Abstract
Yersinia pestis genome sequencing projects have revealed six intact uncharacterized chaperone/usher systems with the potential to play roles in plague pathogenesis. We cloned each locus and expressed them in the Δfim Escherichia coli strain AAEC185 to test the assembled Y. pestis surface structures for various activities. Expression of each chaperone/usher locus gave rise to specific novel fibrillar structures on the surface of E. coli. One locus, y0561-0563, was able to mediate attachment to human epithelial cells (HEp-2) and human macrophages (THP-1) but not mouse macrophages (RAW264.7), while several loci were able to facilitate E. coli biofilm formation. When each chaperone/usher locus was deleted in Y. pestis, only deletion of the previously described pH 6 antigen (Psa) chaperone/usher system resulted in decreased adhesion and biofilm formation. Quantitative RT-PCR (qRT-PCR) revealed low expression levels for each novel chaperone/usher system in vitro as well as in mouse tissues following intravenous infection. However, a Y. pestis mutant in the chaperone/usher locus y1858-1862 was attenuated for virulence in mice via the intravenous route of infection, suggesting that expression of this locus is, at some stage, sufficient to affect the outcome of a plague infection. qRT-PCR experiments also indicated that expression of the chaperone/usher-dependent capsule locus, caf1, was influenced by oxygen availability and that the well-described chaperone/usher-dependent pilus, Psa, was strongly induced in minimal medium even at 28 °C rather than 37 °C, a temperature previously believed to be required for Psa expression. These data indicate several potential roles for the novel chaperone/usher systems of Y. pestis in pathogenesis and infection-related functions such as cell adhesion and biofilm formation.
INTRODUCTION
Yersinia pestis, the causative agent of plague, is believed to have evolved from Yersinia pseudotuberculosis 1500–20 000 years ago (Achtman et al., 1999). Plague is a devastating infectious disease that has decimated populations throughout history (Cantor, 2001; Perry & Fetherston, 1997). While all pathogenic Yersinia species carry the 70 kb Yop-encoding virulence plasmid (pCD1 in Y. pestis), Y. pestis carries two additional plasmids that contribute to its flea-transmitted lifestyle: pMT1 (encoding the F1 capsule and several other factors) and pPCP1 (encoding pesticin, pesticin immunity and plasminogen activator). Six different Yersinia outer proteins (Yops) have been described in Y. pestis and these are translocated into host cells via a pCD1-encoded type III secretion system. Yops inhibit phagocytosis by disrupting the host cell cytoskeleton, blocking the pro-inflammatory cytokine response or inducing apoptosis of macrophages (Cornelis et al., 1998; Cornelis, 2002).
While a clear role for Yop-mediated cytotoxicity and plasminogen activator-dependent dissemination of an infection from the site of flea inoculation have been described (Beesley et al., 1967; Goguen et al., 1986; Higuchi & Smith, 1961; Sodeinde et al., 1992; Straley & Cibull, 1989; Straley & Bowmer, 1986; Welkos et al., 1997), we hypothesized that other Y. pestis surface proteins may be important during a plague infection.
Chaperone/usher systems often mediate assembly of fimbrial surface structures that confer adhesive and pathogenic capacity to bacteria (Hoschützky et al., 1989; Kuehn et al., 1992; Thanassi et al., 1998). Chaperone/usher-associated fimbrial subunits cross the inner membrane of Gram-negative bacteria via the Sec secretion system. Fimbrial subunits that eventually make up the surface structure are bound in the periplasm by a co-expressed chaperone molecule. The region of the fimbrial subunit bound by the chaperone is composed of an immunoglobulin-like β-sheet fold that is missing one β-strand. The chaperone molecule binds to the fimbrial subunit destined for secretion by providing the missing β-strand in a process known as donor-strand complementation (Barnhart et al., 2000; Choudhury et al., 1999; Sauer et al., 1999). The chaperone then delivers the monomeric fimbrial subunit to a co-expressed outer-membrane usher protein. The usher protein functions as an assembly and secretion platform, catalysing polymerization of the subunit proteins into fimbriae (pili) and providing the channel for secretion of the fimbriae across the outer-membrane to the cell surface (Nishiyama et al., 2008; Remaut et al., 2008). Thus, each chaperone/usher system is composed of a collection of genes encoding one or multiple chaperone proteins, an usher protein and one or more secreted subunit proteins destined for secretion to the surface of the bacterium.
Upon sequencing of two Y. pestis strains, KIM10+ [biovar Mediaevalis (Deng et al., 2002)] and CO92 [biovar orientalis (Parkhill et al., 2001)], eight potentially functional chaperone/usher systems were identified. To date, two chaperone/usher systems have been well studied in Y. pestis: Caf1 (F1) capsule and pH 6 antigen (Psa). The Caf1 capsule has antiphagocytic activity against macrophages in tissue culture and it is an excellent protective antigen against bubonic plague, especially when combined with V antigen (LcrV) (Anderson et al., 1996; DeBord et al., 2006; Zavialov et al., 2003). However, the capsule appears to be dispensable for Y. pestis virulence (Drozdov et al., 1995; Quenee et al., 2008). In fact, it was recently observed that Y. pestis escape mutants can spontaneously arise in mice immunized with Caf1 antigen due to IS1541 element insertion within the caf1 locus (Cornelius et al., 2009). Thus, relying on anti-Caf1 antibodies for protection against Y. pestis infections is not advisable. The other well-studied chaperone/usher system in Y. pestis mediates elaboration of Psa on the surface of the bacterium (Ben-Efraim et al., 1961). Psa carries out a number of activities, including haemagglutination of red blood cells from several animal species (Bichowsky-Slomnicki & Ben-Efraim, 1963; Lindler & Tall, 1993), adhesion to cultured cells (Yang et al., 1996) and macrophages (Makoveichuk et al., 2003), and binding to the Fc region of human IgG (Zav'yalov et al., 1996). Psa has also been shown to have antiphagocytic activity for macrophages, similar to the Caf1 capsule (Huang & Lindler, 2004). Disruption of the Psa locus results in attenuation of Y. pestis infection via the intravenous route of inoculation (Felek et al., 2010; Lindler et al., 1990), indicating a role in pathogenesis for this antiphagocytic adhesin.
Given the important activities of both Caf1 and Psa during host infection, we anticipated that other chaperone/usher systems encoded within Y. pestis may also play roles in virulence. These roles could include adhesion, invasion, biofilm formation, antiphagocytosis or other activities. This study investigates functions and expression patterns of the six novel chaperone/usher loci (Fig. 1), using Psa as a positive control for some known activities.
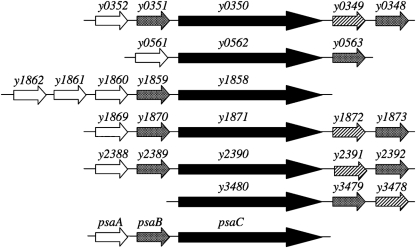
Fig. 1.
Chaperone/usher systems in the Y. pestis KIM genome investigated in this study (Deng et al., 2002). White arrows, putative fimbrial (secreted) proteins; grey arrows, chaperones; black arrows, usher proteins; hatched arrows, large potentially secreted proteins.
We demonstrate that the structure encoded by locus y0561-0563 mediates adhesion of recombinant Escherichia coli to human epithelial and macrophage-like cells, but not murine macrophages. In addition, four novel chaperone/usher loci of Y. pestis facilitate E. coli biofilm formation. Finally, deletion of the Y. pestis y1858-1662 locus results in a virulence defect following intravenous inoculation of mice.
METHODS
Bacterial strains and plasmids.
Characteristics and sources of the bacterial strains and plasmids used in this study are given in Supplementary Table S1 (available with the online version of this paper). Gene deletions in Y. pestis KIM5 were constructed by using PCR products as described previously (Datsenko & Wanner, 2000; Felek et al., 2008; Yu et al., 2000). For cloning, the genes encoding chaperone/usher systems were amplified by PCR. PCR products and plasmid pMMB207 were cut by using appropriate enzymes, purified by gel purification and ligated into plasmid pMMB207. Because strain KIM5 is pgm−, it does not have the y2388-2392 locus (Buchrieser et al., 1999). We cloned this locus from Y. pseudotuberculosis strain IP2666. Y. pestis KIM and Y. pseudotuberculosis IP2666 have 100 % base sequence identity for effector proteins of this system. E. coli DH5α was transformed with the ligation products and chloramphenicol-resistant colonies were selected. Clones were confirmed by DNA sequencing.
SDS-PAGE.
E. coli AAEC185 strains were cultured overnight in LB medium supplemented with 10 μg chloramphenicol ml−1 and 100 μM IPTG at 28 °C and outer-membrane preparations were obtained as described previously (Hantke, 1984). Isolated outer-membranes were subjected to 15 % SDS-PAGE followed by Coomassie blue staining. Whole-cell lysates and outer-membrane preparations were also run on SDS-PAGE and transferred to nitrocellulose membrane for Western blotting using a 1 : 1000 dilution of an anti-RNA polymerase α-subunit antibody (Neoclone) to verify that the outer membrane preparations were free of cytosolic contents as described previously (Felek et al., 2008).
Preparation of E. coli cells for electron microscopy (EM).
E. coli cultures were inoculated from single colonies, grown overnight at 30 °C, diluted 1 : 40 into fresh medium, and then grown at 37 °C for ∼2 h to OD600 0.8 (exponential phase). IPTG was added to 50 μM final concentration to induce expression of the chaperone/usher pathways. For transmission EM, the bacteria were harvested, washed once with PBS, adsorbed onto polyvinyl formal-carbon-coated grids (Ernest F. Fullam) for 2 min, and fixed with 1 % glutaraldehyde (Sigma-Aldrich) for 1 min. The grids were washed twice with PBS and twice with water, and then negatively stained with 0.5 % phosphotungstic acid (Ted Pella) for 35 s. The grids containing the negatively stained bacteria were examined on an FEI TECNAI BioTwin G02 microscope (FEI) at 80 kV accelerating voltage. Digital images were acquired with an AMT XR-60 CCD digital camera system (Advanced Microscopy Techniques).
Adhesion assays.
Twenty-four-well cell culture plates of HEp-2 and RAW264.7 cells were prepared at ≥80 % confluency as described previously (Felek et al., 2008). THP-1 cells were cultured and prepared for infection as described previously (Felek & Krukonis, 2009), with the alteration that after 3 days differentiation in the presence of the phorbol ester 12-myristate 13-acetate, THP-1 were removed from a T75 flask by trypsinization and plated into 24-well plates overnight prior to infection.
E. coli AAEC185 strains containing pMMB207- or pMMB207-encoded chaperone/usher systems were cultured overnight at 28 °C with shaking in Luria–Bertani (LB) medium supplemented with 10 μg chloramphenicol ml−1 and 100 μM IPTG. Since some recombinant proteins were toxic for E. coli AAEC185 at 37 °C under inducing conditions, strains were cultured at 28 °C. Bacterial cultures were centrifuged and resuspended in MEM for HEp-2 cells, DMEM for RAW264.7 cells or RPMI 1640 for THP-1 cells. Host cells were infected with bacteria at an m.o.i. of about 1–3 bacteria per cell after the host cells were washed twice with MEM or DMEM lacking serum. RAW264.7 cells and THP-1 cells were treated with 5 μg cytochalasin-D ml−1 for 45 min to paralyse macrophages before the addition of bacteria. Plates were incubated at 37 °C, 5 % CO2 for 2 h and wells were washed twice with PBS. Cells were lysed with 500 μl double distilled water+0.1 % Triton X-100 (Bio-Rad) for 10 min at room temperature and samples were saved. Wells were washed once more with PBS and washes were saved. Some wells were not washed to remove unbound bacteria in order to calculate total bacteria per well. For this purpose, all media and washes were saved. Ten-fold dilutions in PBS were plated on LB agar plates containing 10 μg chloramphenicol ml−1. The plates were incubated at 37 °C overnight and the output c.f.u. were enumerated.
For Yersinia adhesion, Y. pestis KIM5, KIM5 pCD1− and various KIM5 deletion mutants were cultured overnight at 28 °C pH 7, 37 °C pH 7 or 37 °C pH 6 in heart infusion broth (HIB), with shaking. Cultures were diluted to OD620 0.15 in HIB pH 7.0 and incubated for 4 h under the same culture conditions with shaking. Cultures were pelleted and resuspended in tissue culture medium without serum to OD620 1.5. Cells were infected at an m.o.i. of 1–3 bacteria per host cell for 2 h as for the E. coli binding assays. Binding efficiency was assessed by a c.f.u. plating assay as described previously (Felek & Krukonis, 2009).
Biofilm assays.
Crystal violet staining was used to detect cells attached to polystyrene as described previously (O'Toole et al., 1999). Briefly, overnight cultures were diluted 1 : 100 into LB plus 10 μg chloramphenicol ml−1 and 100 μM IPTG in flat-bottom polystyrene culture plates (Costar Corning) and incubated for 24 h at 28 °C without shaking. The OD595 of cultures was read by using a microplate reader (Perkin Elmer Lambda Reader). Plates were washed three times with PBS. Crystal violet (0.01 %) was added and cells were stained for 15 min at room temperature. Wells were washed three times with distilled water and crystal violet was solubilized with 80 % ethanol and 20 % acetone mixture. The optical density of the mixture was detected by using a microplate reader at 595 nm. Results were normalized with the OD595 of crystal violet/bacterial density of AAEC185+pMMB207 set equal to 1.0. For Y. pestis biofilms, KIM5 derivatives were grown in PMH2 minimal medium (Gong et al., 2001) at 28 °C pH 7, 37 °C pH 7 and 37 °C pH 6 and processed as described for E. coli. For Y. pestis, raw data for OD595/bacterial density are presented without normalization.
Mouse experiments.
Female Swiss Webster mice (6–8-weeks-old) were obtained from Harlan Sprague–Dawley. Y. pestis strains were cultured overnight at 28 °C in HIB pH 7, pelleted and resuspended in PBS to approximately 500 bacteria ml−1. Each mouse was intravenously inoculated with 100 μl of this suspension. At the same time, c.f.u. assays were performed to determine the inoculum c.f.u. for each group. Measured inocula varied from 50 to 56 bacteria per mouse for all KIM5 derivatives except for the KIM5 pCD1-negative strain, for which we used 10 400 bacteria per mouse. Mice were checked daily for survival and signs of illness. Statistical differences in survival between groups were assessed using the logrank test. Animal experiments were performed according to approvals provided by the University of Michigan UCUCA office.
Quantitative RT-PCR (qRT-PCR) expression calculations.
To determine the expression of various KIM5 loci relative to the gene encoding the RNA polymerase β subunit under various growth conditions, Y. pestis mRNA was isolated as described previously in vitro (Felek et al., 2008) or following homogenization of harvested tissues. mRNA (500 ng) was converted to cDNA using Superscript II RT (Invitrogen) and 5 ng cDNA was used for 50 cycles of qRT-PCR using a Bio-Rad iCycler. Once the cycles at which detectable qRT-PCR signals were determined, the mean cycle at which rpoB was detected was calculated and the cycle at which each other locus was detected (n=5–6 measurements over two experiments) was subtracted from the mean detection cycle for rpoB. This gave the number of cycles difference in detection between rpoB and a locus of interest. If this difference was positive, the locus was expressed to higher levels than rpoB (detected at an earlier qRT-PCR cycle number). To measure the fold increase in expression, the number of cycles difference, X, was used as a power function in the equation 2X, since each cycle of qRT-PCR represents a doubling of the PCR targets. Using this equation, a locus that was detected five cycles prior to rpoB would be expressed 25=32 times higher relative to rpoB. The signal from qRT-PCR in the absence of reverse transcriptase was subtracted and standard deviations of the expression measurements were calculated. Primers for qRT-PCR analysis are listed in Supplementary Table S2.
RESULTS
Expression of chaperone/usher system proteins in the outer membrane
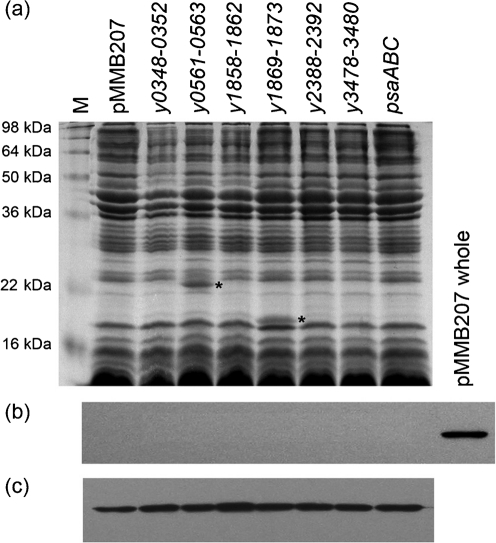
We took two approaches to visualize expression of the various chaperone/usher system proteins from Y. pestis in E. coli. First, we prepared outer-membrane extracts from E. coli strain AAEC185 (Δfim) expressing each of the six novel Y. pestis chaperone/usher loci from the IPTG-inducible plasmid pMMB207 (Morales et al., 1991). Outer membranes would be expected to contain the usher protein from each system and the secreted protein(s) [if the secreted protein is not shed from the surface during preparation of the membranes, a common issue for fimbriae and pili (Felek et al., 2008; Hoschützky et al., 1989)]. Coomassie staining of outer-membrane preparations from AAEC185 demonstrated that clearly visible novel proteins are expressed from two of six loci (Fig. 2; y0561-0563 and y1869-1873). The novel bands are of the predicted size for each anticipated secreted protein and are labelled with an asterisk (Fig. 2; Supplementary Table S3, available with the online version of this paper). Some chaperone/usher loci contain multiple putative secreted proteins. However, in all but one of these cases, one of the putative secreted proteins is between 38 and 48.5 kDa (Y0349, Y1872, Y2391 and Y3478, Supplementary Table S3) and thus is potentially obscured by the major outer-membrane proteins of E. coli. Chaperone/usher systems typically direct assembly of pili or fimbriae on the bacterial surface and these structures are often fragile (Hoschützky et al., 1989; Kuehn et al., 1992; Thanassi et al., 1998). In fact, shearing off from the surface due to centrifugation has been demonstrated for the Y. pestis chaperone/usher-dependent adhesin Psa (Felek et al., 2008). For this reason, we also looked at supernatants from AAEC185 strains expressing each of the Y. pestis chaperone/usher loci. We noticed an 18 kDa protein associated with the y1869-1873 locus that was released into the culture supernatants (data not shown) and this protein was confirmed as the protein encoded by the y1869 gene by mass spectrometry (data not shown). No other chaperone/usher-dependent surface appendage appeared to accumulate at high levels in the culture supernatants except Psa, as demonstrated previously (Felek et al., 2008). It is possible that several of the chaperone/usher-encoded pili are resistant to boiling in SDS and failed to enter the gel (Barnhart & Chapman, 2006; Thanassi & Hultgren, 2000). For all of the chaperone/usher systems, we were unable to distinguish the 87–96 kDa usher protein from other endogenous E. coli outer-membrane proteins in a similar size range.
Fig. 2.
Expression of chaperone/usher proteins in the outer membrane of E. coli AAEC185. Strains were cultured overnight under inducing conditions. Outer membranes were prepared and analysed by using Coomassie-blue-stained SDS-PAGE (a). Anti-RNA polymerase antibody demonstrated that the outer-membrane preparations were not contaminated with cytosolic contents: outer membranes (b) or whole-cell lysate (c). As a positive control on the outer-membrane protein Western blot (b), we included one lane of AAEC185+pMMB207 whole-cell extract (pMMB207 whole). Western blot lanes are aligned with lanes in the Coomassie gel. Target bands are labelled with asterisks. Predicted sizes: Y0561, 21.8 kDa; Y1869, 18.3 kDa.
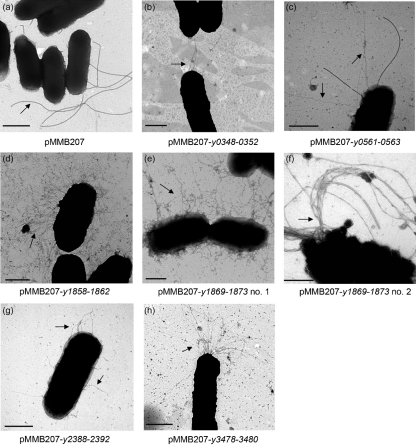
In addition to preparing outer membranes of E. coli to determine chaperone/usher system expression levels, we also prepared E. coli cells for EM. We anticipated that each novel chaperone/usher Y. pestis locus would result in elaboration of a particular structure on the surface of E. coli. All six novel chaperone/usher loci encode chaperone proteins predicted to have a short loop between β-strands F1–G1 and lack disulfide bond-forming cysteines within those strands. These features are indicative of chaperone/usher systems that assemble rigid pili (Hung et al., 1996; Runco et al., 2008). For the EM analysis, bacteria were grown overnight at 30 °C and then subcultured into fresh medium and grown at 37 °C with 50 μM IPTG to induce expression of the chaperone/usher pathways (Methods). Cells were prepared for EM and compared with AAEC185 induced with the empty vector pMMB207. The negative control strain AAEC185+pMMB207 expressed thick appendages on its surface indicative of flagella expression (Fig. 3a). However, strains expressing the various Y. pestis chaperone/usher loci elaborated additional distinct structures on their surface. Expression of locus y0348-0352 resulted in the assembly of a small number of elongated fimbriae-like structures on the surface of E. coli (Fig. 3b). Locus y0561-0563 also expressed thin fimbriae-like filaments (Fig. 3c). Note the difference in thickness between the flagellum and the y0561-0563-encoded fimbriae-like structures (Fig. 3c). Expression of the y3478-3480 locus resulted in long filaments that were polarly localized in many instances (Fig. 3h). Expression of locus y1858-1862 resulted in the bacteria being coated with abundant, thin, peritrichous fimbriae (Fig. 3d). The neighbouring locus, y1869-1873, led to expression of a large number of thick, tangled filaments on the surface of E. coli (Fig. 3e); some bacteria expressing this locus produced twisted, rope-like filaments (Fig. 3f). Finally, expression of locus y2388-2392 resulted in assembly of a smaller number of thick fibres on the bacterial surface (Fig. 3g). This locus is encoded within the horizontally acquired high pathogenicity region of the Y. pestis chromosome (Buchrieser et al., 1999; Hare et al., 1999).
Fig. 3.
EM images of E. coli AAEC185 (Δfim) expressing various Y. pestis chaperone/usher loci. Bacteria were induced for expression of the chaperone/usher loci at 37 °C with 50 μM IPTG. Arrows indicate structures of interest. In AAEC185+pMMB207 (a), the arrow points to a flagellum; in the remaining panels, arrows point to novel fimbrial structures. Bars, 500 nm.
Adhesion to cultured cells
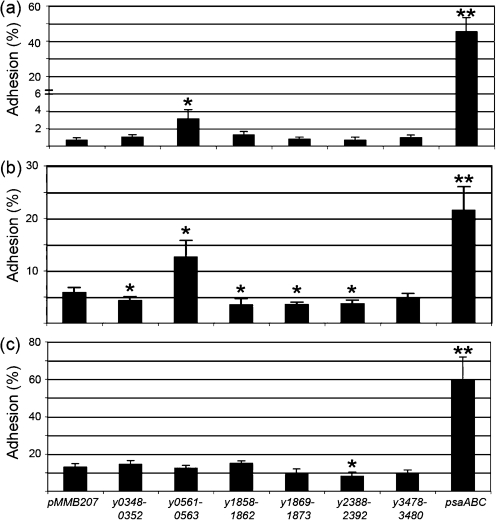
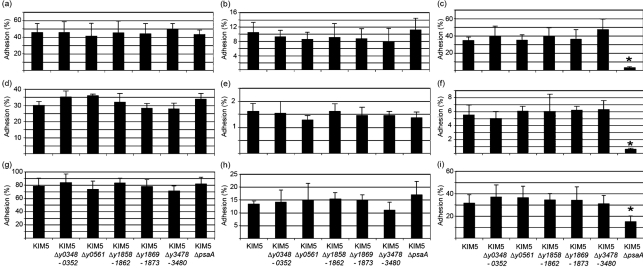
Since many chaperone/usher systems encode adhesive structures (Proft & Baker, 2009; Sauer et al., 2000), we tested the ability of each Y. pestis-encoded structure, when expressed in Δfim E. coli, to adhere to a number of different cell lines including human epithelial cells (HEp-2), human macrophage-like cells (THP-1) and mouse macrophages (RAW264.7 cells). Plate-grown bacteria were used in these studies to prevent shearing of pili from the bacterial cell during broth shaking. Expression of the y0561-0563 locus resulted in a three- or twofold increase in adhesion to HEp-2 (Fig. 4a) and THP-1 (Fig. 4b) cells, respectively, but conferred no increased adhesion to RAW264.7 cells (Fig. 4c). Thus, locus y0561-0563 may encode a surface appendage with adhesive activity specific for human cells. Our positive control for these experiments was the chaperone/usher-encoded surface adhesin, Psa (Galván et al., 2007; Lindler et al., 1990; Payne et al., 1998; Yang & Isberg, 1997). Expression of Psa resulted in a 45-fold increase in HEp-2 cell binding, a fourfold increase in THP-1 cell binding and a fivefold increase in binding to RAW264.7 cells compared with E. coli harbouring pMMB207 alone (Fig. 4).
Fig. 4.
Adhesion of E. coli AAEC185 expressing novel chaperone/usher systems or Psa to HEp-2 (a), THP-1 (b) and RAW264.7 macrophage (c) cells. Strains were cultured overnight at 28 °C on LB agar plates supplemented with 10 μg chloramphenicol ml−1 and 100 μM IPTG. Bacteria were added to tissue culture cells at an m.o.i. of 1–3. Phagocytosis of THP-1 and RAW264.7 cells was inhibited with 5 μg cytochalasin-D ml−1 prior to addition of E. coli. Adhesion was determined by comparing the number of cell-associated c.f.u. with the total number of c.f.u. in the whole tissue culture well. Values are the means±sd of three independent experiments with duplicate assays from each experiment. *P<0.05; **P<0.0005.
Adhesion of Y. pestis to cultured cells
Y. pestis and related pathogenic Yersinia species are known to produce a number of adhesins on their surface (Bliska et al., 1993; El Tahir & Skurnik, 2001; Felek & Krukonis, 2009; Isberg et al., 1987; Kolodziejek et al., 2007; Lähteenmäki et al., 1998; Miller & Falkow, 1988). This redundancy may allow for specific tissue tropism during infection, allow for fine tuned regulation during the progression of an infection, or serve as a backup mechanism to ensure efficient colonization and Yop delivery to host target cells. Although redundancy often obscures the loss of adhesive activity following deletion of a single adhesin, we constructed deletion strains of each novel Y. pestis chaperone/usher locus and the psaA locus and tested those strains for a role in Y. pestis adhesion to various cell lines. None of the novel chaperone/usher loci deletion mutants had a defect in HEp-2, THP-1 or RAW264.7 cell binding (Fig. 5), including a deletion of locus y0561, which was shown to encode an adhesin in E. coli (Fig. 4). For these studies, bacterial cells were grown under three different conditions to mimic different natural environments: 28 °C pH 7 (flea-like), 37 °C pH 7 (host infection) or 37 °C pH 6 [acidified tissues or within a phagolysosome, an environment known to induce Psa (Lindler & Tall, 1993)]. This indicates that other adhesins in addition to y0561 mediate Y. pestis adhesion in KIM5. Alternatively, locus y0561-0563 may not be expressed under the conditions tested. The Psa mutant (ΔpsaA) had an adhesion defect to all three cell lines tested when bacteria were grown at 37 °C pH 6. Thus, under these conditions, Psa was required for maximal binding as expected (Fig. 5).
Fig. 5.
Adhesion of Y. pestis chaperone/usher mutants to tissue culture cells. Y. pestis strains were added to HEp-2 (a–c), THP-1 (d–f) and RAW264.7 (g–i) cells at an m.o.i. of about 10. Cells were cultured at pH 7 28 °C (a, d, g), pH 7 37 °C (b, e, h) or pH 6 37 °C (c, f, i). THP-1 and RAW264.7 cells were blocked for phagocytosis by addition of 5 μg cytochalasin-D ml−1. Results are means±sd of duplicates of three experiments on different days. *P≤0.0003.
Role of chaperone/usher loci in biofilm formation
Another function of some chaperone/usher-encoded structures is to mediate biofilm formation. Biofilm formation can play a role in colonization as well as resistance to innate killing mechanisms of the host (Fux et al., 2005; Jefferson, 2004; Patel, 2005; Stewart & William Costerton, 2001). Each chaperone/usher locus was expressed in the Δfim E. coli strain AAEC185 and early stage biofilm formation was assessed by the development of deposits in polystyrene 96-well plates. For these experiments, we tested biofilm formation at 28 °C (a temperature known to be important for Y. pestis biofilm formation in the flea gut) as well as at 37 °C, to investigate potentially novel roles for biofilms of Y. pestis during human infections.
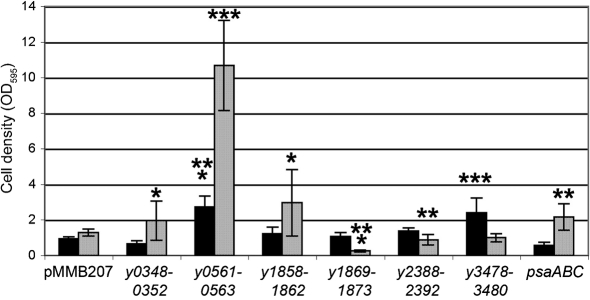
At 28 °C, expression of three novel Y. pestis chaperone/usher loci (y0348-0352, y0561-0563 and y1858-1862) resulted in an increase in E. coli biofilm formation ranging from a 2- to 11-fold (Fig. 6). In addition, the previously characterized psaABC locus was able to mediate early stages of biofilm formation, a newly described function of Psa. When biofilms were allowed to form at 37 °C, locus y0561-0563 was again able to enhance E. coli biofilm formation, as was y3478-3480 (Fig. 6). Thus, many of the structures seen in Fig. 2 are able to facilitate early biofilm formation in E. coli. These may also be important either in the flea vector or during human infection. In these experiments, all loci were expressed from an inducible promoter to allow controlled expression. Related to this point, the ability of Psa to enhance biofilm formation was diminished at 37 °C (the temperature at which psaABC is expressed in Y. pestis). It is unclear why E. coli shows this preference, but as described below, Psa is important for Y. pestis biofilm formation under normal (37 °C pH 6) expression conditions.
Fig. 6.
Formation of biofilm-like deposits in E. coli. Overnight cultures of AAEC185 derivatives were diluted 1 : 100 into LB medium supplemented with 10 μg chloramphenicol ml−1 and 100 μM IPTG in polystyrene culture plates and incubated for 24 h at 28 °C (grey bars) or 37 °C (black bars). Plates were washed three times and attached bacteria were stained with 0.01 % crystal violet. After three washes, crystal violet stain was solubilized with an 80 % ethanol/20 % acetone mixture and optical density was recorded at 595 nm and normalized for bacterial growth (Methods). Results are the means±sd of three independents experiments with at least duplicate assays from each experiment. *P<0.05; **P<0.005; ***P<0.00005.
Role of chaperone/usher systems in hms-independent biofilms in Y. pestis
Biofilm formation in Y. pestis is primarily dependent upon genes of the hms system, including the hmsHFRS genes within the pgm (pigmentation) locus and hmsP and hmsT outside of the pgm locus, that facilitate biofilm formation at ambient temperatures in fleas (Bobrov et al., 2008; Fetherston et al., 1992; Hinnebusch et al., 1996; Kirillina et al., 2004). The KIM5 strain lacks the 102 kb pgm locus due to spontaneous deletion via recombination of two IS100 elements (Fetherston et al., 1992). However, Y. pestis has also been shown to mediate hms-independent biofilm formation (Styer et al., 2005).
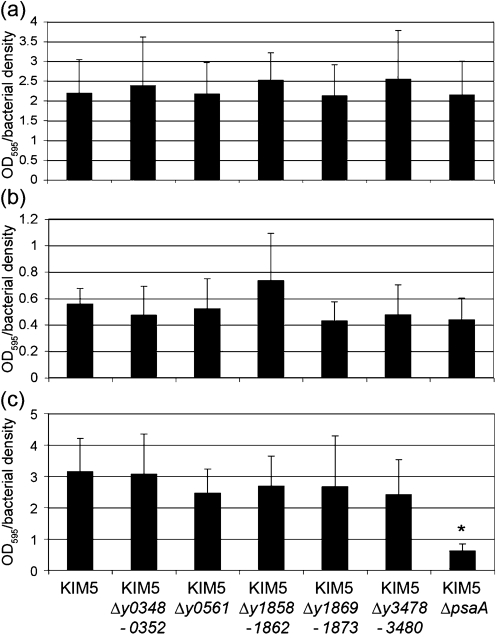
Given the ability of several Y. pestis chaperone/usher loci to facilitate E. coli biofilm formation (Fig. 6), we assessed the effect of each deletion on hms-independent biofilm formation in the pgm-negative strain KIM5. Deletion of each individual novel chaperone/usher system had no effect on KIM5 biofilm formation under three different growth conditions tested (Fig. 7). However, deletion of psaA led to a sixfold decrease in KIM5 biofilm formation when cells were grown at 37 °C pH 6 (Fig. 7c). This is the first report to our knowledge of biofilm-forming activity for Psa in Y. pestis.
Fig. 7.
The ability of Y. pestis chaperone/usher mutants to form hms-independent biofilms. KIM5 derivatives were grown in PMH2 minimal medium (Gong et al., 2001) at 28 °C pH 7 (a), 37 °C pH 7 (b) or 37 °C pH 6 (c) and processed as described for E. coli. For Y. pestis, raw data for OD595/bacterial density are presented without normalization. *P=0.00001.
Expression of chaperone/usher loci in vitro and in vivo
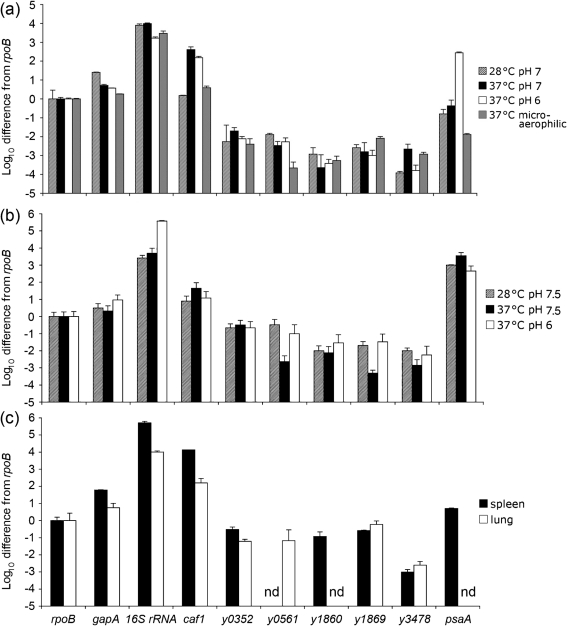
One reason that deletion of novel chaperone/usher loci of Y. pestis had no effect on certain phenotypes may be that the locus of interest was not expressed under that particular growth condition. qRT-PCR was used to assess expression levels of the various chaperone/usher systems under several conditions. Expression under three typical in vitro growth conditions was assessed, including HIB medium at 28 °C pH 7, HIB medium at 37 °C pH 7 and HIB medium at 37 °C pH 6. Cultures were grown overnight and expression levels were compared with the housekeeping gene rpoB (rpoB=1.0). Under each of these three growth conditions, none of the novel chaperone/usher systems was expressed well (all ≤0.02 relative to rpoB; Fig. 8 and Supplementary Table S4). Positive controls in this experiment were caf1, encoding the chaperone/usher-dependent F1 protein capsule of Y. pestis (known to be expressed at 37 °C), and psaA, known to be induced at 37 °C pH <6.7 (Ben-Efraim et al., 1961). Both controls were induced appropriately in HIB, and the 16S rRNA gene was strongly expressed at each condition. As a metabolic gene of intermediate expression, we included gapA (encoding glyceraldehyde 3-phosphate dehydrogenase A), which has been used previously in other qRT-PCR analyses as a normalization control (Alteri & Mobley, 2007). gapA was expressed to slightly higher levels than rpoB under all three standard in vitro conditions.
Fig. 8.
qRT-PCR analysis of chaperone/usher system expression in vitro and in vivo. KIM5 was grown in HIB (a), PMH-3 (b) or harvested from infected tissue 3 days post-infection (c). Expression was normalized to rpoB and plotted on a log scale after subtracting the signal obtained from no reverse transcriptase controls. Transcript levels were also compared to transcripts for gapA and 16S rRNA. qRT-PCR assays were performed twice in duplicate or triplicate (n=5–6). nd, No amplification detected after 50 cycles of qRT-PCR.
As we were unable to detect robust expression of the five novel chaperone/usher systems of KIM5 in standard in vitro conditions, we also assessed expression in oxygen-limited conditions or in minimal medium. To produce microaerophilic conditions, bacteria were grown at 37 °C overnight in a sealed jar without shaking. Under these conditions, none of the novel chaperone/usher systems was expressed to levels higher than those found under aerated conditions previously (Fig. 8 and Supplementary Table S4). However, two unexpected observations were made regarding the well-studied caf1 and psaA loci. Under microaerophilic conditions at 37 °C, caf1 was only modestly expressed, at levels 3.9-fold above rpoB, compared with expression that was 405-fold above rpoB when shaken at 37 °C pH 7 (Fig. 8 and Supplementary Table S4). Thus, oxygen tension strongly affects caf1 expression. Also, while in some cases psaA was poorly expressed under microaerophilic conditions (Fig. 8 and Supplementary Table S4), in some experiments, psaA was expressed as high as 36-fold above rpoB, presumably depending upon whether the cells grown in microaerophilic conditions resulted in extensive acidification of the medium (data not shown).
Growth in minimal medium was another condition we explored to assess expression of novel chaperone/usher systems. PMH3 medium, a Y. pestis minimal medium based on PMH2 (Gong et al., 2001), supplemented with 25 mM HEPES (to stabilize the pH), was used after adjusting the starting pH to pH 7.5 or pH 6.0 as indicated (Methods). In PMH3, the novel chaperone/usher locus y0561 was expressed at levels approaching that of rpoB (0.33 relative expression) when grown at 28 °C and a starting pH of 7.5. The only other novel chaperone/usher locus expressed to levels approaching rpoB was y0352, which was expressed at relative levels between 0.22 and 0.32 under all three conditions tested in PMH3. All other novel chaperone/usher loci were expressed at levels <0.1 (Fig. 8 and Supplementary Table S4). Unexpectedly, caf1 was not very strongly expressed in PMH3 at 37 °C (Fig. 8 and Supplementary Table S4), a temperature known to induce caf1 in rich media (Fig. 8; Fox & Higuchi, 1958). Furthermore, upon growth in PMH3, psaA was altered in its regulation. Rather than being maximally activated at 37 °C pH 6, psaA was strongly activated at all three growth conditions in PMH3 (Fig. 8 and Supplementary Table S4). Over the course of overnight growth, all three cultures were acidified, but the 28 °C pH 7.5 culture was only mildly acidic at pH 6.6 at the end of the experiment, while the 37 °C pH 7.5 and 37 °C pH 6 cultures had final pH readings of 5.8 and 4.6, respectively. These data suggest that either some component of rich media represses psaA expression unless it is grown at 37 °C and pH <6.7 or a component of the minimal medium PMH3 induces expression of the psaABC locus, even at 28 °C. In any event, this result clearly indicates that psaA can be expressed under unconventional growth conditions depending on the medium composition.
Finally, as we were interested in assessing the role of the novel chaperone/usher systems during infection, we assessed the expression levels of each locus from spleen and lung tissue after a 3 day mouse infection with 10 LD50 (100 organisms) of KIM5 administered intravenously. In infected spleen tissue, three loci, y0352, y1860 and y1869, were expressed 15- to 100-fold higher compared with the same loci expressed in the rich medium, HIB (Fig. 8 and Supplementary Table S4), although their absolute expression levels were still modest: 0.12–0.30 relative to rpoB. Two other loci, y0561 and y3478 were undetectable or poorly expressed, respectively (Fig. 8 and Supplementary Table S4).
In lung tissue, four of the novel chaperone/usher loci were expressed to levels of 0.068 or below, relative to rpoB, while y1869 was expressed to nearly the same level as rpoB, with relative expression of 0.59 (Fig. 8 and Supplementary Table S4). Even at their highest expression levels in tissues, none of the novel chaperone/usher loci reached the level of expression of rpoB, suggesting that these systems are expressed to modest levels or only a subpopulation induces expression. As expected, caf1 was strongly expressed in vivo. While psaA was expressed to intermediate levels in the spleen (fivefold above rpoB), it was not detected in lung tissue, as has been described previously (Lathem et al., 2005).
Virulence defects of chaperone/usher mutants
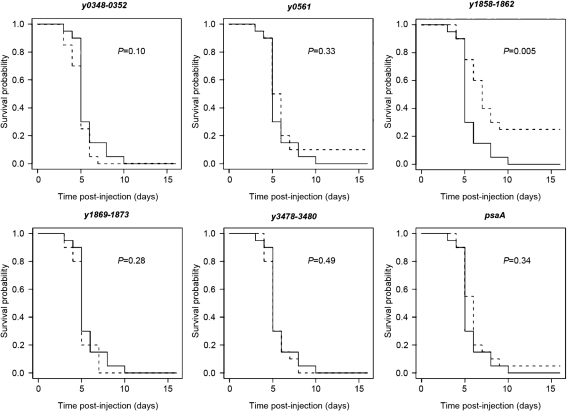
Given the potential roles of the Y. pestis chaperone/usher systems in tissue adhesion, biofilm formation or other virulence-associated activities, we determined whether there was any virulence defect associated with deletion of each Y. pestis chaperone/usher locus. Y. pestis KIM5 and various deletion mutants were tested in a mouse model of plague. Groups of 20 female Swiss Webster mice received 7 LD50 [∼50 bacteria (Felek et al., 2010)] intravenously. Mice were monitored for 14 days to determine whether they succumbed to the infection and a time-course survival curve was generated. While all mice infected with the parental strain (KIM5) had 0 % survival by day 10, one chaperone/usher mutant, Δy1858-1862, showed a statistically significant increase in survival (Fig. 9, P=0.005). Deletion of y0561 also led to 10 % survival; however, this increase was not statistically significant (P=0.33). All mice survived infection with a KIM5 pCD1− strain (lacking the Yop-encoding plasmid) at a dose of 10 400 c.f.u.
Fig. 9.
Mouse survival assay for Y. pestis KIM5 and individual deletion mutants. Overnight cultures of Y. pestis KIM5, KIM5 pCD1− and deletion mutants were intravenously inoculated into mice at a dose of 5–10 LD50 (∼50 bacteria). Mice were observed for 14 days after injection for survival. Ten mice were infected per day and the experiment was repeated once. Data are combined from the two experiments. Solid line, survival curve for KIM5; dashed line, survival curve of chaperone/usher mutant (given above each plot). P values were determined using logrank tests.
Somewhat surprisingly, the ΔpsaA mutant had no defect in virulence (although one mouse did survive infection). This conflicts with the previous report of a similar mutant in KIM5 that was shown to be attenuated for virulence, with a 200-fold increase in LD50 (Lindler et al., 1990). We also observed that the KIM5 ΔpsaA mutant was more strongly attenuated in mice in some experiments, but this defect appeared to be variable (Felek et al., 2010). This may reflect the acidity of the inoculating culture and whether Psa is expressed at the time of injection.
DISCUSSION
Chaperone/usher systems allow for secretion of bacterial surface appendages that can provide various functions to the cell (Proft & Baker, 2009; Sauer et al., 2000). The goal of this study was to identify activities associated with six uncharacterized chaperone/usher systems of Y. pestis, assess their expression patterns and determine their roles in Y. pestis virulence. We found that, of the six novel chaperone/usher systems, y0561-0563 conferred adhesive activity to E. coli (Fig. 4), although deletion of y0561 had no effect on Y. pestis cell attachment (Fig. 5). Given that Y. pestis has several other adhesins including Ail (Felek & Krukonis, 2009; Kolodziejek et al., 2007), plasminogen activator (Pla; Kienle et al., 1992; Lähteenmäki et al., 1998) and Psa (Felek et al., 2008; Galván et al., 2007; Lindler et al., 1990; Yang & Isberg, 1997), we assume they (and other potential adhesins) may mask any binding defect of the y0561 deletion mutant. Some novel Y. pestis chaperone/usher systems had a slight inhibitory effect on adhesion when expressed in E. coli (Fig. 4b). Based on EM of E. coli expressing these structures, at least two novel chaperone/usher pathways, y1858-1862 and y1869-1873, may abundantly coat the bacterium with pilus-like structures potentially occluding other surface adhesins (Fig. 3d and e).
Biofilms can be important virulence factors for bacteria. They can help facilitate colonization and protect bacteria from immune defences of the host (Fux et al., 2005; Jefferson, 2004; Patel, 2005; Stewart & William Costerton, 2001). Attachment of Gram-negative bacteria to a surface via outer-membrane proteins is the first step of biofilm formation, followed by replication to form microcolonies and produce a mature biofilm. In E. coli we found several Y. pestis chaperone/usher systems were able to facilitate biofilm formation upon expression. Among the two growth conditions tested (28 and 37 °C), four novel chaperone/usher systems could enhance E. coli biofilms (Fig. 6). The well-studied chaperone/usher-dependent adhesin Psa was also able to enhance E. coli biofilms. Given that the y0561-0563 locus confers both adhesive and biofilm-forming activities to E. coli, we considered the possibility that strains expressing y0561-0563 are non-specifically sticky to all surfaces. However, we found that E. coli expressing y0561-0563 does not autoaggregate due to bacterial–bacterial interactions over a 6 h time-course, whereas expression of the previously described Y. pestis autotransporter, YapC, led to robust autoaggregation (50 % settling out of solution within 90 min, data not shown) as demonstrated previously (Felek et al., 2008).
Although the Y. pestis strain used in these studies, KIM5, lacks the major biofilm-forming locus (hmsHFRS within the larger pgm locus), we were able to assess the ability of various chaperone/usher system mutants to form previously reported hms-independent biofilms (Styer et al., 2005). Deletion of each individual novel chaperone/usher system had no effect on Y. pestis hms-independent biofilm formation. However, deletion of psaA led to a sixfold decrease in hms-independent biofilms of Y. pestis when the bacteria were grown at a starting pH of 6.0. This is the first report of Psa contributing to biofilm formation in Y. pestis. This activity adds to the growing list of Psa functions, including binding to β1-linked galactosyl residues in glycosphingolipids (Payne et al., 1998) and to phosphatidylcholine (Galván et al., 2007), interaction with plasma lipoproteins (Makoveichuk et al., 2003), acting as an Fc-receptor for human IgG (Zav'yalov et al., 1996) and blocking phagocytosis (Huang & Lindler, 2004).
We also constructed chaperone/usher deletion mutants of Y. pestis KIM6+, a strain fully competent for biofilm formation (Darby et al., 2002; Hinnebusch et al., 1996). Previous studies have shown that KIM6+ derivatives defective for biofilm formation are unable to block Caenorhabditis elegans from feeding on Y. pestis, leading to ingestion of the bacteria and C. elegans development to mature forms (Darby et al., 2002). None of the KIM6+ derivatives deleted for any of the six novel chaperone/usher loci (nor a ΔpsaA mutant) were defective for blockage of C. elegans feeding (Creg Darby, personal communication).
qRT-PCR analysis was used to assess expression levels of the various chaperone/usher systems under a variety of in vitro and in vivo conditions. While a few conditions gave modest levels of expression of some loci – y0352 (spleen and minimal medium), y0561 (minimal medium), y1860 (spleen) and y1869 (spleen and lung) – most conditions tested showed low expression levels (<0.1) of the novel chaperone/usher loci relative to the housekeeping gene rpoB. However, the well-characterized chaperone/usher systems controlling F1 capsule (caf1) and Psa (psaA) production were well expressed under conditions shown previously to induce their expression, 37 °C and 37 °C pH 6, respectively. During our extensive analysis we observed some conditions where caf1 would be predicted to be expressed to high levels (various 37 °C conditions) but was not. These included poor expression of caf1 at 37 °C in microaerophilic conditions (perhaps mimicking the low oxygen tension in non-vascularized necrotic lesions) and in minimal medium (Fig. 8 and Supplementary Table S4). Growth in minimal medium also gave unanticipated results concerning psaA expression. Rather than no expression at 28 °C starting pH 7.5, this condition resulted in a high level of expression of psaA (1000-fold above rpoB; Fig. 8 and Supplementary Table S4). Whereas in rich medium, growing a culture at 37 °C and a starting pH of 6.0 gave the highest levels of psaA expression, in minimal medium, this gave the lowest levels of expression, although psaA was still strongly expressed (450-fold above rpoB; Fig. 8 and Supplementary Table S4). The level of psaA expression in pH 6 PMH-3 medium at 37 °C was significantly different from psaA expression in pH 7 medium at 37 °C (P=0.011), but not pH 7 medium at 28 °C (P=0.096). As psaA is at least partially regulated by the global regulator rovA in Y. pestis (Cathelyn et al., 2006), these changes may reflect effects on rovA expression or RovA activity. Alternatively, these conditions may affect the expression or activity of PsaE, a membrane-bound activator of psaA that is encoded immediately upstream of psaA along with a periplasmically located effector protein PsaF (Yang & Isberg, 1997). While psaA has been shown previously to be induced within phagolysosomes of macrophages in a pH-dependent fashion (requiring acidification; Lindler & Tall, 1993), it remains to be determined whether psaA expression in other tissues may reflect the minimal medium situation and not be as strongly dependent upon acidic pH (Fig. 8 and Supplementary Table S4).
Our study demonstrated that deletion of one novel chaperone/usher locus, y1858-1862, led to decreased virulence in mice after intravenous infection, indicating that it has a role in plague pathogenesis (Fig. 9). This locus was able to facilitate biofilm formation in E. coli (Fig. 6) and also interfere with E. coli adhesion to THP-1 (macrophage-like) cells (Fig. 4b). Expression of this locus in E. coli also resulted in abundant, peritrichous fimbriae on the surface of the bacterium. All of these activities may contribute to the role of this locus in plague pathogenesis.
Acknowledgments
We thank Dr James Bliska for supplying Y. pseudotuberculosis strain IP2666, as a source of the y2388-2392 chaperone/usher locus. We thank Susan Van Horn and the Central Microscopy Imaging Center (Stony Brook University) for assistance with EM. We thank Drs Creg Darby and Alexandra Koumoutsi for performing the C. elegans and Congo red experiments. Animal experiments were performed at the University of Michigan School of Dentistry in accordance with the standards of the AALAC-accredited Unit for Laboratory Animal Medicine (ULAM). This work was supported by grants from the University of Michigan Biomedical Research Council (BMRC), Office of the Vice President for Research (OVPR) and the Rackham Graduate School to E. S. K. and National Institutes of Health grant AI055621 to D. G. T.
Abbreviations
EM, electron microscopy
qRT-PCR, quantitative RT-PCR
Footnotes
Four supplementary tables are available with the online version of this paper.
References
- Achtman, M., Zurth, K., Morelli, G., Torrea, G., Guiyoule, A. & Carniel, E. (1999). Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 96, 14043–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri, C. J. & Mobley, H. L. (2007). Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect Immun 75, 2679–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, G. W. J., Leary, S. E., Williamson, E. D., Titball, R. W., Welkos, S. L., Worsham, P. L. & Friedlander, A. M. (1996). Recombinant V antigen protects mice against pneumonic and bubonic plague caused by F1-capsule-positive and -negative strains of Yersinia pestis. Infect Immun 64, 4580–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart, M. M. & Chapman, M. R. (2006). Curli biogenesis and function. Annu Rev Microbiol 60, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart, M. M., Pinkner, J. S., Soto, G. E., Sauer, F. G., Langermann, S., Waksman, G., Frieden, C. & Hultgren, S. J. (2000). PapD-like chaperones provide the missing information for folding of pilin proteins. Proc Natl Acad Sci U S A 97, 7709–7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley, E. D., Brubaker, R. R., Janssen, W. A. & Surgalla, M. J. (1967). Pesticins III. Expression of coagulase and mechanism of fibrinolysis. J Bacteriol 94, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim, S., Aronson, M. & Bichowsky-Slomnicki, L. (1961). New antigenic component of Pasteurella pestis formed under specified conditions of pH and temperature. J Bacteriol 81, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichowsky-Slomnicki, L. & Ben-Efraim, S. (1963). Biological activities in extracts of Pasteurella pestis and their relation to the “pH 6 antigen”. J Bacteriol 86, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska, J. B., Copass, M. C. & Falkow, S. (1993). The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect Immun 61, 3914–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov, A. G., Kirillina, O., Forman, S., Mack, D. & Perry, R. D. (2008). Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol 10, 1419–1432. [DOI] [PubMed] [Google Scholar]
- Buchrieser, C., Rusniok, C., Frangeul, L., Couve, E., Billault, A., Kunst, F., Carniel, E. & Glaser, P. (1999). The 102-Kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect Immun 67, 4851–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor, N. (2001). In the Wake of the Plague. New York. : Perennial.
- Cathelyn, J. S., Crosby, S. D., Lathem, W. W., Goldman, W. E. & Miller, V. L. (2006). RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc Natl Acad Sci U S A 103, 13514–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, D., Thompson, A., Stojanoff, V., Langermann, S., Pinkner, J., Hultgren, S. J. & Knight, S. D. (1999). X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Cornelis, G. R. (2002). The Yersinia Ysc–Yop ‘type III’ weaponry. Nat Rev Mol Cell Biol 3, 742–752. [DOI] [PubMed] [Google Scholar]
- Cornelis, G. R., Boland, A., Boyd, A. P., Geuijen, C., Iriarte, M., Neyt, C., Sory, M.-P. & Stainier, I. (1998). The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev 62, 1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius, C. A., Quenee, L. E., Elli, D., Ciletti, N. A. & Schneewind, O. (2009). Yersinia pestis IS1541 transposition provides for escape from plague immunity. Infect Immun 77, 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby, C., Hsu, J. W., Ghori, N. & Falkow, S. (2002). Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417, 243–244. [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A. & Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBord, K. L., Anderson, D. M., Marketon, M. M., Overheim, K. A., DePaolo, R. W., Ciletti, N. A., Jabri, B. & Schneewind, O. (2006). Immunogenicity and protective immunity against bubonic plague and pneumonic plague by immunization of mice with the recombinant V10 antigen, a variant of LcrV. Infect Immun 74, 4910–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W., Burland, V., Plunkett, G., III, Boutin, A., Mayhew, G. F., Liss, P., Perna, N. T., Rose, D. J., Mau, B. & other authors (2002). Genome sequence of Yersinia pestis KIM. J Bacteriol 184, 4601–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdov, I. G., Anisimov, A. P., Samoilova, S. V., Yezhov, I. N., Yeremin, S. A., Karlyshev, A. V., Krasilnikova, V. M. & Kravchenko, V. I. (1995). Virulent non-capsulate Yersinia pestis variants constructed by insertion mutagenesis. J Med Microbiol 42, 264–268. [DOI] [PubMed] [Google Scholar]
- El Tahir, Y. & Skurnik, M. (2001). YadA, the multifaceted Yersinia adhesin. Int J Med Microbiol 291, 209–218. [DOI] [PubMed] [Google Scholar]
- Felek, S. & Krukonis, E. S. (2009). The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect Immun 77, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felek, S., Lawrenz, M. B. & Krukonis, E. S. (2008). The Yersinia pestis autotransporter YapC mediates host cell binding, autoaggregation and biofilm formation. Microbiology 154, 1802–1812. [DOI] [PubMed] [Google Scholar]
- Felek, S., Tsang, T. M. & Krukonis, E. S. (2010). Three Yersinia pestis adhesins facilitate Yop delivery to eukaryotic cells and contribute to plague virulence. Infect Immun 78, 4134–4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetherston, J. D., Schuetze, P. & Perry, R. D. (1992). Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol 6, 2693–2704. [DOI] [PubMed] [Google Scholar]
- Fox, E. N. & Higuchi, K. (1958). Synthesis of the fraction I antigenic protein by Pasteurella pestis. J Bacteriol 75, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux, C. A., Costerton, J. W., Stewart, P. S. & Stoodley, P. (2005). Survival strategies of infectious biofilms. Trends Microbiol 13, 34–40. [DOI] [PubMed] [Google Scholar]
- Galván, E. M., Chen, H. & Schifferli, D. M. (2007). The Psa fimbriae of Yersinia pestis interact with phosphatidylcholine on alveolar epithelial cells and pulmonary surfactant. Infect Immun 75, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goguen, J. D., Walker, W. S., Hatch, T. P. & Yother, J. (1986). Plasmid-determined cytotoxicity in Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun 51, 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, S., Bearden, S. W., Geoffroy, V. A., Fetherston, J. D. & Perry, R. D. (2001). Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect Immun 69, 2829–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke, K. (1984). Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol Gen Genet 197, 337–341. [DOI] [PubMed] [Google Scholar]
- Hare, J. M., Wagner, A. K. & McDonough, K. A. (1999). Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol Microbiol 31, 291–303. [DOI] [PubMed] [Google Scholar]
- Higuchi, K. & Smith, J. L. (1961). Studies on the nutrition and physiology of Pasteurella pestis. VI. A differential plating medium for the estimation of the mutation rate to avirulence. J Bacteriol 81, 605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, B. J., Perry, R. D. & Schwan, T. G. (1996). Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273, 367–370. [DOI] [PubMed] [Google Scholar]
- Hoschützky, H., Lottspeich, F. & Jann, K. (1989). Isolation and characterization of the alpha-galactosyl-1,4-beta-galactosyl-specific adhesin (P adhesin) from fimbriated Escherichia coli. Infect Immun 57, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X.-Z. & Lindler, L. E. (2004). The pH 6 antigen is an antiphagocytic factor produced by Yersinia pestis independent of yersinia outer proteins and capsule antigen. Infect Immun 72, 7212–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, D. L., Knight, S. D., Woods, R. M., Pinkner, J. S. & Hultgren, S. J. (1996). Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J 15, 3792–3805. [PMC free article] [PubMed] [Google Scholar]
- Isberg, R. R., Voorhis, D. L. & Falkow, S. (1987). Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50, 769–778. [DOI] [PubMed] [Google Scholar]
- Jefferson, K. K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236, 163–173. [DOI] [PubMed] [Google Scholar]
- Kienle, Z., Emody, L., Svanborg, C. & O'Toole, P. (1992). Adhesive properties conferred by the plasminogen activator of Yersinia pestis. J Gen Microbiol 138, 1679–1687. [DOI] [PubMed] [Google Scholar]
- Kirillina, O., Fetherston, J. D., Bobrov, A. G., Abney, J. & Perry, R. D. (2004). HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol 54, 75–88. [DOI] [PubMed] [Google Scholar]
- Kolodziejek, A. M., Sinclair, D. J., Seo, K. S., Schnider, D. R., Deobald, C. F., Rohde, H. N., Viall, A. K., Minnich, S. S., Hovde, C. J. & other authors (2007). Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology 153, 2941–2951. [DOI] [PubMed] [Google Scholar]
- Kuehn, M. J., Heuser, J., Normark, S. & Hultgren, S. J. (1992). P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature 356, 252–255. [DOI] [PubMed] [Google Scholar]
- Lähteenmäki, K., Virkola, R., Saren, A., Emody, L. & Korhonen, T. K. (1998). Expression of plasminogen activator Pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect Immun 66, 5755–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem, W. W., Crosby, S. D., Miller, V. L. & Goldman, W. E. (2005). Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc Natl Acad Sci U S A 102, 17786–17791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindler, L. E. & Tall, B. D. (1993). Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages. Mol Microbiol 8, 311–324. [DOI] [PubMed] [Google Scholar]
- Lindler, L. E., Klempner, M. & Straley, S. (1990). Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect Immun 58, 2569–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoveichuk, E., Cherepanov, P., Lundberg, S., Forsberg, A. & Olivecrona, G. (2003). pH 6 antigen of Yersinia pestis interacts with plasma lipoproteins and cell membranes. J Lipid Res 44, 320–330. [DOI] [PubMed] [Google Scholar]
- Miller, V. L. & Falkow, S. (1988). Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun 56, 1242–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, V. M., Backman, A. & Bagdasarian, M. (1991). A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97, 39–47. [DOI] [PubMed] [Google Scholar]
- Nishiyama, M., Ishikawa, T., Rechsteiner, H. & Glockshuber, R. (2008). Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379. [DOI] [PubMed] [Google Scholar]
- O'Toole, G. A., Pratt, L. A., Watnick, P. I., Newman, D. K., Weaver, V. B. & Kolter, R. (1999). Genetic approaches to study of biofilms. Methods Enzymol 310, 91–109. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C. & other authors (2001). Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413, 523–527. [DOI] [PubMed] [Google Scholar]
- Patel, R. (2005). Biofilms and antimicrobial resistance. Clin Orthop Relat Res 41–47. [DOI] [PubMed]
- Payne, D., Tatham, D., Williamson, E. D. & Titball, R. W. (1998). The pH 6 antigen of Yersinia pestis binds to beta 1-linked galactosyl residues in glycosphingolipids. Infect Immun 66, 4545–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R. D. & Fetherston, J. D. (1997). Yersinia pestis – etiologic agent of plague. Clin Microbiol Rev 10, 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft, T. & Baker, E. N. (2009). Pili in Gram-negative and Gram-positive bacteria – structure, assembly and their role in disease. Cell Mol Life Sci 66, 613–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenee, L. E., Cornelius, C. A., Ciletti, N. A., Elli, D. & Schneewind, O. (2008). Yersinia pestis caf1 variants and the limits of plague vaccine protection. Infect Immun 76, 2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut, H., Tang, C., Henderson, N. S., Pinkner, J. S., Wang, T., Hultgren, S. J., Thanassi, D. G., Waksman, G. & Li, H. (2008). Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runco, L. M., Myrczek, S., Bliska, J. B. & Thanassi, D. G. (2008). Biogenesis of the fraction 1 capsule and analysis of the ultrastructure of Yersinia pestis. J Bacteriol 190, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, F. G., Futterer, K., Pinkner, J. S., Dodson, K. W., Hultgren, S. J. & Waksman, G. (1999). Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061. [DOI] [PubMed] [Google Scholar]
- Sauer, F. G., Mulvey, M. A., Schilling, J. D., Martinez, J. J. & Hultgren, S. J. (2000). Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol 3, 65–72. [DOI] [PubMed] [Google Scholar]
- Sodeinde, O. A., Subrahmanyam, Y., Stark, K., Quan, T., Bao, Y. & Goguen, J. (1992). A surface protease and the invasive character of plague. Science 258, 1004–1007. [DOI] [PubMed] [Google Scholar]
- Stewart, P. S. & William Costerton, J. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. [DOI] [PubMed] [Google Scholar]
- Straley, S. C. & Bowmer, W. S. (1986). Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun 51, 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straley, S. C. & Cibull, M. (1989). Differential clearance and host-pathogen interactions of YopE− and YopK− YopL− Yersinia pestis in BALB/c mice. Infect Immun 57, 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styer, K. L., Hopkins, G. W., Bartra, S. S., Plano, G. V., Frothingham, R. & Aballay, A. (2005). Yersinia pestis kills Caenorhabditis elegans by a biofilm-independent process that involves novel virulence factors. EMBO Rep 6, 992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi, D. G. & Hultgren, S. J. (2000). Assembly of complex organelles: pilus biogenesis in Gram-negative bacteria as a model system. Methods 20, 111–126. [DOI] [PubMed] [Google Scholar]
- Thanassi, D. G., Saulino, E. T. & Hultgren, S. J. (1998). The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr Opin Microbiol 1, 223–231. [DOI] [PubMed] [Google Scholar]
- Welkos, S. L., Friedlander, A. M. & Davis, K. J. (1997). Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain C092. Microb Pathog 23, 211–223. [DOI] [PubMed] [Google Scholar]
- Yang, Y. & Isberg, R. R. (1997). Transcriptional regulation of the Yersinia pseudotuberculosis pH 6 antigen adhesin by two envelope-associated components. Mol Microbiol 24, 499–510. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Merriam, J., Mueller, J. & Isberg, R. (1996). The psa locus is responsible for thermoinducible binding of Yersinia pseudotuberculosis to cultured cells. Infect Immun 64, 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., Ellis, H. M., Lee, E.-C., Jenkins, N. A., Copeland, N. G. & Court, D. L. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zav'yalov, V. P., Abramov, V. M., Cherepanov, P. G., Spirina, G. V., Chernovskaya, T. V., Vasiliev, A. M. & Zav'yalova, G. A. (1996). pH 6 antigen (PsaA protein) of Yersinia pestis, a novel bacterial Fc-receptor. FEMS Immunol Med Microbiol 14, 53–57. [DOI] [PubMed] [Google Scholar]
- Zavialov, A. V., Berglund, J., Pudney, A. F., Fooks, L. J., Ibrahim, T. M., MacIntyre, S. & Knight, S. D. (2003). Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113, 587–596. [DOI] [PubMed] [Google Scholar]