Abstract
Chlamydia trachomatis is an obligate intracellular pathogen that replicates within a parasitophorous vacuole termed an inclusion. The chlamydial inclusion is isolated from the endocytic pathway but fusogenic with Golgi-derived exocytic vesicles containing sphingomyelin and cholesterol. Sphingolipids are incorporated into the chlamydial cell wall and are considered essential for chlamydial development and viability. The mechanisms by which chlamydiae obtain eukaryotic lipids are poorly understood but require chlamydial protein synthesis and presumably modification of the inclusion membrane to initiate this interaction. A polarized cell model of chlamydial infection has demonstrated that chlamydiae preferentially intercept basolaterally directed, sphingomyelin-containing exocytic vesicles. Here we examine the localization and potential function of trans-Golgi and/or basolaterally associated soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins in chlamydia-infected cells. The trans-Golgi SNARE protein syntaxin 6 is recruited to the chlamydial inclusion in a manner that requires chlamydial protein synthesis and is conserved among all chlamydial species examined. The localization of syntaxin 6 to the chlamydial inclusion requires a tyrosine motif or plasma membrane retrieval signal (YGRL). Thus in addition to expression of at least two inclusion membrane proteins that contain SNARE-like motifs, chlamydiae also actively recruit eukaryotic SNARE-family proteins.
INTRODUCTION
Chlamydiae are significant human pathogens responsible for a number of distinct diseases. Chlamydia trachomatis comprises 15 serologically defined variants or serovars associated with diverse disease states including endemic blinding trachoma, sexually transmitted diseases, and a more invasive granulomatous disease, lymphogranuloma venereum (Schachter, 1999). Chlamydia psittaci causes zoonotic diseases that occasionally are transmitted to humans. Chlamydia pneumoniae contributes to the two to five million cases of respiratory pneumonia per year, although the actual incidence of C. pneumoniae-induced disease is unknown (Schachter, 1999).
Chlamydiae have evolved a unique biphasic developmental cycle. The infectious, metabolically dormant form, termed the elementary body (EB), is endocytosed by the host cell and remains within a vesicle termed the inclusion, where it differentiates into a metabolically active but non-infectious reticulate body. The inclusion membrane grows to accommodate the increasing number of organisms, while allowing the organisms to acquire essential amino acids, nucleotides and lipids from the host cell (Hackstadt et al., 1995; Hatch, 1975a, b; McClarty, 1994; Moulder, 1991; Wylie et al., 1997). A fundamental question of chlamydial biology relates to the mechanisms that allow the inclusion to create a unique intracellular organelle permitting survival and replication of the parasite.
Upon infection, the nascent inclusion membrane surrounding the infectious EB is plasma membrane derived, but within a few hours, chlamydial type III secreted proteins modify the inclusion membrane (Fields et al., 2003; Rockey et al., 1995, 2002; Shaw et al., 2000). These modifications are evidenced by the initiation of a number of interactions with the host cell, including dynein-dependent trafficking to the microtubule-organizing centre (Clausen et al., 1997; Grieshaber et al., 2003), and separation of the inclusion from the classical endosomal pathway, including restricted fusion with lysosomes (Al-Younes et al., 1999; Fields & Hackstadt, 2002; Hackstadt, 1999; Taraska et al., 1996; van Ooij et al., 1997; Wyrick, 2000), and fusion with Golgi-derived vesicles delivering sphingomyelin and cholesterol to the developing chlamydiae (Carabeo et al., 2003; Hackstadt et al., 1996; Scidmore et al., 1996b).
The properties of lipid acquisition suggest that this trafficking is vesicular in nature (Hackstadt, 1999, Carabeo et al., 2003; Hackstadt et al., 1996; Scidmore et al., 1996b). The specificity of this trafficking only to the chlamydial inclusion (Heinzen et al., 1996), a requirement for chlamydial modification of the inclusion membrane (Scidmore et al., 1996b), and the lack of disruption of normal Golgi processing and export of protein (Scidmore et al., 1996a) suggest a unique trafficking pathway. The acquisition of sphingomyelin, but not glucosylceramide, by chlamydiae further implies specificity of this lipid-trafficking pathway (Moore et al., 2008). Development of a polarized epithelial cell model of chlamydial infection demonstrated that in chlamydia-infected polarized cells, the sphingomyelin retained by the chlamydiae is derived predominantly from the basolateral trafficking pathway, indicating that the chlamydial inclusion preferentially intercepts Golgi-derived, basolaterally targeted exocytic vesicles (Moore et al., 2008). This finding has led us to focus on proteins that govern fusion along basolateral trafficking pathways. Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins constitute the predominant mechanism of membrane fusion (Parlati et al., 2002). In this study we examine syntaxins, a family of SNARE proteins, which are associated with trans-Golgi and basolaterally directed membrane fusion events. We demonstrate a specific interaction between syntaxin 6, a trans-Golgi SNARE, and the chlamydial inclusion membrane.
METHODS
Organisms and cell culture.
HeLa 229 cells [American Type Culture Collection (ATCC; Manassas, VA; CCL-2.1)], cultivated in RPMI 1640 (Gibco-BRL) supplemented with 10 % fetal bovine serum (FBS) (Hyclone) and 10 μg gentamicin ml−1 (Gibco-BRL), were used to propagate Chlamydia trachomatis serovar L2 (LGV 434), C. muridarum (MoPn/Weiss strain), C. pneumoniae (AR-39) and C. psittaci (caviae) GPIC (HC/BW). Infectious EBs were purified from HeLa cells using a Renografin (Braco Diagnostics) gradient, as described by Caldwell et al. (1981). Chlamydial titres were determined as described by Furness et al. (1960), by utilizing indirect immunofluorescence with a polyclonal rabbit anti-C. trachomatis L2 EB, followed by an anti-rabbit Alexa Fluor-conjugated secondary antibody (Molecular Probes). Multiplicities of infection (m.o.i.) for all experiments are based on inclusion-forming units (i.f.u.) determined in HeLa cells. Coxiella burnetii Nine Mile phase II was propagated and purified from Vero cells (ATCC; CCL-81) as previously described (Hackstadt et al., 1992).
HeLa and C2BBe1 cell lines were cultured at 37 °C in 5 % CO2. C2BBe1 (ATCC CRL-2102) cells were cultivated in DMEM+2 mM GlutaMax (Invitrogen) supplemented with 10 % FBS, 4 mM l-glutamine, 0.01 mg human transferrin ml−1 (Invitrogen) and 10 μg gentamicin ml−1. All eukaryotic cells were passaged based on ATCC-suggested protocols using a 0.25 % trypsin, 0.53 mM EDTA solution (ATCC).
Examination of localization of syntaxin 6 to the chlamydial inclusion
Endogenous syntaxins.
To examine localization of endogenous syntaxins to the chlamydial inclusion, C2BBe1 cells were seeded onto glass coverslips in 24-well plates, 48 h prior to infection with C. trachomatis L2. At 18 h post-infection, cells were fixed in absolute ethanol at −20 °C for 30 min. Samples were then processed for indirect immunofluorescence using rabbit anti-IncG (inclusion membrane protein), mouse anti-syntaxin 4 (BD Biosciences), mouse anti-syntaxin 16 (Synaptic Systems) or mouse anti-syntaxin 6 (BD Biosciences). All secondary antibodies were conjugated to Dylight Fluors and obtained from Jackson ImmunoResearch Laboratories. Coverslips were mounted to slides using ProLong Gold antifade reagent (Invitrogen). Samples were visualized with an LSM 510 Laser Module Zeiss Axiovert 200M confocal microscope (Carl Zeiss MicroImaging).
eGFP-syntaxin 6.
To examine the localization of eGFP-syntaxin 6 (kindly proved by Jeffrey Pessin, Albert Einstein College of Medicine, Bronx, NY, USA) (Watson & Pessin, 2000) to chlamydial inclusions, C2BBe1 cells were diluted and plated onto glass coverslips in 24-well plates the day before the transfection. DNA was diluted to 250 ng per 100 μl of Optimem (Invitrogen), and the PLUS and Lipofectimine-LTX reagents (Invitrogen) were used according to the manufacturer's protocol. Cells were incubated with the DNA–lipid complexes for a minimum of 4 h prior to recovery in culture medium. Cells were then infected with either C. trachomatis L2, C. muridarum or C. caviae for an additional 18 h prior to fixation in 3 % paraformaldehyde, permeabilized with 0.1 % Triton X-100 and 0.5 % SDS in PBS, and processed for indirect immunofluorescence to detect intracellular bacteria. To examine localization of eGFP-syntaxin 6 to C. pneumoniae or Coxiella burnetii Nine Mile Phase II, cells were infected for 36–72 h prior to transfection with the eGFP-syntaxin 6 construct. An antibody made in rabbits against whole paraformaldehyde-fixed C. trachomatis serovar L2 or C. caviae EBs was used to detect C. trachomatis serovar L2 and C. muridarum, or C. caviae, respectively. Detection of C. burnetii was achieved using an antibody against whole paraformaldehyde-fixed organisms raised in rabbits. A mouse monoclonal antibody raised against C. pneumoniae was kindly provided by Harlan Caldwell (NIAID, Rocky Mountain Laboratories, Hamilton, MT, USA).
3XFLAG-syntaxin 6 wild-type and mutants.
To examine which domain of syntaxin 6 is involved in localizing the protein to the chlamydial inclusion, eGFP-syntaxin 6 (Watson & Pessin, 2000) was used as a template to make the following syntaxin 6 deletion constructs: ΔH1 (helical domain, encoding amino acids 47–71), ΔH2 (helical/SNARE domain, encoding amino acids 166–225) and ΔYGRL (tyrosine motif encoding amino acids 140–143). The GeneTailor Site-Directed Mutagenesis System (Invitrogen) was used in the production of all constructs, with the primers listed in Table 1. To complete the construction of the ΔH1 and ΔH2 syntaxin 6 mutants, PCR products were digested with HindIII (New England Biolabs), followed by ligation with T4 DNA ligase (New England Biolabs) and transformed into One-Shot MAX Efficiency DH5α-T1R (Invitrogen). All deletion constructs and wild-type syntaxin 6 were subcloned into p3XFLAG-CMV 7.1 expression vector (Sigma Aldrich) using Phusion High Fidelity Polymerase (New England Biolabs) and primers 7 and 8 (Table 1). All mutations were confirmed by sequencing (SeqWright). 3XFLAG-syntaxin 6 constructs were transformed into C2BBe1 cells as described above.
Table 1.
Primers used in cloning syntaxin 6 (stx6)
| Primer | Sequence | Purpose |
|---|---|---|
| 1 | for 5′-GCTGGAGTGACGGATCGAGACCGGGAGC-3′ | Delete YGRL sequence from pEGFP stx6 |
| 2 | rev 5′-TCGATCCGTCACTCCAGCATCCCAATTCTGGC-3′ | |
| 3 | for 5′-GGGGGAAGCTTGGTCCAGTCGATCTCTTCCC-3′ | Delete H1 domain from pEGFP stx6 |
| 4 | rev 5′-GGGGGAAGCTTGAAGCAATCCTAGAAAATTCAACC-3′ | |
| 5 | for 5′-GGGGGAAGCTTTCTCACATGACCAGTGATCGG-3′ | Delete H2 domain from pEGFP stx6 |
| 6 | rev 5′-GGGGGAAAGCTTAATCAACTGCTGCTGTGCCTG-3′ | |
| 7 | for 5′-GGGGAATTCAATGTCCATGGAGGACCCC-3′ | Clone stx6 constructs into p3XFLAG-CMV 7.1 expression vector |
| 8 | rev 5′-TCTAGATCCGGTGGATCCCGGGCCCGCGG-3′ |
mCherry-syntaxin 6.
Syntaxin 6 was subsequently cloned into mCherry (Clontech) and transfected into HeLa cells as described above.
RESULTS
Co-localization of syntaxin 6 with the chlamydial inclusion
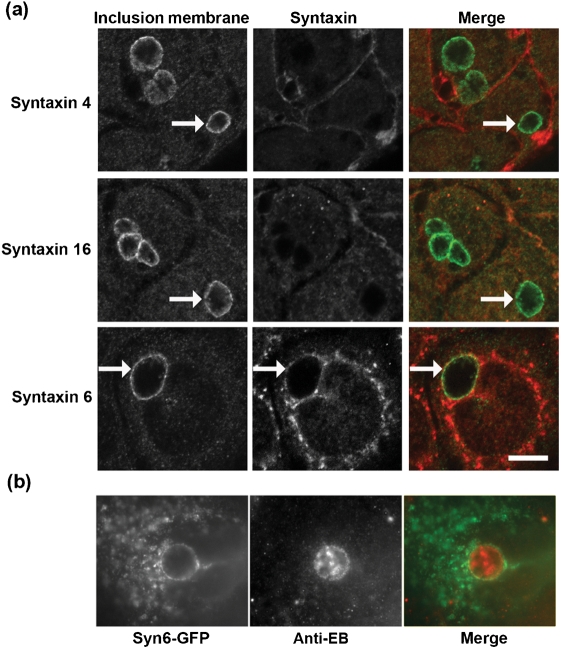
C. trachomatis serovar L2 obtains sphingomyelin from a Golgi-derived basolaterally targeted pathway via an unknown mechanism (Moore et al., 2008). Given the integral role of SNARE proteins in mediating vesicular trafficking, we examined whether syntaxin family members co-localized with the chlamydial inclusion in C. trachomatis-infected C2BBe1 cells by indirect immunofluorescence. The trans-Golgi-associated syntaxin 6 was found to associate with the chlamydial inclusion (Fig. 1). Syntaxin 6 displays a distinct ring-like staining pattern around the chlamydial inclusion similar to the inclusion membrane protein IncG. Syntaxin 4, known to control fusion to the basolateral plasma membrane (Low et al., 1996; Teng et al., 2001), and syntaxin 16, a ubiquitious trans-Golgi-associated syntaxin which mediates retrograde endosomal-Golgi transport (Mallard et al., 2002), did not colocalize with the chlamydial inclusion but can be observed on the plasma membrane (Fig. 1a).
Fig. 1.
Localization of endogenous syntaxin proteins to the chlamydial inclusion. (a) C2BBe1 cells were seeded onto glass coverslips for 48 h prior to infection with C. trachomatis serovar L2 (m.o.i. 6 : 1) for an additional 18 h. Cells were fixed in absolute ethanol for 30 min at −20 °C and processed essentially as described in Methods. Samples were visualized with an LSM 510 Laser Module Zeiss Axiovert 200M confocal microscope. Arrows indicate chlamydial inclusions. (b) C2BBe1 cells were transfected with eGFP-syntaxin 6 and infected with C. trachomatis L2 for 18 h. Cells were fixed with methanol and counterstained with an anti-EB antiserum. Bars, 10 μm.
To confirm this interaction, HeLa cells were transiently transfected with eGFP-syntaxin 6 and infected with C. trachomatis. Syntaxin 6 was found to localize to the chlamydial inclusion membrane (Fig. 1b). These findings suggest that syntaxin 6 is specifically recruited to the chlamydial inclusion membrane.
Syntaxin 6 recruitment is conserved among chlamydia species
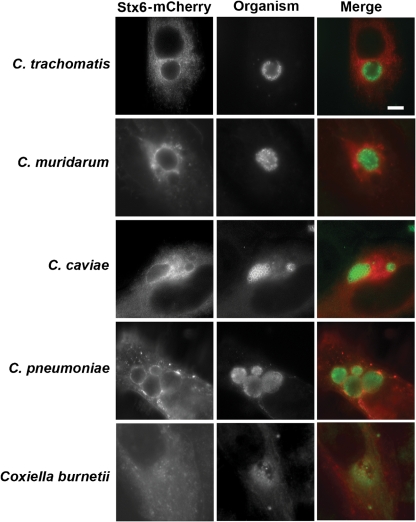
Because sphingomyelin trafficking to the inclusion is a conserved feature among chlamydial species (Hackstadt et al., 1995; Rockey et al., 1996; Wolf & Hackstadt, 2001), we next examined whether mCherry-syntaxin 6 would colocalize to the inclusion membrane of other Chlamydia species. mCherry-syntaxin 6 was recruited to inclusions formed by C. trachomatis serovar L2, C. muridarum, C. pneumoniae and C. caviae (Fig. 2). Syntaxin 6 localized to the inclusion membrane of C. muridarum and C. pneumoniae inclusions in a similar manner as seen for C. trachomatis serovar L2 (Fig. 2). Syntaxin 6 clustered amongst the multiple lobed inclusions formed by C. caviae; however, the morphology was distinct from the ring-like pattern of syntaxin 6 surrounding the inclusions of chlamydial species that form a single inclusion within the host cell. Syntaxin 6 was not recruited to the parasitophorous vacuole formed by the unrelated intracellular bacterium, Coxiella burnetii Nine Mile Phase II (Fig. 2). The interaction of syntaxin 6 with the chlamydial inclusion therefore appears to be chlamydia-specific and is conserved across chlamydial species.
Fig. 2.
Localization of syntaxin 6 to the inclusions of multiple chlamydial species. C2BBe1 cells were seeded onto coverslips and transfected with mCherry-syntaxin 6 or with eGFP-syntaxin 6, then infected with C. trachomatis serovar L2 (m.o.i. 6 : 1), C. muridarum (m.o.i. 0.1 : 1), C. caviae (m.o.i. 0.2 : 1), C. pneumoniae (m.o.i. 13 : 1) or Coxiella burnetii Nile Mile phase II (m.o.i. 50 : 1) as described in Methods. To terminate the infections, cells were fixed in methanol and processed for indirect immunofluorescence to detect the organisms (green). Samples were visualized with an LSM 510 Laser Module Zeiss Axiovert 200M confocal microscope. Bar, 10 μm.
Requirement of chlamydial protein synthesis for syntaxin 6 colocalization
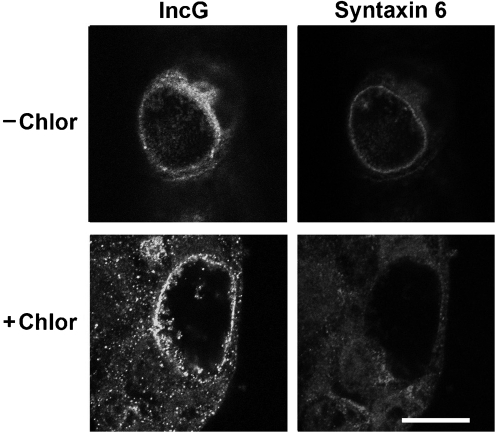
Because sphingomyelin trafficking to the chlamydial inclusion requires chlamydial protein synthesis (Hackstadt et al., 1996; Scidmore et al., 1996b, 2003), we examined whether chlamydial protein synthesis was also required for syntaxin 6 colocalization. C2BBe1 cells were infected with C. trachomatis and allowed to develop for 18 h, then treated with chloramphenicol for an additional 24 h (Fig. 3). C2BBe1 cells fixed at 18 h post-infection displayed the characteristic ring-like pattern of syntaxin 6 localization to the inclusion membrane. However, no inclusion-membrane-associated staining of syntaxin 6 was visible in chloramphenicol-treated cells (Fig. 3). These results suggest that syntaxin 6 localization to the inclusion and retention at the inclusion membrane are likely mediated by a chlamydial protein. Treatment of C. trachomatis-infected cells at 18 h post-infection with brefeldin A, which collapses the Golgi (Lippincott-Schwartz et al., 1989), or nocodozole, which disrupts microtubules and fragments the Golgi apparatus (Cheung & Terry, 1980; Tassin et al., 1985), did not inhibit syntaxin 6 retention or recruitment to the inclusion membrane (data not shown).
Fig. 3.
Chlamydial protein synthesis requirement for syntaxin 6 localization to the inclusion. C2BBe1 cells were seeded onto glass coverslips in 24-well plates 48 h prior to infection with C. trachomatis serovar L2 (m.o.i. 9 : 1). After 18 h, cells were either fixed in absolute ethanol (−Chlor) or treated with 200 μg ml−1 for an additional 24 h (+Chlor), then fixed in absolute ethanol and processed for indirect immunofluorescence essentially as described in Methods. IncG staining was used to identify the inclusion. Samples were visualized with an LSM 510 Laser Module Zeiss Axiovert 200M confocal microscope. Bar, 10 μm.
Characterization of the syntaxin 6 domain responsible for localization to the chlamydial inclusion
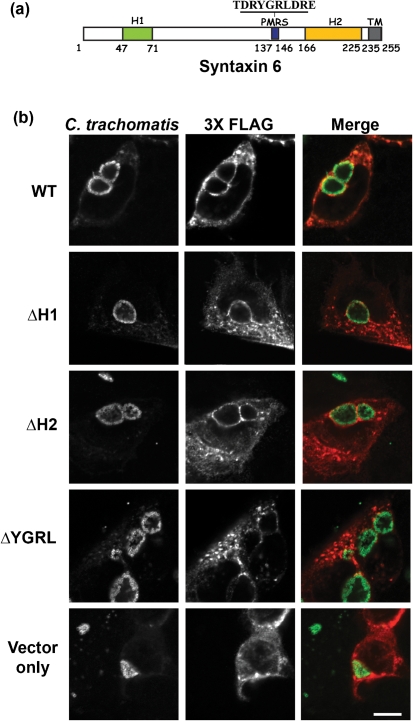
Syntaxin 6 contains two helical domains (H1 and H2) and a plasma membrane retrieval signal (PMRS) which are required for syntaxin 6 cycling and function in eukaryotic cells (Fig. 4a). The H1 domain resides within the N-terminus of syntaxin 6 and is responsible for protein–protein interactions as characterized by its interaction with α-SNAP (Bock et al., 2001). The H2 domain resides proximal to the C-terminal transmembrane domain and comprises the Q-SNARE activity, which is similar to the SNARE activity of SNAP25 (Bock et al., 2001; Watson & Pessin, 2000). Within the C-terminal portion of syntaxin 6 is a 10 amino acid domain, which is characterized as the PMRS, with residues YGRL being absolutely required for activity (Watson & Pessin, 2000). Because syntaxin 6 is trafficked within vesicles from the Golgi apparatus to the plasma membrane, the YGRL signal sequence is required for the recycling of syntaxin 6 from the plasma membrane to the trans-Golgi region. Deletion of this domain causes an accumulation of syntaxin 6 in the plasma membrane (Watson & Pessin, 2000). Both the H2 and PMRS domains contribute to the trans-Golgi localization of syntaxin 6 . To understand which signalling or protein–protein binding domains facilitate the trafficking of syntaxin 6 to the chlamydial inclusion, we constructed a series of syntaxin 6 mutants from which the H1, H2 or PMRS domains from syntaxin 6 were deleted. Subsequently, C2BBe1 cells were transfected with 3XFLAG wild-type and mutant syntaxin 6 constructs and their subcellular localization relative to C. trachomatis serovar L2 inclusions was examined (Fig. 4b). Deletion of either the H1 or H2 domain did not have any appreciable effect on syntaxin 6 localization to the chlamydial inclusion. However, deletion of the PMRS domain resulted in loss of recruitment of syntaxin 6 to the chlamydial inclusion membrane (Fig. 4b). These results suggest that the PMRS may be acting as signal sequence which targets eukaryotic proteins to the chlamydial inclusion, and/or that the chlamydial inclusion membrane may be mimicking the trans-Golgi membrane.
Fig. 4.
Identification of the syntaxin 6 protein domain mediating localization to the chlamydial inclusion. (a) Functional domains of syntaxin 6. These include the 24 amino acid H1 domain, the 10 amino acid plasma membrane retrieval signal (PMRS), including the YGRL tyrosine motif and the 59 amino acid H2 domain. Also depicted is the C-terminal transmembrane domain (TM); this domain anchors syntaxin 6 in vesicular membranes (Wendler & Tooze, 2001). (b) Examination of the involvement of syntaxin 6 functional domains in localization to the chlamydial inclusion. C2BBe1 cells were transfected with the indicated 3XFLAG-syntaxin 6 (syn6) constructs, followed by infection with C. trachomatis serovar L2 (m.o.i. 4 : 1). At 18 h post-infection, cells were fixed in absolute ethanol and processed for indirect immunofluorescence. Chlamydial inclusions were labelled with an antibody that recognizes the inclusion membrane (IncG), and the syntaxin 6 constructs were detected with an anti-M2 FLAG tag antibody (Sigma-Aldrich). Slides were visualized with an LSM 510 Laser Module Zeiss Axiovert 200M confocal microscope. Bar, 10 μm.
DISCUSSION
Recent studies have indicated that the chlamydial inclusion preferentially intercepts sphingomyelin from a basolaterally directed pathway (Moore et al., 2008). The recognition of a specific pathway targeted by chlamydiae stimulated a search for host proteins that may serve to regulate this pathway. Syntaxin 6 is recruited to the chlamydial inclusion in a process that requires chlamydial protein synthesis and is conserved across chlamydial species. Interestingly, the PMRS signal of syntaxin 6 is required for syntaxin 6 colocalization to the inclusion.
Vesicle fusion in eukaryotic cells is of necessity a highly regulated process designed to maintain the integrity and distinction of intracellular compartments. Specificity in vesicle fusion with target membranes is conferred by integral membrane proteins termed v-SNAREs and t-SNAREs for vesicle- and target-specific soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) receptors, respectively (Rothman & Wieland, 1996). Binding of t-SNAREs with v-SNAREs on opposing membrane faces causes the vesicles to dock with the acceptor membrane. v-SNAREs and t-SNAREs have more recently been termed Q-SNAREs and R-SNAREs, respectively, based on conserved amino acids within the SNARE protein fusion complex. Once the complex is formed, two soluble proteins, N-ethylmaleimide-sensitive factor (NSF) and SNAP, bind the complex. Subsequent ATP hydrolysis by NSF promotes actual membrane fusion. Several other host proteins are also involved in regulation of vesicle trafficking. Specific small GTPases of the Rab family are localized to the surface of the various compartments of the endocytic and exocytic pathways where, depending upon the concentration of the GTP-bound state, they positively or negatively regulate the rates of SNARE complex assembly and membrane fusion (Novick & Zerial, 1997; Schimmöller et al., 1998).
Microbial manipulation of host SNARE machinery is an emerging theme in cellular microbiology. Mycobacterium tuberculosis-containing vacuoles (MCVs) transiently acquire syntaxin 3, acquire and retain syntaxins 4 and 8, but exclude syntaxin 6 (Fratti et al., 2003; Parlati et al., 2002; Perskvist et al., 2002). Each step of syntaxin acquisition or exclusion marks a critical step in the MCV maturation from a plasma-membrane-derived vacuole. Simlarly, syntaxin 13 appears to play a role in maturation of the Salmonella-containing vacuole (Smith et al., 2005). Virulent Legionella pneumophila require syntaxins 2, 3 and 4 for proper vacuolar biogenesis and fusion with endoplasmic-reticulum-derived vesicles (Arasaki & Roy, 2010). As shown here, C. trachomatis excludes syntaxins 4 and 16, but recruits syntaxin 6, a trans-Golgi SNARE protein. While the SNARE domain of syntaxin 6 is not involved in localizing the protein to the chlamydial inclusion, we hypothesize that the SNARE domain plays an important role at the inclusion membrane and may mediate specific vesicle fusion events. The role(s) of syntaxin 6, however, remain undefined as siRNA depletion did not dramatically diminish inclusion development or trafficking of sphingomyelin to the inclusion (data not shown).
Chlamydiae are known to extensively modify the inclusion membrane very early in infection by the insertion of type III secreted intrinsic membrane proteins collectively known as Incs. The Inc proteins show little similarity to known host proteins but display a predicted, bi-lobed hydrophobic domain approximately 40 amino acids in length. C. trachomatis encodes up to 50 Inc proteins (Rockey et al., 2002; Shaw et al., 2000). In addition to recruiting eukaryotic SNARE proteins to the inclusion membrane, Inc proteins may mimic eukaryotic SNAREs. Computer modelling has identified two chlamydial proteins, IncA and CT813, as having SNARE-like domains (Delevoye et al., 2004, 2008). Additionally, in vitro studies utilizing reconstituted liposomes with recombinant proteins have found that IncA can bind the SNARE protein vamp 3 in vitro (Delevoye et al., 2008). Interestingly, vamp 3 operates along a basolateral trafficking pathway in polarized epithelial cells (Pocard et al., 2007).
C. trachomatis IncA is required for homotypic vesicle fusion of multiple C. trachomatis inclusions within the same cell. Other chlamydia species also carry genes annotated as IncA; however, C. caviae IncA does not mediate fusion with other inclusions formed by C. caviae or other chlamydial species. It is likely that host factors are also required for C. trachomatis homotypic vesicle fusion (Delevoye et al., 2008). In addition to promoting fusion of C. trachomatis inclusions, IncA has also been proposed to act as an inhibitory SNARE by blocking specific SNARE-mediated membrane fusion events in vitro (Paumet et al., 2009). Interestingly, C. trachomatis IncA had no inhibitory effect on exocytic complexes examined but was specific for endocytic SNAREs (Paumet et al., 2009). This is particularly relevant since chlamydial inclusions are non-fusogenic with endocytic compartments but are believed to intercept sphingolipids and cholesterol from exocytic vesicles.
Several Rab-family GTPases are also recruited to the chlamydial inclusion membrane but not in patterns common throughout the genus. For example, Rab1, Rab4, Rab11 and Rab14 are recruited to C. trachomatis, C. muridarum and C. pneumoniae inclusions. Rab 6 is recruited to C. trachomatis exclusively, however, and Rab10 is recruited only to C. muridarum and C. pneumoniae inclusions (Brumell & Scidmore, 2007; Rzomp et al., 2003). The endocytic Rabs, Rab5, Rab7 and Rab9, are excluded from the chlamydial inclusion. Rab4 recruitment to C. trachomatis inclusions is mediated by Inc229 (Rzomp et al., 2006) whereas C. pneumoniae Cpn585 interacts with Rabs 1, 10 and 11, but not Rab 4 (Cortes et al., 2007). How this combination of Rab proteins associated with different compartments and pathways affects inclusion membrane fusion events remains to be fully defined, although recent studies have implicated a role in mediating the phosphoinositide composition of the inclusion membrane (Moorhead et al., 2010).
Syntaxin 6 is recruited to the chlamydial inclusion membrane protein by a mechanism that requires both an unknown chlamydial protein, presumably one localized to the inclusion membrane, and a plasma membrane retrieval signal on syntaxin 6. SNARE complexes consist of three proteins, which overall contribute three Q-SNARE motifs and one R-SNARE motif (Fasshauer et al., 1998). In a typical SNARE complex, a syntaxin supplies one Q-SNARE motif, a vamp supplies the single R-SNARE motif, and cytosolic SNAP 23 supplies the additional two Q-SNARE motifs (Sutton et al., 1998). SNARE proteins, such as vamps and syntaxins, remain membrane bound whether or not they are found within a complex, and control fusion events within distinct subcellular compartments (Teng et al., 2001). An improved understanding of the chlamydial proteins involved in conferring specificity to the cellular interactions of the chlamydial inclusion is critical to elucidating chlamydial pathogenesis.
Acknowledgments
This work was supported by the Intramural Research Program of the NIAID/NIH. We thank Dr Jeffrey Pessin (Albert Einstein College of Medicine, Bronx, NY) for the eGFP-syntaxin 6 construct and Dr Harlan Caldwell (NIAID Rocky Mountain Laboratories, Hamilton, MT) for the antibody against C. pneumoniae. We would also like to thank Tina Clark for excellent technical assistance.
Abbreviations
EB, elementary body
FBS, fetal bovine serum
PMRS, plasma membrane retrieval signal
SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor
References
- Al-Younes, H. M., Rudel, T. & Meyer, T. F. (1999). Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell Microbiol 1, 237–247. [DOI] [PubMed] [Google Scholar]
- Arasaki, K. & Roy, C. R. (2010). Legionella pneumophila promotes functional interactions between plasma membrane syntaxins and Sec22b. Traffic 11, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, J. B., Matern, H. T., Peden, A. A. & Scheller, R. H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839–841. [DOI] [PubMed] [Google Scholar]
- Brumell, J. H. & Scidmore, M. A. (2007). Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol Mol Biol Rev 71, 636–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, H. D., Kromhout, J. & Schachter, J. (1981). Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31, 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo, R. A., Mead, D. J. & Hackstadt, T. (2003). Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc Natl Acad Sci U S A 100, 6771–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, H. T. & Terry, D. S. (1980). Effects of nocodazole, a new synthetic microtubule inhibitor, on movement and spreading of mouse peritoneal macrophages. Cell Biol Int Rep 4, 1125–1129. [DOI] [PubMed] [Google Scholar]
- Clausen, J. D., Christiansen, G., Holst, H. U. & Birkelund, S. (1997). Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol Microbiol 25, 441–449. [DOI] [PubMed] [Google Scholar]
- Cortes, C., Rzomp, K. A., Tvinnereim, A., Scidmore, M. A. & Wizel, B. (2007). Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases. Infect Immun 75, 5586–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye, C., Nilges, M., Dautry-Varsat, A. & Subtil, A. (2004). Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J Biol Chem 279, 46896–46906. [DOI] [PubMed] [Google Scholar]
- Delevoye, C., Nilges, M., Dehoux, P., Paumet, F., Perrinet, S., Dautry-Varsat, A. & Subtil, A. (2008). SNARE protein mimicry by an intracellular bacterium. PLoS Pathog 4, e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer, D., Sutton, R. B., Brunger, A. T. & Jahn, R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, K. A. & Hackstadt, T. (2002). The chlamydial inclusion: escape from the endocytic pathway. Annu Rev Cell Dev Biol 18, 221. [DOI] [PubMed] [Google Scholar]
- Fields, K. A., Mead, D., Dooley, C. A. & Hackstadt, T. (2003). Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol Microbiol 48, 671–683. [DOI] [PubMed] [Google Scholar]
- Fratti, R. A., Chua, J., Vergne, I. & Deretic, V. (2003). Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci U S A 100, 5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness, G., Graham, D. M. & Reeve, P. (1960). The titration of trachoma and inclusion blennorrhoea viruses in cell cultures. J Gen Microbiol 23, 613–619. [DOI] [PubMed] [Google Scholar]
- Grieshaber, S. S., Grieshaber, N. & Hackstadt, T. (2003). Chlamydia trachomatis uses host cell dynein to traffic to the microtube organizing center in a p50 dynamitin-independent process. J Cell Sci 116, 3793–3802. [DOI] [PubMed] [Google Scholar]
- Hackstadt, T. (1999). Cell Biology. In Chlamydia: Intracellular Biology, Pathogenesis, and Immunity, pp. 101–138. Edited by Stephens, R. S.. Washington, DC. : American Society for Microbiology.
- Hackstadt, T., Messer, R., Cieplak, W. & Peacock, M. G. (1992). Evidence for the proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect Immun 60, 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt, T., Scidmore, M. A. & Rockey, D. D. (1995). Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci U S A 92, 4877–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt, T., Rockey, D. D., Heinzen, R. A. & Scidmore, M. A. (1996). Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J 15, 964–977. [PMC free article] [PubMed] [Google Scholar]
- Hatch, T. P. (1975a). Utilization of L-cell nucleoside triphosphates by Chlamydia psittaci for ribonucleic acid synthesis. J Bacteriol 122, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, T. P. (1975b). Competition between Chlamydia psittaci and L cells for host isoleucine pools: a limiting factor in chlamydial multiplication. Infect Immun 12, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen, R. A., Scidmore, M. A., Rockey, D. D. & Hackstadt, T. (1996). Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 64, 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S. & Klausner, R. D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, S. H., Chapin, S. J., Weimbs, T., Kornuves, L. G., Bennett, M. K. & Mostov, K. E. (1996). Differential localization of syntaxin isoforms in polarized Madin-Darby canine kidney cells. Mol Biol Cell 7, 2007–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard, F., Tang, B. L., Galli, T., Tenza, D., Saint-Pol, A., Yue, X., Antony, C., Hong, W., Goud, B. & Johannes, L. (2002). Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol 156, 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarty, G. (1994). Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol 2, 157–164. [DOI] [PubMed] [Google Scholar]
- Moore, E. R., Fischer, E. R., Mead, D. J. & Hackstadt, T. (2008). The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic 9, 2130–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead, A. M., Jung, J. Y., Smirnov, A., Kaufer, S. & Scidmore, M. A. (2010). Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect Immun 78, 1990–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder, J. W. (1991). Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55, 143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, P. & Zerial, M. (1997). The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol 9, 496–504. [DOI] [PubMed] [Google Scholar]
- Parlati, F., Varlamov, O., Paz, K., McNew, J. A., Hurtado, D., Sollner, T. H. & Rothman, J. E. (2002). Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc Natl Acad Sci U S A 99, 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumet, F., Wesolowski, J., Garcia-Diaz, A., Delevoye, C., Aulner, N., Shuman, H. A., Subtil, A. & Rothman, J. E. (2009). Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS ONE 4, e7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perskvist, N., Roberg, K., Kulyte, A. & Stendahl, O. (2002). Rab5a GTPase regulates fusion between pathogen-containing phagosomes and cytoplasmic organelles in human neutrophils. J Cell Sci 115, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Pocard, T., Bivic, A. L., Galli, T. & Zurzolo, C. (2007). Distinct v-SNAREs regulate direct and indirect apical delivery in polarized epithelial cells. J Cell Sci 120, 3309–3320. [DOI] [PubMed] [Google Scholar]
- Rockey, D. D., Heinzen, R. A. & Hackstadt, T. (1995). Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol 15, 617–626. [DOI] [PubMed] [Google Scholar]
- Rockey, D. D., Fischer, E. R. & Hackstadt, T. (1996). Temporal analysis of the developing Chlamydia psittaci inclusion by use of fluorescence and electron microscopy. Infect Immun 64, 4269–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey, D. D., Scidmore, M. A., Bannantine, J. P. & Brown, W. J. (2002). Proteins in the chlamydial inclusion membrane. Microbes Infect 4, 333–340. [DOI] [PubMed] [Google Scholar]
- Rothman, J. E. & Wieland, F. T. (1996). Protein sorting by transport vesicles. Science 272, 227–234. [DOI] [PubMed] [Google Scholar]
- Rzomp, K. A., Scholtes, L. D., Briggs, B. J., Whittaker, G. R. & Scidmore, M. A. (2003). Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect Immun 71, 5855–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzomp, K. A., Moorhead, A. R. & Scidmore, M. A. (2006). The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect Immun 74, 5362–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter, J. (1999). Infection and disease epidemiology. In Chlamydia; Intracellular Biology, Pathogenesis, and Immunity, pp. 139–169. Edited by Stephens, R. S.. Washington, DC. : American Society for Microbiology.
- Schimmöller, F., Simon, I. & Pfeffer, S. R. (1998). Rab GTPases, directors of vesicle docking. J Biol Chem 273, 22161–22164. [DOI] [PubMed] [Google Scholar]
- Scidmore, M. A., Fischer, E. R. & Hackstadt, T. (1996a). Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J Cell Biol 134, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore, M. A., Rockey, D. D., Fischer, E. R., Heinzen, R. A. & Hackstadt, T. (1996b). Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun 64, 5366–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore, M. A., Fischer, E. R. & Hackstadt, T. (2003). Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun 71, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, E. I., Dooley, C. A., Fischer, E. R., Scidmore, M. A., Fields, K. A. & Hackstadt, T. (2000). Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37, 913–925. [DOI] [PubMed] [Google Scholar]
- Smith, A. C., Cirulis, J. T., Casanova, J. E., Scidmore, M. A. & Brumell, J. H. (2005). Interaction of the Salmonella-containing vacuole with the endocytic recycling system. J Biol Chem 280, 24634–24641. [DOI] [PubMed] [Google Scholar]
- Sutton, R. B., Fasshauer, D., Jahn, R. & Brunger, A. T. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Taraska, T., Ward, D. M., Ajioka, R. S., Wyrick, P. B., Davis-Kaplan, S. R., Davis, C. H. & Kaplan, J. (1996). The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect Immun 64, 3713–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin, A. M., Paintrand, M., Berger, E. G. & Bornens, M. (1985). The Golgi apparatus remains associated with microtubule organizing centers during myogenesis. J Cell Biol 101, 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, F. Y. H., Wang, Y. & Tang, B. L. (2001). The syntaxins. Genome Biol 2, REVIEWS3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij, C., Apodaca, G. & Engel, J. (1997). Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect Immun 65, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R. T. & Pessin, J. E. (2000). Functional cooperation of two independent targeting domains in syntaxin 6 is required for its efficient localization in the trans-Golgi network of 3T3L1 adipocytes. J Biol Chem 275, 1261–1268. [DOI] [PubMed] [Google Scholar]
- Wendler, F. & Tooze, S. (2001). Syntaxin 6: the promiscuous behavior of a SNARE protein. Traffic 2, 606–611. [DOI] [PubMed] [Google Scholar]
- Wolf, K. & Hackstadt, T. (2001). Sphingomyelin trafficking in Chlamydia pneumoniae-infected cells. Cell Microbiol 3, 145–152. [DOI] [PubMed] [Google Scholar]
- Wylie, J. L., Hatch, G. M. & McClarty, G. (1997). Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J Bacteriol 179, 7233–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, P. B. (2000). Intracellular survival by Chlamydia. Cell Microbiol 2, 275–282. [DOI] [PubMed] [Google Scholar]