Abstract
Transcription factors (TFs) are key nodes of regulatory networks in eukaryotic organisms, including filamentous fungi such as Neurospora crassa. The 178 predicted DNA-binding TFs in N. crassa are distributed primarily among six gene families, which represent an ancient expansion in filamentous ascomycete genomes; 98 TF genes show detectable expression levels during vegetative growth of N. crassa, including 35 that show a significant difference in expression level between hyphae at the periphery versus hyphae in the interior of a colony. Regulatory networks within a species genome include paralogous TFs and their respective target genes (TF regulon). To investigate TF network evolution in N. crassa, we focused on the basic leucine zipper (bZIP) TF family, which contains nine members. We performed baseline transcriptional profiling during vegetative growth of the wild-type and seven isogenic, viable bZIP deletion mutants. We further characterized the regulatory network of one member of the bZIP family, NCU03905. NCU03905 encodes an Ap1-like protein (NcAp-1), which is involved in resistance to multiple stress responses, including oxidative and heavy metal stress. Relocalization of NcAp-1 from the cytoplasm to the nucleus was associated with exposure to stress. A comparison of the NcAp-1 regulon with Ap1-like regulons in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans and Aspergillus fumigatus showed both conservation and divergence. These data indicate how N. crassa responds to stress and provide information on pathway evolution.
INTRODUCTION
The fungal kingdom is an excellent model for the study of transcription factor (TF) diversity and evolution, primarily due to the functional characterization and identification of direct target genes of TFs in the yeast Saccharomyces cerevisiae. The duplication of TFs (paralogues) and subsequent regulatory network divergence are major forces in the evolution of phenotypic complexity. In S. cerevisiae, the basic leucine zipper (bZIP) family of TFs is an excellent example of gene duplication and divergence of function. In S. cerevisiae, the bZIP TF family contains 14 genes and can be divided into three groups. The largest is the YAP1 group, which includes eight members. Five YAP1 family members (YAPs 1, 2, 4, 5 and 6) have been implicated in the oxidative stress and DNA-damage responses (Tan et al., 2008; Workman et al., 2006). The differences in gene regulation among the Yap members can be attributed to two important mechanisms: sequence differences in the DNA-binding motifs that are targeted by the different Yap proteins (Cohen et al., 2002), and variations in the regulation of the Yap proteins themselves, including post-translational modifications such as phosphorylation and disulfide bond formation (Kuge et al., 2001). The integration of expression profiles and chromatin-binding studies of the Yap proteins in response to DNA damage have shown a significant overlap of Yap regulons, especially between Yap1 and Yap2 (Tan et al., 2008). The second largest group of bZIP members in S. cerevisiae includes SKO1, ACA1 and CST6 (ACA2). Strains containing a mutation in SKO1 are affected in osmotic and oxidative stress, while mutations in ACA1 and CST6 result in strains that are affected in the utilization of alternative carbon sources (Rep et al., 2001). The third group of bZIP TFs in S. cerevisiae contains a single gene encoding Gcn4, which regulates genes in response to amino acid starvation (Hinnebusch, 2005).
Genome sequences of about 50 additional fungal genomes are currently available, and a number of these organisms, such as Aspergillus nidulans, Magnaporthe grisea and Neurospora crassa, have genetic tools available to assess the functions of predicted TF genes. In the filamentous fungus N. crassa, there are about 200 predicted general TFs (with and without DNA-binding domains) (Borkovich et al., 2004; Galagan et al., 2003; Shiu et al., 2005). The predicted DNA-binding TFs in N. crassa mostly fall into six families, i.e. the basic helix–loop–helix (bHLH), bZIP, C2H2 zinc finger, GATA factor, Myb and Zn binuclear cluster families. Recently, whole-genome microarrays for N. crassa have been validated and have provided expression data associated with asexual spore germination and vegetative growth (Kasuga et al., 2005; Kasuga & Glass, 2008). From a National Institutes of Health (NIH)-funded Neurospora program project, a large number of gene deletion strains have been constructed, including a viable deletion set of 103 predicted TF genes (Colot et al., 2006; Dunlap et al., 2007). The availability of such tools makes it possible to dissect TF networks and to compare regulatory networks that have evolved among paralogous TFs (homologues within one species) and between orthologous TFs (homologues between different species). Previously, we showed that the regulons of the bZIP TF Gcn4 of S. cerevisiae, which is involved in regulating genes in response to amino acid starvation (Hinnebusch, 2005), and its orthologue in N. crassa, CPC-1 (Ebbole et al., 1991), show conservation in the functional categories of genes that respond to amino acid starvation and which are dependent on functional Gcn4 and CPC-1 (Tian et al., 2007). Although the overall cellular response to amino acid starvation is similar between S. cerevisiae and N. crassa, there is substantial divergence in the direct target genes of Gcn4 in S. cerevisiae versus the predicted CPC-1 target genes in N. crassa.
In addition to cpc-1, there are eight genes in N. crassa with predicted bZIP DNA-binding domains, including the sulfur metabolism regulator cys-3 (Fu & Marzluf, 1990). While this study was in progress, an additional bZIP family member, NCU03905 (nap-1; here named NcAp-1, see Results), was shown by Takahashi et al. (2010) to be involved in menadione resistance and was postulated to be a YAP1 orthologue. The functions of the remaining members of this family are currently unknown. In an effort to understand the bZIP regulatory network in N. crassa, we determined the transcriptional profiles of seven viable bZIP mutants during vegetative growth. This study revealed a genome-wide map of transcriptional regulation via members of the bZIP family in N. crassa and showed both cooperativity and specificity among the bZIP TF family in a filamentous ascomycete species. We subsequently assessed the sensitivity of the bZIP mutants to a variety of environmental stresses and also identified NCU03905 (Neurospora crassa AP 1-like TF, NcAp-1) as the functional orthologue of S. cerevisiae YAP1. By performing transcriptional profiling experiments of both the wild-type (WT) and a ΔNcAp-1 mutant in response to oxidative stress, we identified the NcAp-1 regulon. Functional and transcriptional profiling comparisons of the S. cerevisiae Yap1, Schizosaccharomyces pombe Pap1, C. albicans Cap1 and N. crassa NcAP1 regulatory networks in response to oxidative stress (this study) and in response to exposure to menadione (Takahashi et al., 2010) revealed both conservation and divergence of TF regulons.
METHODS
Strains and culture conditions.
Strains containing deletion mutations in the predicted bZIP TF genes included FGSC 11121 (ΔNCU00499), FGSC 11343 (ΔNCU01994), FGSC 11131 (ΔNCU03905), FGSC 11133 (ΔNCU04211) and FGSC 11269 (ΔNCU08055) (Colot et al., 2006), and were kindly provided by Dr K. Borkovich (University of California, Riverside), while the cpc-1 mutant (FGSC 4264), cys-3 mutant (FGSC 4028) and the WT sequenced strain (FGSC 2489) were obtained from the Fungal Genetics Stock Center (FGSC) (McCluskey, 2003). Strains were inoculated onto slants containing Vogel's minimal medium (VMM) (Vogel, 1956) and grown at 30 °C for 2 days, followed by incubation at 25 °C under constant light for 7 days. Conidia were harvested with water and inoculated as a horizontal strip onto 150 ml Bird's minimal medium (BMM) (Metzenberg, 2004) on a large (25 cm×25 cm) Petri plate layered with a sheet of cellophane. Conidia were germinated under constant light at 25 °C for 24 h. A 4 mm wide mycelial section was subsequently inoculated as a horizontal strip onto fresh solid BMM. Twenty-four hours post-inoculation, a 1 cm wide section of mycelia at the periphery of the colony, which corresponded to hyphae 1–3 h old, and a 1 cm internal section of the colony (corresponding to hyphae 12–15 h old) were harvested, immediately plunged into liquid nitrogen and stored at −80 °C for RNA isolation.
For transcriptional profiling of the WT and the ΔNcAp-1 strain in response to hydrogen peroxide (H2O2), strains were treated as above, with the exception that large (25 cm×25 cm) Petri plates containing a solid physical barrier in the middle of the plate were used. One half of the plate contained BMM, while the other half contained BMM supplemented with 6 mm H2O2 (added when the medium was at about 40 °C). A preliminary comparison of expression data for the WT exposed to 2 and 6 mM H2O2 versus minimal medium revealed that there were no significant differences in expression when N. crassa was exposed to 2 mM H2O2, but significant differences were detected upon exposure to 6 mM H2O2. A horizontal strip containing hyphae was inoculated on the BMM, 3 cm from the barrier. After 12 h, the periphery of the colony reached the barrier and grew into the BMM+H2O2. After 6 h of growth, an ∼1 cm strip of the colony periphery was isolated from the BMM+H2O2 plate, plunged into liquid nitrogen and stored at −80 °C until use.
For transcriptional profiling of the WT and ΔNcAp-1 mutant in response to cadmium chloride (CdCl2) treatment, conidia were harvested and inoculated in 100 ml liquid minimal medium for 24 h at 30 °C with shaking (225 r.p.m.). CdCl2 was added to the cultures to a final concentration of 10 mM. After 30 min treatment, the mycelium-containing cultures were spun down, filtered through Whatman No. 1 filter paper and frozen immediately in liquid nitrogen.
RNA isolation and cDNA labelling.
Total RNA from frozen samples was isolated using zirconia/silica beads (0.2 g, 0.5 mm diameter, Biospec) and a Mini-BeadBeater (Biospec) with 1 ml TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. Total RNA (100 μg) was further purified using an RNeasy kit (Qiagen). RNA integrity was checked with a Nanodrop spectrophotometer and by agarose gel electrophoresis. cDNA synthesis and labelling followed the protocol of the ChipShot Indirect Labeling kit from Promega (catalogue no. Z4000). The dyes Cy3 and Cy5 (Amersham, catalogue no. RPN5661) were incorporated into cDNA by adding Cy3 or Cy5 monofunctional N-hydroxysuccinimide ester dye to the cDNA solution for 1 h at 22 °C. The cDNA was subsequently cleaned by using a ChipShot membrane column and dried under vacuum.
Microarray hybridization and data analysis.
The methods for microarray hybridization and data analysis were as described in Tian et al. (2007). Briefly, 70-mer oligonucleotides representing all predicted N. crassa genes and additional intergenic regions were printed onto GAP slides (Corning, catalogue no. 40005). We used a closed loop for experimental design that increases statistical power (Townsend & Taylor, 2005). Each RNA sample was used in at least four individual hybridizations. Images from the Axon 4000B microarray scanner were analysed by Genepix 6.0 software; statistical support for differential expression levels was obtained by bagel analyses (Townsend & Hartl, 2002). cis-Element analysis was performed as previously described (Tian et al., 2007). The 1000 bp region upstream of the predicted translational start site of nine common targets between H2O2 and CdCl2 conditions was retrieved and evaluated by MDscan (Liu et al., 2002), BioProspector (Liu et al., 2001) and meme (Bailey & Elkan, 1994) for motif prediction.
Quantitative real-time RT-PCR (qRT-PCR).
qRT-PCR was performed on a 7300 Real-Time PCR system (Applied Biosystems) using a Qiagen SYBR kit (catalogue no. 204143). Amplification of mRNA from the actin gene was used as an internal control for normalization. Methods were as described previously (Dementhon et al., 2006).
NcAp-1–GFP plasmid construction and Neurospora transformation.
The ORF of NCU03905 was inserted into the pMF272 vector, which includes a GFP cassette (Freitag et al., 2004). For the construct using the NcAp-1 native promoter to drive expression, the NcAp-1 upstream sequence and ORF were amplified using primers containing NotI and PacI sites (forward primer 5′-CTG CGG CCG CCC TTC CCT TCT CAG ATC TGA ATC G-3′, reverse primer 5′-CTT TAA TTA ACA GTG TGG CAG CTG CTT TTC G-3′). For the construct using the ccg1 promoter, the NcAP-1 gene was amplified using primers containing SpeI and PacI sites (forward primer 5′-CTA CTA GTA TGA CGT CGA CTC AGA ACC CTC-3′, reverse primer 5′-CTT TAA TTA ACA GTG TGG CAG CTG CTT TTC G-3′). The NcAp-1–gfp constructs were transformed to a ΔNCU03905 strain containing a his-3 marker (R21-07) using Neurospora transformation methods (Margolin et al., 1997). Verification of full-length NcAp-1–gfp insertion at the his-3 locus was confirmed by PCR. To assess localization of NcAp-1–GFP upon oxidative stress, 16 h-old hyphae from the ΔNCU03905 (NcAp-1–gfp) strains (CT-C17 and CT-N9) were treated with 30 mm H2O2 and 10 mM CdCl2. After 30 min of exposure, NcAp-1–GFP localization was assessed via fluorescence microscopy using a Zeiss Axioskop II microscope and GFP fluorescence filter. Photographs of hyphae were taken using a Zeiss Axioskop II microscope with a Hamamatsu digital charge-coupled device (CCD) camera, and further processed with Adobe Photoshop 7.0.
Stress screening of conidia.
Fresh conidia were harvested from 7 day-old slants and suspended in 500 μl H2O to a final concentration of 106 μl−1. Seven microlitres of a conidial dilution series (105, 104, 103, 102 and 101 μl−1) was spotted onto a BDES plate (Davis & De Serres, 1970) or BDES plates plus one of the following compounds: H2O2, 1 or 2 mM; CdCl2, 50 or 100 μM; sorbitol, 0.8 M; β-mercaptoethanol (β-ME), 1 or 4 mM; 3-aminotriazole (3-AT), 1 or 2 mM; NaCl, 0.8 mM. Plates were incubated at 25 °C for 2 days and then growth was evaluated.
Identification of orthologues and regulogs in N. crassa and other fungi.
The sequences of predicted ORFs in the N. crassa genome were downloaded from Broad Institute version 7 (http://www.broad.mit.edu/annotation/genome/neurospora/Downloads.html). Neurospora discreta and Neurospora tetrasperma genomic data were obtained from http://genome.jgi-psf.org/Neudi1/Neudi1.download.html and http://genome.jgi-psf.org/Neute1/Neute1.download.html, respectively. S. cerevisiae ORFs were downloaded from the SGD database (http://downloads.yeastgenome.org/sequence/genomic_sequence/orf_dna/). Aspergillus fumigatus genome data were downloaded from TIGR (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/a_fumigatus/annotation_dbs/). Candida albicans genome data were downloaded from http://www.candidagenome.org/DownloadContents.shtml. Sch. pombe ORFs were downloaded from http://www.sanger.ac.uk/Projects/S_pombe/protein_download.shtml.
A. nidulans and M. grisea ORF protein sequences were downloaded from The Broad Institute (http://www.broad.mit.edu/annotation/fgi/). Yarrowia lipolytica ORF protein sequences were downloaded from http://www.genolevures.org/download.html. Orthologous genes were identified as best bidirectional hits using blastp, with a cut-off value of 1e-6. The bZIP member prediction was performed by hidden Markov model (HMM) searches; the bZIP TF HMM model (PF00170.10) was downloaded from Pfam (http://pfam.sanger.ac.uk/). The sequence was defined as a DNA-binding motif if the HMM search P value was <1×104.
Transcriptional profiling data for S. cerevisiae in response to oxidative stress were obtained from Gasch et al. (2000), and the chromatin-immunoprecipitation (ChIP) data were downloaded from Harbison et al. (2004). C. albicans oxidant stress response data were downloaded from Wang et al. (2006). The Cap1 ChIP data were downloaded from Znaidi et al. (2009). Sch. pombe oxidant stress profiling data were downloaded from Chen et al. (2008). The A. fumigatus proteomics data for the H2O2 stress response were downloaded from Lessing et al. (2007). The N. crassa profiling data on menadione were downloaded from Takahashi et al. (2010).
RESULTS
TF gene expression in N. crassa during vegetative growth
In N. crassa, there are about 200 predicted general TFs (with and without a DNA-binding domain) (Borkovich et al., 2004; Galagan et al., 2003; Shiu et al., 2005). The class of TFs with predicted DNA-binding domains comprises six families, i.e. the bHLH, bZIP, C2H2 zinc finger, Myb, GATA factor and Zn (II)2 binuclear cluster groups, plus a group of miscellaneous TFs. We used an HMM to revise this list based on the sequence of the DNA-binding domain (see Methods). We identified 178 TFs (Supplementary Table S1, worksheet p1) with a clear DNA-binding motif; 166 of these belonged to one of the six defined gene families listed above. The remaining 12 genes encoded TFs classified as miscellaneous (various DNA-binding domains). These 178 TFs can be thought of as the core set for N. crassa, although additional TF genes may be present that contain a non-typical DNA-binding motif(s).
Since TFs play a critical role in the regulation of gene expression patterns, we evaluated the expression pattern of the 178 predicted TFs during vegetative growth of N. crassa at two developmental time points. We evaluated the expression of predicted TFs using full-genome Neurospora microarrays (Kasuga et al., 2005; Kasuga & Glass, 2008; Tian et al., 2007) from hyphae at the periphery of a colony (a section of the colony that is 0–3 h old) and a second section from the interior of a 12–15 h-old colony. Different sections of a colony of N. crassa have been shown to vary in expression profile (Kasuga & Glass, 2008), and in addition the hyphae in these sections show morphological and developmental differences. For example, hyphae at the periphery exhibit foraging behaviour, while hyphae in the interior of a colony form asexual reproductive structures, including aerial hyphae and conidia. Expression patterns for 98 (55 %) of the predicted TFs were detected. With a minimum difference of 1.5-fold, 35 TFs showed a significantly different expression pattern between the periphery and the interior of a colony (Table 1). Most of these TFs (33/35) showed an increase in relative expression level in the colony periphery, while only two increased in expression level in the interior of a colony [NCU01459 and NCU08726; NCU08726 encodes fl (fluffy), a TF involved in asexual spore development] (Bailey & Ebbole, 1998). Strains containing mutations in 15 of the 35 predicted TFs showed phenotypes different from a WT strain (Table 1), including nine with vegetative growth defects [Δpp-1 (NCU00340), ΔNCU02671, ΔNCU06503, Δcre-1 (NCU08807), Δada-6 (NCU04866), Δcol-26 (NCU07788), Δvad-3 (NCU06407), Δkal-1 (NCU03593) and the bZIP TF gene, Δada-1 (NCU00499)] (Colot et al., 2006).
Table 1.
Genes encoding TFs with significant differences in expression between the periphery and the interior of a colony
| TF | Family | Tip/interior* | Growth (mm day−1)† | Annotation |
|---|---|---|---|---|
| NCU00749 | BHLH | 2.0 | 60–65 | Predicted protein |
| NCU04731 | BHLH | 1.9 | 65–70 | sah-2, short aerial hyphae-2 |
| NCU02724 | BHLH | 1.6 | 70–75 | Helix–loop–helix DNA-binding domain |
| NCU00499 | bZIP | 1.6 | 25–30 | ada-1, all development altered-1 |
| NCU01459 | bZIP | 0.6 | asl-2, ascospore lethal-2 | |
| NCU00090 | C2H2 | 6.0 | 60–65 | Related to PacC protein |
| NCU00340 | C2H2 | 3.5 | 20–30 | pp-1, Ste12-like TF |
| NCU00694 | C2H2 | 3.0 | 70–75 | Putative protein |
| NCU02671 | C2H2 | 2.4 | 20–25 | Cutinase G-box binding protein |
| NCU03043 | C2H2 | 2.0 | 60–65 | Related to Krüppel protein |
| NCU03421 | C2H2 | 7.7 | 60–65 | Conserved hypothetical protein |
| NCU03699 | C2H2 | 2.4 | 70–75 | Conserved hypothetical protein |
| NCU06487 | C2H2 | 2.1 | 75–80 | Predicted protein |
| NCU06907 | C2H2 | 1.8 | 70–75 | Conserved hypothetical protein |
| NCU06503 | C2H2 | 1.8 | 55–60 | Related to DNA-binding protein |
| NCU08807 | C2H2 | 1.7 | cre-1, carbon repressor protein 1 | |
| NCU09252 | C2H2 | 1.5 | 75–80 | Conserved hypothetical protein |
| NCU01154 | GATA | 2.3 | sub1, submerged protoperithecia-1 | |
| NCU09068 | GATA | 2.3 | 60–65 | Conserved hypothetical protein |
| NCU05685 | Myb | 1.6 | Conserved hypothetical protein | |
| NCU00155 | Zn2Cys6 | 2.0 | 70–75 | Transcriptional activator Mut3p |
| NCU00289 | Zn2Cys6 | 2.5 | 65–70 | tah-1, tall aerial hyphae-1 |
| NCU01097 | Zn2Cys6 | 3.6 | 70–75 | Related to NITA protein |
| NCU04866 | Zn2Cys6 | 2.0 | 50–55 | Related to FacB DNA-binding protein |
| NCU06799 | Zn2Cys6 | 3.0 | 60–65 | Hypothetical protein |
| NCU07139 | Zn2Cys6 | 5.0 | 80–85 | Conserved hypothetical protein |
| NCU07788 | Zn2Cys6 | 2.7 | 55–60 | col-26, colonial-26 |
| NCU08063 | Zn2Cys6 | 2.1 | 65–70 | Putative protein |
| NCU08307 | Zn2Cys6 | 3.0 | Hypothetical protein | |
| NCU02576 | Zn2Cys6 | 1.9 | Related to cutinase TF | |
| NCU07392 | Zn2Cys6 | 1.5 | 60–65 | adv-1, all development altered |
| NCU06407 | Zn2Cys6 | 1.5 | 55–60 | vad-3, vegetative asexual development-3 |
| NCU02307 | Zn2Cys6 | 1.5 | 70–75 | Conserved hypothetical protein |
| NCU08726 | Zn2Cys6 | 0.2 | 20–25 | fl, conidial development protein fluffy |
| NCU03593 | 1.7 | 10–15 | Probable homeoprotein |
*Significance cut-off (bagel) of at least 1.5-fold.
†Growth data from the Broad Institute (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
Phylogenetic analysis of N. crassa bZIP family members
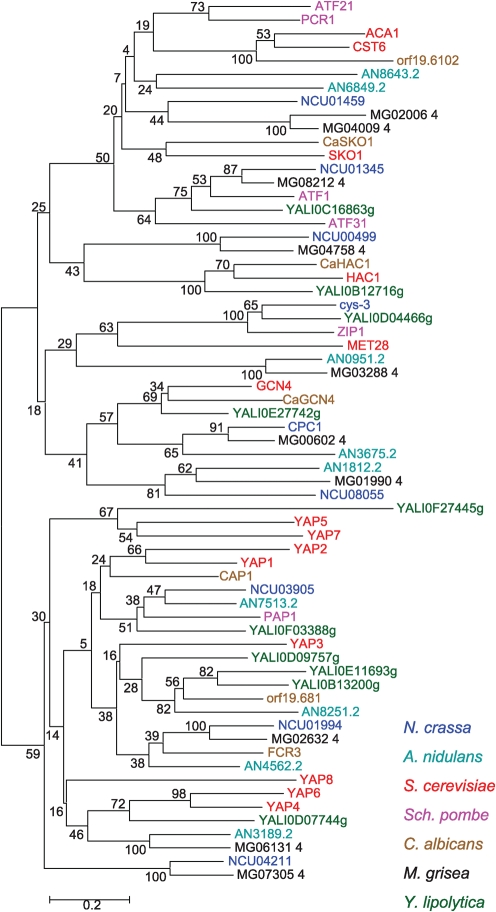
In S. cerevisiae, the bZIP TF family has been well studied and consists of 14 members. The YAP family of bZIP TFs, of which there are eight members, are thought to be a result of at least six duplication events from a common ancestor (GCN4) (Kellis et al., 2004), and can be divided into four subfamilies: YAP1 and YAP2, YAP4 and YAP6, YAP5 and YAP7, and YAP3 and YAP8 (Tan et al., 2008) (Fig. 1). We performed phylogenetic analysis of predicted bZIP domain proteins from the hemiascomycete species S. cerevisiae (14 members) and C. albicans (seven members), a species at the base of the hemiascomycete clade, Yarrowia lipolytica (10 members), the archiascomycete species Sch. pombe (six members), and the euascomycete species A. nidulans (nine members), M. grisea (10 members) and N. crassa (nine members). The phylogenetic tree of the bZIP domains showed divergence into two separate clades (Fig. 1). The lower clade includes all known members of the YAP family (YAP clade), while the upper clade includes all other bZIP domain proteins, including S. cerevisiae GCN4, SKO1, HAC1, MET28, ACA1 and CST6 (Fig. 1) (GCN4 clade). In N. crassa, the GCN4 clade bZIP TFs include cpc-1 (NCU04050), cys-3 (NCU03536), ada-1 (NCU00499), asl-1 (NCU01345) and NCU01459. While the YAP family clade includes eight members from S. cerevisiae, Sch. pombe contains only one homologue, pap1 (Fig. 1). The four YAP-like TFs in A. nidulans and the three YAP-like TFs in M. grisea and N. crassa clustered within the YAP bZIP clade, although none of the predicted euascomycete YAP-like proteins clustered closely with the S. cerevisiae Yap proteins.
Fig. 1.
Phylogenetic analysis of the bZIP TF gene family in ascomycete species. The bZIP member prediction was performed by HMM searches using the bZIP domain matrix downloaded from Pfam (PF00170.10). The tree was generated by the mega2 program using neighbour joining with bootstrap=100.
Although phylogenetic analysis clearly showed that gene duplication of bZIP genes occurred in S. cerevisiae (ACA1 and CST6, and YAP members) and Sch. pombe (ATF21 and PCR1), within the euascomycete species only a single recent gene duplication event was detected in M. grisea (MG02006.4 and MG04009.4), which clustered with a bZIP protein of unknown function (NCU01459) in N. crassa. Viable homokaryotic deletion mutations in NCU01459 are not recoverable (Colot et al., 2006). A more ancient duplication within the GCN4 clade was identified in A. nidulans (AN8643.2 and AN6849.2); these bZIP domains do not cluster closely with any other bZIP proteins. No recent duplication of any bZIP domain gene was detected in N. crassa, consistent with the observation that gene duplications are not tolerated (Galagan & Selker, 2004). Thus, the major expansion of the bZIP gene family in fungi occurred before the divergence of euascomycete species from Sch. pombe and S. cerevisiae, approximately 300 million years ago.
Transcriptional profiling of bZIP mutants
In N. crassa, two bZIP genes have been well characterized. The first encodes an orthologue of GCN4, called cpc-1, which is involved in regulating genes in response to amino acid starvation (Ebbole et al., 1991; Hinnebusch, 2005; Tian et al., 2007). The second bZIP gene encodes a transcriptional regulator, CYS-3, which regulates genes in response to sulfur sources (Paietta, 1992). While this study was in progress, a third member, NCU03905 (nap-1), was shown to be important for resistance to treatment with menadione and proposed to be a YAP1 orthologue (Takahashi et al., 2010). Here we refer to NCU03905 as NcAp-1, as nap refers to a neutral amino acid permease locus in N. crassa (Perkins et al., 2001; Rao & DeBusk, 1973). Homokaryotic mutant strains are available for seven of nine predicted bZIP genes [Δada-1 (NCU00499), ΔNCU01994, ΔNcAp-1 (NCU03905), ΔNCU04211, cpc-1 (NCU04050), ΔNCU08055 and cys-3 (NCU03536)]. The Δada-1 mutant (all development altered; NCU00499), showed a reduced growth rate (2.5 cm day−1 compared with 7.5 cm day−1 for the WT) and very short aerial hyphae. Strains containing mutations in the other six bZIP TF genes (ΔNCU01994, ΔNcAp-1, ΔNCU04211, cpc-1, ΔNCU08055 and cys-3) showed growth and asexual reproduction phenotypes indistinguishable from those of the WT strain. Strains containing mutations in two remaining bZIP genes, asl-1 (NCU01345) and asl-2 (NCU01459), showed an ascospore lethality phenotype; homokaryotic strains containing mutations in these genes are not available (Colot et al., 2006).
To identify putative target genes of the bZIP TF mutants, we compared transcriptional profiles of the seven bZIP mutants from hyphae at the colony periphery (0–3 h) with those from the interior of a colony (12–15 h). In the WT, Functional Category (FunCat) analysis (Ruepp et al., 2004) of 509 genes that had higher expression levels in the colony periphery showed an enrichment for genes involved in cell type differentiation, cell wall proteins, carbon compound and polysaccharide metabolism, cell growth/morphogenesis (regulation of directional cell growth), cellular export and secretion, and cell signalling (P<0.001; Supplementary Table S2, worksheet p1). FunCat analysis of the 363 genes that had an increased expression level in the interior of the colony showed an enrichment for genes involved in stress response and detoxification, energy-related genes (glycolysis and pentose phosphate pathway), and amino acid and carbon metabolism (P<0.001; Supplementary Table S2, worksheet p4). In the colony periphery, the number of genes in the seven bZIP mutants that showed altered expression profiles compared with the WT ranged from 21 to 509 (Table 2 and Supplementary Table S3), including genes that increased in relative expression level (from three in ΔNcAp-1 to 290 in Δada-1) and genes that showed a decrease in relative expression level (from five in Δcys-3 to 219 in Δada-1) (Table 2). Expression patterns of genes in the interior of a colony in the bZIP TF deletion strains also showed differences from the WT (Table 2 and Supplementary Table S4), ranging from three affected genes in ΔNcAp-1 to 139 in Δcys-3. The strain carrying a deletion of ada-1 showed the greatest number of expression differences from the WT, consistent with its growth defect.
Table 2.
Summary of expression profiling data for seven bZIP mutants
| TF mutant | Genes downregulated in periphery | Genes upregulated in periphery | Genes downregulated in interior | Genes upregulated in interior |
|---|---|---|---|---|
| ΔNCU00499 | 219 | 290 | 80 | 56 |
| ΔNCU01994 | 12 | 9 | 49 | 77 |
| ΔNCU03905 | 47 | 3 | 6 | 3 |
| ΔNCU04211 | 45 | 5 | 65 | 28 |
| cpc-1 | 22 | 19 | 5 | 52 |
| ΔNCU08055 | 32 | 6 | 50 | 107 |
| cys-3 | 5 | 56 | 24 | 139 |
FunCat analysis of affected genes in the Δada-1 mutant that showed reduced expression in the colony periphery showed enrichment for carbon compound and carbohydrate metabolism (P=2.3×10−7). Genes involved in carbon compound and carbohydrate metabolism also showed enrichment in other bZIP mutants, including ΔNCU01994, ΔNCU04211 and ΔNCU08055 (P=0.001 to 0.01).
Similar to data from the colony periphery, affected genes displaying decreased expression in the colony interior in the Δada-1 mutant showed an enrichment for carbon compound and carbohydrate metabolism, and the pentose phosphate pathway (P=1.03×10−6). In the ΔNCU01994 and cys-3 strains, genes involved in amino acid metabolism, carbon compound and carbohydrate metabolism, lipid, fatty acid and isoprenoid metabolism, and metabolism of vitamins, cofactors and prosthetic groups, were enriched (P=0.002 to 6.3×10−9), while NAD/NADP binding was enriched in the ΔNCU04211 and ΔNCU08055 strains. Enriched categories of genes that showed an increased expression level in the colony interior included transport facility and cell defence genes in Δada-1, polysaccharide metabolism and cell wall genes in ΔNCU01994 and ΔNCU08055, and virulence and disease factor genes in ΔNCU04211.
Consistent with their phylogenetic relationships, strains containing mutations in bZIP TF genes that were more closely related showed an overlap in affected gene sets, with an overlap of up to 24 % in strains containing mutations in the closely related paralogues NCU08055 and cys-3 (Supplementary Tables S3 and S4). By contrast, only a 1 % overlap in expression profiles was identified between strains containing deletions of distant bZIP paralogues, for example, NcAp-1 (NCU03905) and ada-1 (NCU00499). These data suggest that the regulation of some genes and processes has been conserved in the bZIP TF regulons following duplication and divergence. Similarly, in S. cerevisiae, the number of genes co-regulated by YAP family members ranges between 9 and 15 %, and increases to 25 % upon exposure to DNA-damaging agents (Tan et al., 2008).
NcAp-1 functional characterization and localization
To elucidate the molecular function of the bZIP TFs in N. crassa, we screened the bZIP TF mutants for sensitivity to a variety of chemical stresses, including exposure to sorbitol, NaCl, 3-AT, cadmium, β-ME and H2O2. The ΔNcAp-1 mutant was sensitive to all the stresses tested except for β-ME (Supplementary Fig. S1). However, only slight sensitivity to H2O2, and no sensitivity to NaCl has been observed in an independently derived NCU03905 (nap-1) mutant during hyphal growth (Takahashi et al., 2010). The Δcys-3 mutant showed a mild sensitivity to sorbose (Supplementary Fig. S1). The other bZIP TF deletion mutants did not show any obvious sensitivity under any of the conditions tested.
The predicted NcAp-1 protein (NCU03905) has a PAP1 domain (Fig. 2c), which is an important H2O2 response domain in Sch. pombe (Chen et al., 2008; Cohen et al., 2002; Vivancos et al., 2006). In N. crassa, the closest paralogue to NcAp-1 is NCU01994, although the predicted protein product of NCU01994 lacks a PAP1 domain (Fig. 2c). However, a strain containing both ΔNcAp-1 and ΔNCU01994 deletions resembles the ΔNcAp-1 mutant with respect to sensitivity to stress (Supplementary Fig. S1).
Fig. 2.
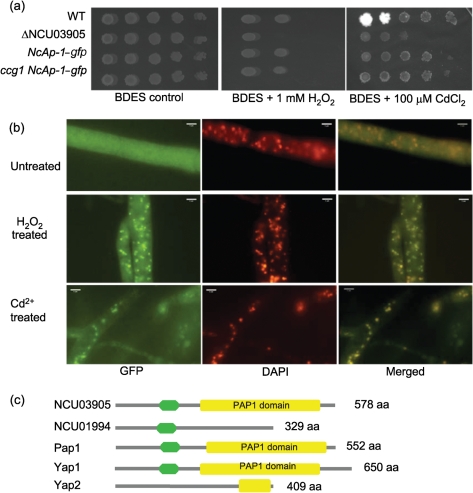
Complementation of ΔNcAp-1 and localization of NcAp-1–GFP. (a) Comparison of the phenotype of the WT (FGSC 2489), the NcAp-1 mutant (ΔNCU03905) and ΔNcAp-1 complemented strains (NcAp-1–gfp and ccg1 NcAp-1–gfp) exposed to 1 mM H2O2 or 100 μM Cd2+ using a dilution series of conidial suspensions (from left to right: 105, 104, 103, 102, 101). Plates were incubated at 25 °C for 2 days and then evaluated for growth. (b) NcAp-1–GFP localization in a ΔNcAp-1 (his-3 : : NcAp-1–gfp) strain grown on minimal medium (untreated, upper panels), after treatment with 30 mM H2O2 (H2O2 treated, centre panels) or 10 mM CdCl2 (Cd2+ treated, lower panels). The left-hand panels show GFP fluorescence (NCAp-1–GFP), while the centre panels show nuclear 4,6-diamidino-2-phenylindole (DAPI) staining. The right-hand panels show merged GFP and DAPI images. (c) Domain prediction of Yap1, Yap2, Pap1, NCU03905 (NcAp-1) and its closest paralogue NCU01994. Green, bZIP motif; yellow, PAP1 domain.
The redistribution of S. cerevisiae Yap1 from the cytoplasm to the nucleus in response to oxidative stress is a major regulatory mechanism (Kuge et al., 1997), and also occurs in Sch. pombe (PAP1) (Vivancos et al., 2004), C. albicans (CAP1) (Zhang et al., 2000) and A. fumigatus (AfYap1) (Lessing et al., 2007). In N. crassa, NcAp-1–GFP in a WT strain shows redistribution from the cytoplasm to the nucleus upon exposure to menadione (Takahashi et al., 2010). To observe the behaviour of NcAp-1 during oxidative and heavy metal stress in N. crassa, we tagged NcAp-1 with GFP at the C terminus. The introduction of the NcAp-1–gfp construct into the ΔNcAp-1 strain restored cadmium and H2O2 resistance, indicating that the construct encodes a functional NcAp-1 protein (Fig. 2a). Under conditions of no (or low) stress (minimal medium), NcAp-1 localized to the cytoplasm (Fig. 2b). However, when the ΔNcAp-1 (NcAp-1–gfp) strain was exposed to stress (30 mM H2O2 for 5–30 min or 10 mM CdCl2 for 5–10 min), NcAp-1–GFP accumulated in nuclei across the fungal colony (Fig. 2b). These data indicate that the redistribution of YAP-1-like proteins to the nucleus upon exposure to oxidants or heavy metals is a highly conserved mechanism of transcriptional regulation in ascomycete fungi in response to a variety of stresses.
Identification of the NcAp-1 regulon under oxidative stress
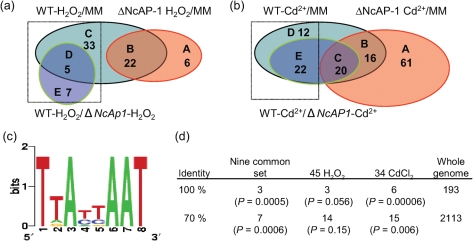
To determine the NcAp-1 stress regulon, we performed transcriptional profiling of the WT and the ΔNcAp-1 mutant grown in minimal medium with and without oxidative stress imposed by treatment with 6 mM H2O2 (see Methods). There were 45 genes induced by H2O2 (Fig. 3a, sets C, D and E) that were dependent on NcAp-1 for transcription. FunCat analysis (Ruepp et al., 2004) of these 45 putative target genes showed that stress responses, oxygen detoxification and metabolism of peptide-derived compounds (such as thioredoxin) were enriched (Table 3). Of the 45 genes (Supplementary Table S5, worksheet P1), 11 have an annotated function in relation to the stress response, especially the oxidant stress response pathway, including two glutathione S-transferases, gst-1 (NCU05780) and gst-2 (NCU04109); four NADP(H)-dependent dehydrogenase/flavin oxidoreductases, mig-2 [menadione-induced gene (Takahashi et al., 2010)] (NCU07452), mig-3 (NCU04452), mig-6 (NCU09285) and NCU08402; an oxidase (NCU05164); catalase-2 (NCU05770); a probable reductase (mig-4, NCU09040); a glutathione peroxidase (NCU09534); and a predicted ABC transporter (NCU03776) (Table 4). We confirmed the dependence of expression on NcAp-1 for four genes, cat-2 (NCU05770), gpx3 (NCU09534), NADH oxidase (mig-3, NCU04452) and glutathione transferase-1 (gst-1, NCU05780) by qRT-PCR (Supplementary Fig. S2). Five genes from this 11-gene oxidant stress set have been shown to be NcAp-1-dependent upon exposure to menadione (Takahashi et al., 2010), including two predicted glutathione S-transferases [gst-1 (NCU05780) and gst-2 (NCU04109)] and three NADP(H)-dependent dehydrogenase/flavin oxidoreductases [mig-2 (NCU07452), mig-3 (NCU04452) and mig-6 (NCU09285)]. These data indicate that the glutathione S-transferase and NADPH dehydrogenases are important for resistance to multiple types of oxidant stress in N. crassa.
Fig. 3.
The NcAp-1 regulon. (a) Venn diagram showing the overlap among the genes that showed statistically significantly increased expression levels in the WT (FGSC 2489) as compared with the ΔNcAp-1 mutant when exposed to H2O2. Genes within C, D and E represent genes that were induced by exposure to H2O2 and were NcAp-1-dependent. (b) Venn diagram showing the overlap among the genes that showed statistically significantly increased expression levels in the WT (FGSC 2489) as compared with the ΔNcAp-1 mutant when exposed to CdCl2. Genes within D and E represent genes that were induced by exposure to CdCl2 and were NcAp-1-dependent. (c) WebLogo of the cis-element that showed enrichment in the putative direct target genes of NcAp-1 (see Methods) (http://weblogo.berkeley.edu/logo.cgi). (d) Enrichment analysis of predicted cis-elements in the NcAp-1 target gene set. Using the predicted cis-element as an input matrix, the 1000 bp upstream of the predicted target genes was evaluated by paster (Hertz & Stormo, 1999). The perfect match and 70 % match to the cis-element are shown; statistical analysis (P values) was conducted by Fisher's exact test.
Table 3.
FunCat analysis of NcAp-1 targets
Enriched groups with P <0.001 are shown; groups enriched under both Cd2+ and H2O2 conditions are in bold type.
| Functional category | Cd2+P value | H2O2P value |
|---|---|---|
| 01 METABOLISM | 3.99×10−6 | 0.075951 |
| 01.02 Nitrogen, sulfur and selenium metabolism | 1.13×10−5 | 0.029173 |
| 01.02.07 Regulation of nitrogen, sulfur and selenium metabolism | 7.41×10−5 | 0.005066 |
| 01.20 Secondary metabolism | 6.53×10−9 | 0.000718 |
| 01.20.37 Metabolism of peptide-derived compounds | 3.13×10−9 | 6.50×10−5 |
| 01.20.37.01 Metabolism of thioredoxin, glutaredoxin, glutathione | 1.20×10−7 | 0.001718 |
| 02 ENERGY | 3.35×10−5 | 0.085526 |
| 02.16 Fermentation | 1.27×10−6 | 0.025830 |
| 02.16.01 Alcohol fermentation | 7.41×10−5 | 0.005066 |
| 32 CELL RESCUE, DEFENCE AND VIRULENCE | 0.000144 | 0.004342 |
| 32.07 Detoxification | 0.000216 | 0.000716 |
| 32.07.07 Oxygen and radical detoxification | 6.01×10−8 | 1.00×10−5 |
| 32.07.07.03 Glutathione conjugation reaction | 2.53×10−8 | 9.80×10−6 |
Table 4.
NcAp-1 common targets under H2O2 and Cd2+ exposure
Bold type indicates genes that have been shown to be a direct target of the AP-1 protein.
| NcAp-1 target | Orthologue in S. cerevisiae | Orthologue in Sch. pombe | Orthologue in C. albicans | Orthologue in A. fumigatus | Annotation |
|---|---|---|---|---|---|
| NCU00847.2 | NO | NO | NO | Afu3g15010 | Methoxylase |
| NCU04109.2 | YNL229C | SPCC191.09c | orf19.2693 | Afu3g10830 | Glutathione S-transferase |
| NCU04452.2 | OYE3 | SPAC5H10.10 | orf19.3443 | Afu2g04060 | NADPH dehydrogenase chain OYE2 |
| NCU05164.2 | YKL071W | SPCC24B10.20 | orf19.6502 | Afu4g00910 | Protoporphyrinogen oxidase |
| NCU05780.2 | YNL229C | SPCC965.07c | NO | Afu4g14530 | Glutathione S-transferase |
| NCU07452.2 | OYE2 | SPBC23G7.10c | orf19.3131 | Afu5g01450 | Flavin oxidoreductase |
| NCU08402.2 | YCR105W | SPCC13B11.01 | orf19.5517 | Afu8g02430 | Alcohol dehydrogenase |
| NCU09285.2 | ADH5 | SPBC1773.06c | orf19.5113 | Afu4g08240 | Alcohol dehydrogenase |
| NCU09534.2 | Yir037w | SPBC32F12.03c | orf19.86 | Afu3g12270 | Peroxiredoxin |
Identification of the NcAp-1 regulon under cadmium stress
In addition to sensitivity to H2O2, the ΔNcAp-1 mutant was also sensitive to other stress conditions, including exposure to heavy metals such as cadmium (Supplementary Fig. S1). We therefore compared expression profiles of the WT and ΔNcAp-1 mutant upon exposure to CdCl2 versus H2O2 (see Methods and Supplementary Table S1). A total of 34 genes induced by exposure to Cd2+ were dependent on NcAp-1 for transcription (Fig. 3b, sets D and E, Supplementary Table S5, worksheet P2). Nine of these 34 genes overlapped with the 45 targets identified upon exposure to H2O2 and were dependent upon functional NcAp-1. These nine genes form the core targets of NcAp-1 (Table 4) and include the two glutathione S-transferase genes, gst-1 and gst-2 (NCU04109 and NCU05780, respectively), four NAD(P)H-dependent dehydrogenase/oxidoreductase genes, mig-2, mig-3, mig-6 and NCU08402, a predicted methoxylase (NCU00847), an oxidase (NCU05164) and a glutathione peroxidase (NCU09534). Six of these 34 genes are induced by menadione treatment (Takahashi et al., 2010) (Supplementary Table S5, worksheet P2), including a predicted glutathione S-transferase, gst-4 (NCU10521), plus five of the nine core genes identified above (gst-1, gst-2, mig-2, mig-3 and mig-6). The 24 Cd2+ condition-specific target genes included several stress/oxidant stress-related genes, including a predicted thioredoxin (NCU06556) and uricase (NCU07853). Interestingly, the bZIP TF cys-3 and nitrogen and sulfur metabolism genes were also enriched in this list (Supplementary Table S5, worksheet P2), suggesting that sulfur metabolism regulated by CYS-3 may cross-talk with the metal response pathway regulated by NcAp-1.
cis-Element analysis of NcAp-1 targets
As indicated by FunCat analysis, the NcAp-1 target gene set (45 under H2O2 and 34 under Cd2+) was enriched for genes involved in oxidative stress responses (Table 4). An earlier study evaluating promoter regions of genes that were induced upon treatment with menadione and which were NcAp-1-dependent failed to identify upstream regulatory elements (Takahashi et al., 2010). To identify potential NcAp-1 cis-regulatory elements in this study, we analysed 1000 bp upstream of the nine common target genes identified between the H2O2 and Cd2+ responses (see Methods). We identified the N. crassa cis-regulatory element TTA(t/c)(t/c)AAT, which was similar to cis-regulatory elements identified for Yap1 (TTACTAA), Cap1 (TTAc/gTAA), AfYap1 [TTA(G/C)TAA] and Pap1 (TTACGTAA), and which occurred in seven of the nine common target genes (P=0.0006), 14 of 45 H2O2-responsive target genes (P=0.15), and 15 of 34 cadmium-responsive target genes (P=0.006) (Fig. 3c, d). This cis-regulatory element was enriched in NcAp-1 putative target genes relative to its prevalence in the N. crassa genome. This set of genes is predicted to be directly regulated by NcAp-1. Of the nine common target genes, conservation of the NcAp-1 cis sequence was identified in the promoter regions of four orthologues in the related species N. discreta (NCU00847, NCU04452, NCU09285 and NCU05164) and in five orthologues in N. tetrasperma (NCU00847, NCU04452, NCU07452, NCU09285 and NCU09534).
A common response gene set between cpc-1 and NcAp-1
The bZIP TF CPC-1 regulates the response to amino acid starvation in N. crassa (Flint, 1985). Previously, we determined the cpc-1 regulon and the response of N. crassa to amino acid starvation (Tian et al., 2007). To determine whether a common set of genes is coordinately regulated under stress responses, we compared the cpc-1 gene set (443 genes) with the NcAp-1 H2O2/Cd2+ datasets (79 genes). There were 17 genes that were shared between the cpc-1 and NcAp-1 H2O2 or Cd2+ regulons, a significant enrichment (P=0.02), suggesting that crosstalk occurs between the H2O2, Cd2+ and 3-AT stress responses in N. crassa. Three of the 17 genes showed increased expression levels under all three conditions (Cd2+, H2O2 and 3-AT), including NCU04109 (glutathione S-transferase), NCU05164 (protoporphyrinogen oxidase) and NCU09285 (alcohol dehydrogenase). Four of 17 shared targets between the CPC-1 and NcAp-1 H2O2 regulons encode hypothetical proteins (NCU00995, NCU09507, NCU09874 and NCU10970), while the remaining three genes that overlap include cat-2 (NCU05770), a predicted serine protease spr-7 (NCU07159) and mig-4 (NCU09040). Two of the seven genes shared between the CPC-1 and NcAp-1 Cd2+ regulons encode hypothetical proteins (NCU02888 and NCU07503), two encode predicted reductases (NCU04510 and NCU09821), one encodes a predicted pyruvate decarboxylase (NCU02397), one encodes a probable urate oxidase (NCU07853), and the last encodes a predicted epimerase/dehydratase (NCU00884).
DISCUSSION
In this study, we evaluated both the phenotype and the expression patterns associated with strains containing mutations in predicted bZIP TF genes in N. crassa. We identified a single bZIP mutant, containing a deletion of NCU03905, which showed sensitivity to multiple stresses. We further characterized the transcriptional response of strain ΔNcAp-1 (NCU03905) upon exposure to oxidative and heavy metal stresses in comparison with the WT, and identified a number of core stress-response genes. We subsequently compared our data with those from a recent report of an independently derived Δnap-1 mutant and its effect on transcription in response to exposure to menadione (Takahashi et al., 2010). We further compared our datasets with those obtained under different growth conditions and with expression profiling datasets from hemiascomycete species. These comparisons indicated that a small number of genes, which are primarily involved in an oxidant response, are regulated by AP1-like TFs and have been evolutionarily conserved.
Our screening showed that the ΔNcAp-1 strain was sensitive to multiple stresses, including oxidant, heavy metal, osmotic and amino acid starvation stress, while strains containing mutations in six additional bZIP genes did not show increased sensitivity to the tested conditions. These data indicate that NcAp-1 is a critical stress response regulator in N. crassa. Our expression profiling results of the WT versus the NcAp-1 mutant under H2O2 and Cd2+ exposure support this prediction, as multiple pathways were affected in the NcAp-1 mutant, including, as expected, the oxidant response pathway, but also nitrogen metabolism and energy-related genes. In addition, cys-3 (which is a sulfur metabolism regulator) and sulfur metabolism genes were induced by exposure to Cd2+ and were NcAp-1-dependent, suggesting crosstalk between the NcAp-1 and cys-3 regulons. In support of this hypothesis, genes involved in sulfate metabolism are also induced by Cd2+ exposure in C. albicans (Enjalbert et al., 2006).
Genome-wide stress-response profiling data have been published for the genes orthologous to NcAp-1 in S. cerevisiae (Yap1) (Gasch et al., 2000; Cohen et al., 2002), Sch. pombe (Pap1) (Chen et al., 2008) and C. albicans (Cap1) (Wang et al., 2006). In addition, ChIP data are also available for Yap1 (Harbison et al., 2004) and Cap1 (Znaidi et al., 2009), and proteomics data are available for AfYap1 in A. fumigatus (Lessing et al., 2007). The stress response, and in particular, the oxidant stress response, was enriched in the Yap1, Cap1, Pap1, NcAp-1 and AfYap1 datasets. Out of the nine core NcAp-1-dependent stress response genes (Table 4), eight have orthologues in S. cerevisiae, and four of them (OYE3, YKLO71W, OYE2 and ADH5) are regulated by Yap1 (Cohen et al., 2002; Gasch et al., 2000; Harbison et al., 2004). Furthermore, YAP1 binds to the promoter region of OYE2, based on the ChIP data (Harbison et al., 2004). Three of seven orthologues of the NcAp-1-dependent genes are bound and regulated by CAP1 in C. albicans (Wang et al., 2006; Znaidi et al., 2009), while in Sch. pombe, the regulation of six of the eight orthologues is dependent on PAP1 (Table 4). In A. fumigatus, the protein abundance of protoporphyrinogen oxidase (Afu4g00910) is dependent on AfYap1 (Lessing et al., 2007) and the protein is orthologous to the NcAp-1 target NCU05164. In summary, eight of nine NcAp-1 core target genes (Table 4) have been shown to be regulated by Ap1-like proteins in at least one other ascomycete species. The ninth gene, NCU00847 (related to 7-alpha-cephem-methoxylase), has no clear orthologue in S. cerevisiae, Sch. pombe and C. albicans, and is possibly a filamentous fungal-specific oxidant stress response gene. In addition, genes encoding catalases are another group that show regulation under H2O2 conditions by Ap1-like TFs, including catalase-2 (NCU05770) (NcAp-1), catalase-2 (AfYap1), catalase-1 (Cap1) and catalase-T (Yap1). In a recent publication, Takahashi and co-workers found that catalase-3 (NCU00355) rather than catalase-2 is induced by menadione treatment in N. crassa, and is nap-1 (NcAp-1)-dependent (Takahashi et al., 2010). However, in our study, upon treatment with H2O2, NCU00355 was induced in both the WT (3.3-fold) and the ΔNcAp1 mutant (2.5-fold).
The identification of the NcAp-1 regulon and pathway comparisons among ascomycete species suggest that the oxidant stress response pathway in fungi has been conserved over 300 million years of evolution and that AP1-like TFs are the main regulators of this response pathway. Both catalases and flavin oxidoreductases are regulated by AP1-like TFs in S. cerevisiae, C. albicans, Sch. pombe and N. crassa. We identified a common gene set that was induced by amino acid starvation, and oxidant and heavy metal stress, and whose predicted function was oxygen detoxification. These data suggest that reactive oxygen species (ROS) might be a general signal for stress. In all five ascomycete species (S. cerevisiae, Sch. pombe, C. albicans, A. fumigatus and N. crassa), the AP1-like protein localized to nuclei from the cytoplasm following exposure to a variety of stresses.
However, there is also evidence that the AP1 pathway has diverged among the ascomycete species. In S. cerevisiae, the functions of the duplicated Yap proteins are partially redundant, especially those of Yap1 and Yap2 (Cohen et al., 2002). In C. albicans, the oxidant stress response has multiple distinct pathways: the Cap1 pathway and the Hog1 mitogen-activated protein kinase (MAP kinase) pathway (Enjalbert et al., 2006). The Cap1 pathway is activated upon exposure to both low and high concentrations of H2O2, while the Hog1 pathway is only activated at higher concentrations of H2O2. In Sch. pombe, the Pap1 pathway is activated upon exposure to low H2O2 concentrations, while the Sty1 (Hog1 orthologue)–Atf1 (Sko1 orthologue) pathway is activated upon exposure to high concentrations of H2O2 (Chen et al., 2008; Vivancos et al., 2006). However, the Hog1 and Sko1 pathways in S. cerevisiae mainly regulate responses to osmotic stress (Rep et al., 2001), although Skn7 is also involved in oxidative stress, independently of the Yap1 pathway (Estruch, 2000). In N. crassa, the HOG1 orthologue is os-2 (NCU07024) and the SKN7 orthologue is rrg-2 (NCU02413); both genes may be involved in the oxidant stress response pathway, as mutants are sensitive to H2O2 (Banno et al., 2007). The Atf1/Sko1 orthologue in N. crassa is NCU01345, but is ascospore-lethal, and so homokaryotic strains are not available. Based on these data, in N. crassa, at least three oxidant stress response pathways may exist, the OS-2, RRG-2 and NcAp-1 pathways. In the future, it will be informative to study the relationships between various stresses and the pathways regulated by these three TFs.
Evolution of the bZIP TF family regulatory network
How an organism coordinates its transcriptional repertoire in response to developmental or environmental signals, and how evolution affects the topology of these TF networks, are still mostly unanswered questions. One way to study the regulatory networks is to focus on a single biological function. For example, over 30 TFs are involved in DNA damage in S. cerevisiae (Workman et al., 2006). An alternative is to examine regulatory relationships using a phylogenetic framework, such as analysing the regulatory network within the Yap subfamily (Tan et al., 2008). In our study, we analysed seven of the nine members of the bZIP TF family in N. crassa. Phylogenetic analyses indicated that the bZIP TF family is an ancient duplicated gene family, with no recent genome duplications in any fungal species, with the exception of S. cerevisiae. Although our data showed that the regulons of the bZIP TFs have diverged, up to 25 % of the target genes were shared between some TF pairs, for example, the affected gene sets from the colony interiors of the cys-3 and ΔNCU08055 mutants. However, profile comparisons showed differences between the peripheral and interior sections of the colony among the different bZIP mutants. For example, although 25 % of the affected genes in the interior of a colony were identical between the ΔNCU08055 and cys-3 mutants, only 6 % were identical between the ΔNCU08055 and cys-3 datasets from the periphery of the colony. From our stress screening and phenotype analyses, most bZIP TF mutants did not show increased sensitivity to oxidant, metal or osmotic stress. In addition, none of the mutants showed a growth phenotype under minimal media conditions, with the exception of the Δada-1 (NCU00499) mutant. Transcriptional profiling of four bZIP mutants (Δada-1, ΔNCU01994, ΔNCU04211 and ΔNCU08055) revealed a role for these TF genes in carbon compound and carbohydrate metabolism, providing a further testable hypothesis for metabolic processes regulated by these TFs. Three bZIP TF mutants could not be assayed, because homokaryotic mutants are not available, suggesting that these bZIP TFs regulate essential processes in N. crassa. Further genetic, phenotypic and molecular analyses to identify direct targets of the N. crassa bZIP TF family and epistasis relationships will reveal the commonalities and complexities of transcriptional networks regulated by this group of TFs. These data will also enable further comparative analyses to decipher mechanisms associated with the evolution of regulatory networks in N. crassa and other fungi.
Acknowledgments
We thank Dr Sarah Brown for critical reading of this manuscript and Dr Kathy Borkovich's laboratory (University of California, Riverside) for sending us the bZIP transcription deletion strains. We thank Elizabeth Hutchison (Glass laboratory) for construction of the ΔNCU03905 his-3 strains and Dr Charles Hall for help with the phylogenetic analyses. We thank Drs Takao Kasuga and Andre Flessiner for discussions and suggestions during the course of this project. This work was funded by a National Institutes of Health multi-institutional program project grant (GM068087) to N. L. G.
Abbreviations
3-AT, 3-aminotriazole
bHLH, basic helix–loop–helix
bZIP, basic leucine zipper
ChIP data, chromatin-immunoprecipitation data
FunCat analysis, Functional Category analysis
HMM, hidden Markov model
β-ME, β-mercaptoethanol
qRT-PCR, quantitative real-time RT-PCR
TF, transcription factor
WT, wild-type
Footnotes
The microarray data discussed in this paper are available from the Neurospora Functional Genomics Microarray database (experiment ID no. 141) (http://bioinfo.townsend.yale.edu/browse.jsp).
Five supplementary tables and two supplementary figures are available with the online version of this paper.
References
- Bailey, L. A. & Ebbole, D. J. (1998). The fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 148, 1813–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T. L. & Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36. Menlo Park, CA: AAAI Press. [PubMed]
- Banno, S., Noguchi, R., Yamashita, K., Fukumori, F., Kimura, M., Yamaguchi, I. & Fujimura, M. (2007). Roles of putative His-to-Asp signaling modules HPT-1 and RRG-2, on viability and sensitivity to osmotic and oxidative stresses in Neurospora crassa. Curr Genet 51, 197–208. [DOI] [PubMed] [Google Scholar]
- Borkovich, K. A., Alex, L. A., Yarden, O., Freitag, M., Turner, G. E., Read, N. D., Seiler, S., Bell-Pedersen, D., Paietta, J. & other authors (2004). Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68, 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Wilkinson, C. R., Watt, S., Penkett, C. J., Toone, W. M., Jones, N. & Bähler, J. (2008). Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell 19, 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, B. A., Pilpel, Y., Mitra, R. D. & Church, G. M. (2002). Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol Biol Cell 13, 1608–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., Weiss, R. L., Borkovich, K. A. & Dunlap, J. C. (2006). A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103, 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. H. & De Serres, F. J. (1970). Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol 17A, 79–143. [Google Scholar]
- Dementhon, K., Iyer, G. & Glass, N. L. (2006). VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot Cell 5, 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C., Borkovich, K. A., Henn, M. R., Turner, G. E., Sachs, M. S., Glass, N. L., McCluskey, K., Plamann, M., Galagan, J. E. & other authors (2007). Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet 57, 49–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole, D. J., Paluh, J. L., Plamann, M., Sachs, M. S. & Yanofsky, C. (1991). cpc-1, the general regulatory gene for genes of amino acid biosynthesis in Neurospora crassa, is differentially expressed during the asexual life cycle. Mol Cell Biol 11, 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert, B., Smith, D. A., Cornell, M. J., Alam, I., Nicholls, S., Brown, A. J. & Quinn, J. (2006). Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell 17, 1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch, F. (2000). Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24, 469–486. [DOI] [PubMed] [Google Scholar]
- Flint, H. J. (1985). Changes in gene expression elicited by amino acid limitation in Neurospora crassa strains having normal or mutant cross-pathway amino acid control. Mol Gen Genet 200, 283–290. [DOI] [PubMed] [Google Scholar]
- Freitag, M., Hickey, P. C., Raju, N. B., Selker, E. U. & Read, N. D. (2004). GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol 41, 897–910. [DOI] [PubMed] [Google Scholar]
- Fu, Y. H. & Marzluf, G. A. (1990). cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. J Biol Chem 265, 11942–11947. [PubMed] [Google Scholar]
- Galagan, J. E. & Selker, E. U. (2004). RIP: the evolutionary cost of genome defense. Trends Genet 20, 417–423. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., Calvo, S. E., Borkovich, K. A., Selker, E. U., Read, N. D., Jaffe, D., FitzHugh, W., Ma, L. J., Smirnov, S. & other authors (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859–868. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison, C. T., Gordon, D. B., Lee, T. I., Rinaldi, N. J., Macisaac, K. D., Danford, T. W., Hannett, N. M., Tagne, J. B., Reynolds, D. B. & other authors (2004). Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz, G. Z. & Stormo, G. D. (1999). Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A. G. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59, 407–450. [DOI] [PubMed] [Google Scholar]
- Kasuga, T. & Glass, N. L. (2008). Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot Cell 7, 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, T., Townsend, J. P., Tian, C., Gilbert, L. B., Mannhaupt, G., Taylor, J. W. & Glass, N. L. (2005). Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res 33, 6469–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., Birren, B. W. & Lander, E. S. (2004). Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624. [DOI] [PubMed] [Google Scholar]
- Kuge, S., Jones, N. & Nomoto, A. (1997). Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J 16, 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, S., Arita, M., Murayama, A., Maeta, K., Izawa, S., Inoue, Y. & Nomoto, A. (2001). Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol Cell Biol 21, 6139–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessing, F., Kniemeyer, O., Wozniok, I., Loeffler, J., Kurzai, O., Haertl, A. & Brakhage, A. A. (2007). The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell 6, 2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Brutlag, D. L. & Liu, J. S. (2001). BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127–138. [PubMed]
- Liu, X. S., Brutlag, D. L. & Liu, J. S. (2002). An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20, 835–839. [DOI] [PubMed] [Google Scholar]
- Margolin, B. S., Freitag, M. & Selker, E. U. (1997). Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet Newsl 44, 34–36. [Google Scholar]
- McCluskey, K. (2003). The Fungal Genetics Stock Center: from molds to molecules. Adv Appl Microbiol 52, 245–262. [DOI] [PubMed] [Google Scholar]
- Metzenberg, R. L. (2004). Bird medium: an alternative to Vogel medium. Fungal Genet Newsl 51, 19–20. [Google Scholar]
- Paietta, J. V. (1992). Production of the CYS3 regulator, a bZIP DNA-binding protein, is sufficient to induce sulfur gene expression in Neurospora crassa. Mol Cell Biol 12, 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D., Radford, A. & Sachs, M. S. (2001). The Neurospora Compendium: Chromosomal Loci. San Diego, CA. : Academic Press.
- Rao, T. K. & DeBusk, A. G. (1973). Characteristics of a transport-deficient mutant (nap) of Neurospora crassa. Biochim Biophys Acta 323, 619–626. [DOI] [PubMed] [Google Scholar]
- Rep, M., Proft, M., Remize, F., Tamás, M., Serrano, R., Thevelein, J. M. & Hohmann, S. (2001). The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol 40, 1067–1083. [DOI] [PubMed] [Google Scholar]
- Ruepp, A., Zollner, A., Maier, D., Albermann, K., Hani, J., Mokrejs, M., Tetko, I., Güldener, U., Mannhaupt, G. & other authors (2004). The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32, 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S. H., Shih, M. C. & Li, W. H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M., Yamashita, K., Shiozawa, A., Ichiishi, A., Fukumori, F. & Fujimura, M. (2010). An AP-1-like transcription factor, NAP-1, regulates expression of the glutathione S-transferase and NADH : flavin oxidoreductase genes in Neurospora crassa. Biosci Biotechnol Biochem 74, 746–752. [DOI] [PubMed] [Google Scholar]
- Tan, K., Feizi, H., Luo, C., Fan, S. H., Ravasi, T. & Ideker, T. G. (2008). A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci U S A 105, 2934–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, C., Kasuga, T., Sachs, M. S. & Glass, N. L. (2007). Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot Cell 6, 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, J. P. & Hartl, D. L. (2002). Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol 3, RESEARCH0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, J. P. & Taylor, J. W. (2005). Designing experiments using spotted microarrays to detect gene regulation differences within and among species. Methods Enzymol 395, 597–617. [DOI] [PubMed] [Google Scholar]
- Vivancos, A. P., Castillo, E. A., Jones, N., Ayté, J. & Hidalgo, E. (2004). Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol Microbiol 52, 1427–1435. [DOI] [PubMed] [Google Scholar]
- Vivancos, A. P., Jara, M., Zuin, A., Sansó, M. & Hidalgo, E. (2006). Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol Genet Genomics 276, 495–502. [DOI] [PubMed] [Google Scholar]
- Vogel, H. J. (1956). A convenient growth medium for Neurospora. Microbiol Genet Bull 13, 42–46. [Google Scholar]
- Wang, Y., Cao, Y. Y., Jia, X. M., Cao, Y. B., Gao, P. H., Fu, X. P., Ying, K., Chen, W. S. & Jiang, Y. Y. (2006). Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med 40, 1201–1209. [DOI] [PubMed] [Google Scholar]
- Workman, C. T., Mak, H. C., McCuine, S., Tagne, J. B., Agarwal, M., Ozier, O., Begley, T. J., Samson, L. D. & Ideker, T. (2006). A systems approach to mapping DNA damage response pathways. Science 312, 1054–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., De Micheli, M., Coleman, S. T., Sanglard, D. & Moye-Rowley, W. S. (2000). Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol Microbiol 36, 618–629. [DOI] [PubMed] [Google Scholar]
- Znaidi, S., Barker, K. S., Weber, S., Alarco, A. M., Liu, T. T., Boucher, G., Rogers, P. D. & Raymond, M. (2009). Identification of the Candida albicans Cap1p regulon. Eukaryot Cell 8, 806–820. [DOI] [PMC free article] [PubMed] [Google Scholar]