. N., Ou, C. Y., Boone, L. R., Glover, P. L. & Yang, W. K. (1985). Nucleotide sequence analysis of endogenous murine leukemia virus-related proviral clones reveals primer-binding sites for glutamine tRNA. J Virol 54, 889–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, L. A. (2003). Therapeutic targeting of Toll-like receptors for inflammatory and infectious diseases. Curr Opin Pharmacol 3, 396–403. [DOI] [PubMed] [Google Scholar]
- Phillips, S. M., Stephenson, J. R., Greenberger, J. S., Lane, P. E. & Aaronson, S. A. (1976). Release of xenotropic type C RNA virus in response to lipopolysaccharide: activity of lipid-A portion upon B lymphocytes. J Immunol 116, 1123–1128. [PubMed] [Google Scholar]
- Quandt, K., Frech, K., Karas, H., Wingender, E. & Werner, T. (1995). MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla, R. C. (1997). Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomembr 29, 109–119. [DOI] [PubMed] [Google Scholar]
- Schwacha, M. G. (2003). Macrophages and post-burn immune dysfunction. Burns 29, 1–14. [DOI] [PubMed] [Google Scholar]
- Selten, G., Cuypers, H. T. & Berns, A. (1985). Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J 4, 1793–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick, T. M., Lerner, R. A. & Sutcliffe, J. G. (1981). Nucleotide sequence of Moloney murine leukaemia virus. Nature 293, 543–548. [DOI] [PubMed] [Google Scholar]
- Skalet, A. H., Isler, J. A., King, L. B., Harding, H. P., Ron, D. & Monroe, J. G. (2005). Rapid B cell receptor-induced unfolded protein response in nonsecretory B cells correlates with pro- versus antiapoptotic cell fate. J Biol Chem 280, 39762–39771. [DOI] [PubMed] [Google Scholar]
- Stoye, J. P. & Coffin, J. M. (1987). The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol 61, 2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverdlov, E. D. (2000). Retroviruses and primate evolution. Bioessays 22, 161–171. [DOI] [PubMed] [Google Scholar]
- Tokusumi, Y., Zhou, S. & Takada, S. (2004). Nuclear respiratory factor 1 plays an essential role in transcriptional initiation from the hepatitis B virus x gene promoter. J Virol 78, 10856–10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga, K. & Coffin, J. M. (1998). Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J Virol 72, 8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga, K. & Coffin, J. M. (1999). Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J Virol 73, 4327–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristem, M. (2000). Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol 74, 3715–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusko, S. P., Hoffman, E. K. & George, D. L. (1989). Transcriptional activation of cKi-ras proto-oncogene resulting from retroviral promoter insertion. Nucleic Acids Res 17, 9259–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urisman, A., Molinaro, R. J., Fischer, N., Plummer, S. J., Casey, G., Klein, E. A., Malathi, K., Magi-Galluzzi, C., Tubbs, R. R. & other authors (2006). Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog 2, e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Verstrepen, L., Adib-Conquy, M., Kreike, M., Carpentier, I., Adrie, C., Cavaillon, J. M. & Beyaert, R. (2008). Expression of the NF-κB inhibitor ABIN-3 in response to TNF and Toll-like receptor 4 stimulation is itself regulated by NF-κB. J Cell Mol Med 12, 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M. & other authors (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- Wells, C. A., Ravasi, T., Faulkner, G. J., Carninci, P., Okazaki, Y., Hayashizaki, Y., Sweet, M., Wainwright, B. J. & Hume, D. A. (2003). Genetic control of the innate immune response. BMC Immunol 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Tung, J. W., Ghosn, E. E. & Herzenberg, L. A. (2007). Division and differentiation of natural antibody-producing cells in mouse spleen. Proc Natl Acad Sci U S A 104, 4542–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee, J. K., Friedmann, T. & Burns, J. C. (1994). Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol 43, 99–112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.