Abstract
Development of β-lactam resistance, production of alginate and modulation of virulence factor expression that alters host immune responses are the hallmarks of chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. In this study, we propose that a co-regulatory network exists between these mechanisms. We compared the promoter activities of ampR, algT/U, lasR, lasI, rhlR, rhlI and lasA genes, representing the β-lactam antibiotic resistance master regulatory gene, the alginate switch operon, the las and rhl quorum-sensing (QS) genes, and the LasA staphylolytic protease, respectively. Four isogenic P. aeruginosa strains, the prototypic Alg− PAO1, Alg− PAOampR, the mucoid Alg+ PAOmucA22 (Alg+ PDO300) and Alg+ PAOmucA22ampR (Alg+ PDOampR) were used. We found that in the presence of AmpR regulator and β-lactam antibiotic, the extracytoplasmic function sigma factor AlgT/U positively regulated PampR, whereas AmpR negatively regulated PalgT/U. On the basis of this finding we suggest the presence of a negative feedback loop to limit algT/U expression. In addition, the functional AlgT/U caused a significant decrease in the expression of QS genes, whereas loss of ampR only resulted in increased PlasI and PlasR transcription. The upregulation of the las QS system is likely to be responsible for the increased lasA promoter and the LasA protease activities in Alg− PAOampR and Alg+ PDOampR. The enhanced expression of virulence factors in the ampR strains correlated with a higher rate of Caenorhabditis elegans paralysis. Hence, this study shows that the loss of ampR results in increased virulence, and is indicative of the existence of a co-regulatory network between β-lactam resistance, alginate production, QS and virulence factor production, with AmpR playing a central role.
INTRODUCTION
Pseudomonas aeruginosa, a ubiquitous, versatile saprophytic bacterium, is a major aetiological agent of nosocomial infections and the leading cause of mortality among patients with cystic fibrosis (CF) (Greenberg, 2000; Rahme et al., 1995). This Gram-negative bacillus is equipped with an impressive arsenal of virulence factors to resist host defence mechanisms, counteract antibacterial agents, circumvent nutrient deprivation and withstand harsh environmental changes (Govan & Harris, 1986; Lyczak et al., 2002; Pedersen, 1992). One distinctive feature of P. aeruginosa lung isolates of patients with advanced CF is that a higher proportion of them are mucoid as compared to those from other sites of infection (Doggett, 1969; Fick et al., 1992). This mucoid phenotype is the result of an overproduction of the exopolysaccharide alginate (Evans & Linker, 1973). The activation of genes for alginate overproduction occurs primarily through the deregulation of algT/U or its product, σ22, a member of the extracytoplasmic function (ECF) sigma factors (DeVries & Ohman, 1994; Hershberger et al., 1995; Martin et al., 1993). Genomic, proteomic and microarray analyses have shown that AlgT/U regulates a diverse group of genes, ranging from extracellular proteases, periplasmic proteins like DsbA and intracellular enzymes (Firoved et al., 2002; Firoved & Deretic, 2003; Malhotra et al., 2000). Mucoid P. aeruginosa isolates from CF patients frequently have a defective mucA allele, a gene downstream of algT/U (Martin et al., 1993). The mucA gene product is an anti-sigma factor that negatively regulates the activity of AlgT/U (Hughes & Mathee, 1998).
P. aeruginosa is intrinsically resistant to most β-lactam antibiotics. One of the factors contributing to the resistance is the existence of enzymes that can deactivate β-lactams, known as β-lactamases (Kong et al., 2010; Rolinson, 1998). Two inducible chromosome-encoded β-lactamases, AmpC and PoxB (Oxa-50), have been identified in P. aeruginosa (Girlich et al., 2004; Kong et al., 2005a; Lodge et al., 1990). Expression of the ampC and poxB genes is tightly controlled by AmpR, a global LysR-like transcriptional regulator (Kong et al., 2005b). In addition, inactivation of ampR in the prototypic non-mucoid PAO1 (henceforth referred to as Alg− PAO1) resulted in high constitutive production of β-lactamases and pyocyanin, increased LasA staphylolytic protease activity and decreased LasB elastase activity (Kong et al., 2005b).
The production of virulence factors in P. aeruginosa is under the control of quorum-sensing (QS) systems mediated by diffusible chemical signalling molecules such as acylhomoserine lactones and quinolones. P. aeruginosa has three QS systems – las, rhl and Pseudomonas quinolone system that controls many virulence mechanisms (Ng & Bassler, 2009). Transcriptome studies have led to the identification of a large number of virulence factors that are under QS regulation in P. aeruginosa, these include proteases and toxins (Hentzer et al., 2003; Schuster & Greenberg, 2006; Wagner et al., 2004).
It has long been established that the production of proteases is inversely correlated with alginate production (Mathee et al., 1999; Mohr et al., 1990; Ohman & Chakrabarty, 1982). Previous comparison of Alg− PAO1 and its isogenic ampR mutant strain, Alg− PAOampR, showed differential regulation of virulence factors, including the las QS system (Kong et al., 2005b). In the present study, we sought to understand the regulatory network between alginate production, protease activity, β-lactam resistance and QS in P. aeruginosa. We hypothesized that AmpR may be differentially regulated in alginate-producing strains with consequent effects on the protease activities. To address this, ampR was inactivated in an alginate constitutive producer, Alg+ PDO300, generating an Alg+ PDOampR mutant strain. This mutant produced exceedingly high levels of β-lactamase, extracellular proteases and pyocyanin suggesting that AmpR either directly or indirectly suppresses the expression of many other virulence factors.
METHODS
Bacterial strains, plasmids and media.
Table 1 shows the bacterial strains, plasmids and primers used in this study. The bacterial strains of Escherichia coli and P. aeruginosa were routinely cultured in Luria–Bertani medium. Pseudomonas isolation agar (Difco) was used in triparental mating experiments for the selection of P. aeruginosa. Antibiotics, when used, were at the following concentrations unless indicated otherwise: ampicillin at 50 μg ml−1, tetracycline at 20 μg ml−1, gentamicin at 30 μg ml−1 for E. coli; and carbenicillin at 300 μg ml−1, gentamicin at 300 μg ml−1, tetracycline at 60 μg ml−1 for P. aeruginosa. For induction, 500 μg benzyl-penicillin ml−1 was used.
Table 1.
Bacterial strains, plasmids and primers used in this study
| Strain/plasmid | Genotype | Reference |
|---|---|---|
| E. coli | ||
| DH5α | F−φ80dlacZΔM15 Δ(lacZYA–argF) U169 deoR recA1 endA1 hsdR17(rk−, mk+) phoA supE44 λ- thi-1 gyrA96 relA1 | New England Biolabs |
| TOP10F′ | F′[lacIq, Tn10(TetR)] mcrA Δ(mrr-hsdRMS-mcrBC)φ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 D(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| P. aeruginosa | ||
| PAO1 | Prototype; Alg− | Holloway & Morgan (1986) |
| PDO300 | PAOmucA22; Alg+ | Mathee et al. (1999) |
| PKM805 | PAOmucA22algT24-1; Alg− | PDOalgT; PDO300 derivative; Ramos et al. (2003) |
| PKM300 | PAOampR : : aacCI; Alg− | PAOampR; Kong et al. (2005b) |
| PKM307 | PAOmucA22 ampR : : aacCI; Alg+ | PDOampR; this study |
| PKM308 | PAOmucA22 attB : : PampC–lacZ; TcR, Alg+ | PDO300 derivative; this study |
| PKM309 | PAOmucA22 attB : : PampR–lacZ; TcR, Alg+ | PDO300 derivative; this study |
| PKM310 | PAOmucA22 ampR : : aacCI attB : : PampC-lacZ; GmR, TcR, Alg+ | PDOampR derivative; this study |
| PKM311 | PAOmucA22 ampR : : aacCI attB : : PampR–lacZ; GmR, TcR, Alg+ | PDOampR derivative; this study |
| Plasmids | ||
| Mini-CTX-lacZ | TcR; integration-proficient vector for single-copy chromosomal lacZ fusion | Becher & Schweizer (2000) |
| pEX100T | ApR; sacB oriT | Schweizer & Hoang (1995) |
| pGEMEX-1 | ApR; ColE1ori lacZα | Promega |
| pKMG37 | ApR; pQF50 containing PalgT-lacZ transcriptional fusion | Mathee et al. (1997) |
| pLP170 | ApR; lacZ transcriptional fusion vector that contains an RNase III splice sequence positioned between the MCS and lacZ | Preston et al. (1997) |
| pLPLA | ApR; pLP170 containing PlasA-lacZ transcriptional fusion | Preston et al. (1997) |
| pLPLB | ApR; pLP170 containing PlasB-lacZ transcriptional fusion | Preston et al. (1997) |
| pLPR1 | ApR; pLP170 containing PrhlI-lacZ transcriptional fusion | Van Delden & Iglewski (1998) |
| pME6030 | TcR; oriVpVS1oriVp15AoriT | Heeb et al. (2000) |
| pPCS223 | ApR; pLP170 containing PlasI-lacZ transcriptional fusion | Van Delden & Iglewski (1998) |
| pPCS1001 | ApR; pLP170 containing PlasR-lacZ transcriptional fusion | Pesci et al. (1997) |
| pPCS1002 | ApR; pLP170 containing PrhlR-lacZ transcriptional fusion | Pesci et al. (1997) |
| pQF50 | ApR; broad-host-range vector with promoterless lacZ | Farinha & Kropinski (1990) |
| pRK2013 | KmR; ColE1ori-Tra (RK2)+ | Figurski & Helinski (1979) |
| pSJ01 | ApR; pGEMEX-1 with a 1220 bp EcoRI–BamHI flanked fragment containing ampR | Kong et al. (2005b) |
| pSJ06 | ApR; pME6030 with a 1220 bp EcoRI–BamHI flanked fragment containing ampR (referred to as pAmpR) | Kong et al. (2005b) |
| pSJ07 | ApR; pEX100T derivative with ampR : : aacCI | Kong et al. (2005b) |
| pSJ09 | ApR, GmR; pGEMEX-1 with a 330 bp EcoRI–BamHI flanked fragment containing ampC-ampR intergenic region | Kong et al. (2005b) |
| pSJ10 | TcR; CTX-lacZ fused with ampC promoter, PampC | Kong et al. (2005b) |
| pSJ11 | TcR; CTX-lacZ fused with ampR promoter, PampR | Kong et al. (2005b) |
| pUCGm | ApR, GmR; pUC19 derivative containing gentamicin cassette | Schweizer (1993) |
| Primers | ||
| SBJ01ampRFor* | 5′-GGAATTCTGGCGAACAGCAGTGTGGAAGCGG-3′ | |
| SBJ02ampRRev* | 5′-CGGGATCCATTCCAATCACAACCCCAACGCC-3′ | |
| SBJ03ampCRFor* | 5′-GGAATTCTGAGGCCGCGCGGCAGACGCTTGAACA-3′ | |
| SBJ04ampCRRev* | 5′-CGGGATCCCATGAGGATTGGCGTCCTTTG-3′ | |
| DBS_QRTAmpRF | 5′-CATTGGCCTTCATCACCGGTTGTA-3′ | |
| DBS_QRTAmpRR | 5′-GGTTTCTCATGCAGCCCACGACAA-3′ |
*The italicized portion of the sequence indicates a restriction site in a PCR product prepared with the primer.
DNA manipulations.
All molecular techniques were performed according to standard protocols (Ausubel et al., 1999).
Insertional inactivation of the ampR gene.
Inactivation of ampR in Alg+ PDO300 (PAOmucA22) was performed as previously reported using the same constructs (Kong et al., 2005b). The ampR : : aacCI fragment subcloned into pEX100T (Schweizer & Hoang, 1995) was introduced by conjugation into an alginate-overproducing P. aeruginosa, Alg+ PDO300 (Mathee et al., 1999), with a helper strain harbouring pRK2013 (Figurski & Helinski, 1979). The merodiploids resulting from homologous recombination were selected with Pseudomonas isolation agar containing gentamicin. The colonies were then screened for gentamicin resistance and carbenicillin sensitivity by replica plating. The insertion was confirmed by PCR and restriction analysis of the PCR product. The Alg+ PDO300 isogenic strain with defective ampR (PAOmucA22ampR) is named Alg+ PDOampR (Table 1). Complementation studies were performed using plasmid pSJ06 that contains a PCR-amplified ampR on a low-copy-number, highly stable shuttle vector pME6030 to minimize the effects of gene dosage (Kong et al., 2005b). This plasmid is referred to as pAmpR.
Construction of promoter-lacZ fusions.
A 330 bp ampC–ampR intergenic region with the putative promoters was subcloned into the promoterless lacZ in the mini-CTX-lacZ reporter plasmid (Becher & Schweizer, 2000), creating pSJ10 (PampC-lacZ) and pSJ11 (PampR-lacZ) (Table 1) (Kong et al., 2005b). The resulting clones were mobilized into Alg+ PDO300 and Alg+ PDOampR (Table 1).
Quantitative real-time PCR (qPCR).
RNA extraction was performed with an RNeasy mini kit (Qiagen) following the manufacturer's protocols after treatment of cells with subMIC levels (200 μg ml−1) of penicillin G at OD600 0.6 for 1 h. The samples were stabilized with 5 % phenol/95 % ethanol mixture (pH 4.7) immediately after harvesting and during cell lysis (Brencic et al., 2009). After determining RNA quantity spectrophotometrically (Beckman DU640; Beckman Coulter) and quality by denaturing agarose gel electrophoresis (Northern Max Gly; Ambion), cDNA was synthesized by annealing NS5 random primers to total purified RNA. Subsequent extension was carried out using SuperScript III reverse transcriptase (Invitrogen) (Brencic et al., 2009). The cDNA was quantified and 10 ng cDNA was used per qPCR. We used the ABI 7500 cycler (Applied Biosystems) and Power SYBR Green PCR mastermix with ROX (Applied Biosystems) to test for expression of the ampR gene in these strains. The ATP-binding subunit clpX (PA1802) of the ATP-dependent protease was used as the internal control. Assays were performed in triplicate. Primer specificity was determined from dissociation profiles using melt curves. The cycling conditions for the qPCR were: 95 °C for 2 min (holding); 40 cycles of 95 °C for 15 s, 60 °C for 1 min (cycling); 95 °C for 15 s, 60 °C for 1 min (melt curve conditions).
Quantification of pyocyanin and LasA protease.
Extracellular pyocyanin was quantified as previously described (Kong et al., 2005b). LasA protease activity was measured by determining the ability of P. aeruginosa culture supernatants to lyse boiled Staphylococcus aureus, as described by Kessler et al. (1993).
β-Lactamase assay.
The assay of the P. aeruginosa chromosomal β-lactamase was performed as previously described using nitrocefin as the colorimetric substrate (Kong et al., 2005b).
β-Galactosidase assay.
Assays for β-galactosidase in P. aeruginosa were performed as previously described (Mathee et al., 1997) and adapted into a high-throughput 96-well array (Griffith & Wolf, 2002).
P. aeruginosa–Caenorhabditis elegans paralysis assays.
The P. aeruginosa–C. elegans standard paralysis assay was modified from that of Gallagher & Manoil (2001). Bacterial cultures were grown overnight. A 1 : 1000 dilution was plated onto brain heart infusion agar plates. These plates were incubated for 18–24 h for the formation of bacterial lawns. Meanwhile, a synchronized culture of L4 stage larvae hermaphrodite Bristol N2 C. elegans was washed off an E. coli OP50-seeded nematode growth medium plate (1.7 % agar, 0.35 % peptone, 0.34 % K2HPO3, 0.3 % NaCl, 0.012 % MgSO4, 0.011 % CaCl2, 0.0005 % cholesterol). The worms were centrifuged at 1300 g for 2 min and washed twice with M9 medium to remove residual E. coli bacteria. A total of 30 to 50 worms was then added to the P. aeruginosa bacterial lawns. Both live and paralysed worms were scored at 1, 2 and 4 h by microscopic observation. The analysis was performed in triplicate.
Statistical analysis.
All data were analysed with one-way ANOVA using the statistical software package spss (SPSS).
RESULTS
PampC expression in ampR mutants
We have previously reported that in the non-mucoid strain, AmpR positively regulates ampC expression but negatively controls the expression of poxB (Kong et al., 2005a, b). To test whether such opposing controls remain true in the Alg+ background, strains were constructed with a single copy of the ampC promoter fused to a promoterless reporter gene, lacZ (PampC-lacZ). This was integrated into the Alg+ PDO300 and the Alg+ PDOampR chromosomes via attB-attP site-specific recombination, thus allowing mimicking of the chromosomal regulation. In the absence of inducer, the PampC-lacZ activity remained at a basal level in Alg+ PDO300 and Alg+ PDOampR strains (Table 2). A significant ninefold induction of the ampC promoter was observed in Alg+ PDO300 upon challenge with β-lactams (Table 2). However, the inducibility of the PampC was lost in Alg+ PDOampR.
Table 2.
β-Galactosidase activities of ampC and ampR promoters
| Strain | PampC-lacZ (Miller units) | P value* | PampR-lacZ (Miller units) | P value* | ||
|---|---|---|---|---|---|---|
| Non-induced | Induced | Non-induced | Induced | |||
| Alg− PAO1† | 124.1±11.6 | 1644.2 ± 33.7 | <0.05 | 77.1±8.7 | 123±1.2 | ns |
| Alg− PAOampR† | 113.2±7.5 | 122.3±7.4 | ns | 96.3±15.2 | 106.0±16.0 | ns |
| P value‡ | ns | <0.05 | ns | ns | ||
| Alg+ PDO300 | 104.2±4.5 | 957.5±161.4 | <0.05 | 79.5±26.3 | 332.8±14.3 | <0.05 |
| Alg+ PDOampR | 142.4±4.9 | 143.8±6.9 | ns | 155.8±0.8 | 153.0±2.1 | ns |
| P value§ | ns | <0.05 | ns | <0.05 | ||
ns, Not significant (P values >0.05).
*ANOVA compares the activity values between the presence (+) and absence (−) of inducers.
†These data are presented in a previous paper (Kong et al., 2005b); they are included here for comparison.
‡ANOVA compares the activity values between the Alg− PAO1 and the mutant Alg− PAOampR.
§ANOVA compares the activity values between the Alg+ PDO300 and the mutant Alg+ PDOampR.
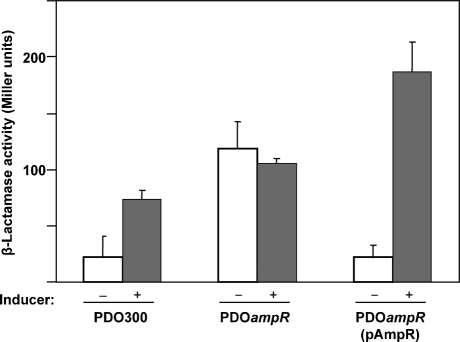
Based on the above analysis, we expected to observe a loss of β-lactamase activity concomitant with the loss of ampR. However, the Alg+ PDOampR expressed a statistically significant sixfold higher β-lactamase compared to the parent Alg+ PDO300 in the absence of antibiotics (Fig. 1). No further induction was demonstrated in the presence of the inducer. This phenotype varied from the parental strain Alg+ PDO300, which showed only a threefold inducible phenotype (Fig. 1). The inducible phenotype was restored in Alg+ PDOampR mutant by complementation with pAmpR. The high β-lactamase activity in an ampR mutant has been shown previously to be due to the uninhibited expression of an oxacillinase poxB gene, rather than the elevated expression of ampC gene (Kong et al., 2005b).
Fig. 1.
β-Lactamase expression in the Alg+ PDOampR mutant. Assays were performed using the parent strain Alg+ PDO300, the mutant Alg+ PDOampR and Alg+ PDOampR (pAmpR) in the absence (−) and presence (+) of an inducer. The plasmid pAmpR carries the wild-type ampR gene on a broad-host-range low-copy-number plasmid pME6030 (Heeb et al., 2000). Fresh cultures of OD600 0.6–0.8 were induced with 100 μg benzylpenicillin ml−1 for 3 h before harvesting. Assays were performed on sonicated lysate using nitrocefin as a chromogenic substrate. Assays were performed in triplicate. One Miller unit of β-lactamase is defined as 1 nmol nitrocefin hydrolysed min−1 (μg protein)−1.
ampR transcription in alginate-overproducing strains
The LysR family of transcriptional regulators is known to repress their own transcription as in the case of Citrobacter freundii AmpR (Lindquist et al., 1989). However, we have previously reported that P. aeruginosa AmpR does not autoregulate in the prototypic strain Alg− PAO1 (Kong et al., 2005b). To determine if there is a change in the AmpR autoregulation in Alg+ strains, a single-copy fusion of PampR-lacZ was introduced at the attP site in Alg+ PDO300 and Alg+ PDOampR. In the absence of inducers, the ampR transcription remained at low levels in both strains (Table 2). In the presence of inducers, a significant increase in PampR expression was seen in Alg+ PDO300 (Table 2). Comparing the genotypes of the isogenic Alg− PAO1 and Alg+ PDO300 strains, this significant increase was likely due to the uninhibited activity of the ECF sigma factor AlgT/U in the latter. This suggests that AlgT/U activates the ampR promoter in the presence of inducers. Due to loss of ampR, no significant induction of PampR was seen in Alg+ PDOampR. In order to test AlgT/U regulation of ampR, mRNA levels of ampR were determined by qPCR with the Alg− PAO1, Alg+ PDO300 and Alg− PDOalgT strains. The Alg+ PDO300 strain showed an increase in the ampR mRNA levels (relative quantity of 2.1±0.2 compared to 1.0±0 in Alg− PAO1) indicating positive regulation of ampR by AlgT/U. Mutation in algT/U in Alg− PDOalgT led to a decrease in this expression (relative quantity 1.4±0.1 compared to 2.1±0.2 in Alg+ PDO300) supporting our hypothesis of positive regulation of ampR by AlgT/U and concurs with the transcriptional fusion assays. These results suggest that both AlgT/U and AmpR are required for the induction of the ampR promoter in the presence of inducers.
ampR mutation affects algT/U transcription
The loss of inducibility of ampR transcription in the Alg+ PDOampR background provided us with the first clue of the existence of a co-regulatory network involving β-lactam resistance and alginate production. To determine if this relationship is bidirectional, a PalgT/U-lacZ fusion construct was introduced into Alg− PAO1, Alg+ PDO300 and the corresponding ampR mutant strains. As expected, the expression of algT/U promoter is constitutive in Alg− PAO1 and increased in Alg+ PDO300 (Fig. 2). Insertional inactivation of ampR in Alg− PAO1 and Alg+ PDO300 resulted in an approximately twofold increase in PalgT/U activity in the absence of inducer. The effect of ampR mutation in Alg− PAOampR is the same as the known AlgT/U repressor mucA mutation (Alg+ PDO300) with respect to PalgT/U expression, indicating negative regulation of PalgT/U by AmpR. The AmpR-regulation of algT/U promoter in these strains was not significantly affected by β-lactam antibiotic. The consistent increase in PalgT/U in the absence of ampR suggests that AmpR is a negative modulator of the ECF sigma factor, AlgT/U.
Fig. 2.
The effects of the ampR mutation on algT/U transcription. The promoter fusion PalgT/U-lacZ was introduced into Alg− PAO1, Alg− PAOampR, Alg+ PDO300 and Alg+ PDOampR. Induction was carried out using 500 μg benzylpenicillin ml−1 and the β-galactosidase activity was determined in Miller units after 30 min incubation. The basal level of expression was detected in the promoterless lacZ vector, pLP170.
AlgT/U-dependent regulation of pyocyanin
Our quantitative analysis showed that the Alg+ PDO300 produced threefold less pyocyanin than Alg− PAO1 in the absence of β-lactam antibiotics (Table 3). This finding confirms that the AlgT/U sigma factor suppresses the production of pyocyanin. The presence of inducer resulted in an increase in pyocyanin production, albeit at low levels in Alg+ PDO300. However, the Alg+ PDOampR mutant produced a significantly high basal level of pyocyanin, which was inducible in the presence of β-lactam antibiotics (Table 3). Expressing ampR in trans in Alg+ PDOampR on a low-copy-number plasmid restored the phenotype to the parental strain, Alg+ PDO300 (data not shown). On the basis of this data we further argue that AmpR acts as a negative regulator of pyocyanin production.
Table 3.
Pyocyanin, LasA and PlasA-lacZ activities
| Inducer* | Pyocyanin [μg (μg total protein)−1]† | LasA [ΔOD600 h−1 (μg protein)−1]‡ | PlasA-lacZ (Miller units)§ | ||
|---|---|---|---|---|---|
| − | + | − | + | − | |
| Alg− PAO1|| | 0.285±0.219 | 2.293±0.216¶ | 0.310±0.065 | 0.317±0.059 | 790.0±128.9 |
| Alg− PAOampR|| | 2.934±0.761# | 3.317±0.638 | 1.109±0.099# | 0.951±0.045# | 1970.5±312.6# |
| Alg+ PDO300 | 0.102±0.017** | 0.324±0.051 | 0.135±0.011 | 0.147±0.024 | 246.5±26.5 |
| Alg+ PDOampR | 0.538±0.026†† | 2.518±0.640†† | 0.268±0.017 | 0.143±0.025 | 536.0±19.0 |
*Induction was carried out using 500 μg benzylpenicillin ml−1 for P. aeruginosa.
†Pyocyanin concentrations were expressed as μg pyocyanin produced (μg total protein)−1.
‡LasA activities were determined as the reduction of OD600 over a period of 1 h (μg total protein)−1.
§β-Galactosidase assays were performed in a high-throughput 96-well array and the results expressed in Miller units.
||These data are presented in a previous paper (Kong et al., 2005b); they are included here for comparison.
¶P<0.05 between non-induced and induced in the same strain.
#P<0.05 between Alg− PAO1 and Alg− PAOampR under the same conditions.
**P<0.05 between Alg− PAO1 and Alg+ PDO300 under the same conditions.
††P<0.05 between Alg+ PDO300 and Alg+ PDOampR under the same conditions.
LasA protease activity and lasA promoter expression in Alg+ PDOampR
The inverse relationship seen between alginate production and proteases is presumed to be AlgT/U-dependent (Mathee et al., 1999; Mohr et al., 1990; Ohman & Chakrabarty, 1982). Thus, a significant increase in algT/U expression in Alg+ PDOampR (Fig. 2) should result in downregulation of LasA protease expression. As expected, in comparison to the wild-type Alg− PAO1, Alg+ PDO300 produced 2.3-fold less LasA protease. However, loss of ampR resulted in a marginal increase in the LasA protease activity (Table 3) in an inducer-independent manner. To further confirm the above hypothesis, a PlasA-lacZ transcriptional fusion plasmid was introduced into Alg− PAO1, Alg+ PDO300 and the ampR mutant strains (Table 3). In concordance to the LasA activity analysis, the PlasA levels were low in all mucoid strains, suggesting that the transcription of these promoters was suppressed (Table 3). Furthermore, the PlasA-lacZ fusion expression was increased twofold to threefold in Alg− PAOampR and Alg+ PDOampR, as compared to their respective parental strains (Table 3). These results suggest that AmpR is a negative regulator of lasA expression.
QS gene expression in mucoid ampR mutants
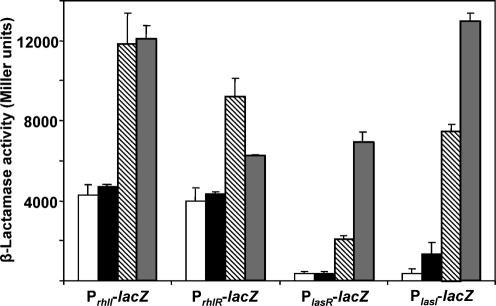
In line with previous observations (Kong et al., 2005b), we postulated that the slight increase of lasA expression in Alg+ PDOampR could be due to upregulation of the las system. To address this, all the four QS promoter fusions, PlasI-lacZ, PlasR-lacZ, PrhlI-lacZ and PrhlR-lacZ were introduced into Alg− PAO1, Alg− PAOampR, Alg+ PDO300 and Alg+ PDOampR. As we postulated, the Alg+ PDO300 exhibited significantly lower QS gene expression as compared to Alg− PAO1. There was no difference in the PlasR expression in Alg+ PDO300 and Alg+ PDOampR (Fig. 3). However, the PlasI activity was significantly increased in Alg+ PDOampR. Similar to the Alg− PAOampR, the loss of ampR in Alg+ PDO300 resulted in minimal alteration of PrhlI and PrhlR expression. Thus, AmpR negatively regulates lasI expression in alginate-overproducing strains.
Fig. 3.
The effects of the ampR mutation on QS las and rhl gene transcription. The alteration in the transcription of the QS systems in Alg− PAO1 (hatched bars), Alg− PAOampR (grey bars), Alg+ PDO300 (white bars) and Alg+ PDOampR (black bars) was monitored using four transcriptional fusions, PlasI-lacZ, PlasR-lacZ, PrhlI-lacZ and PrhlR-lacZ. The promoterless lacZ vector has a low basal level of activity of <20 Miller units.
Role of AmpR in virulence
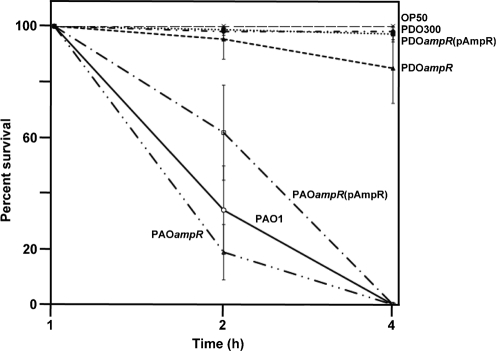
The nematode C. elegans has been used as a bacterial pathogenesis model for the determination of virulence in P. aeruginosa (Gallagher & Manoil, 2001; Sifri et al., 2005; Tan et al., 1999). This simple host–pathogen interaction model was used to ascertain the virulence of Alg− PAO1, Alg− PAOampR, Alg+ PDO300 and Alg+ PDOampR. As expected, there was no observable paralysis in the negative control (E. coli OP50) plates (Fig. 4) and during the first hour of incubation with all the four isogenic P. aeruginosa strains. Consistent with the molecular and biochemical data, Alg− PAOampR paralysed C. elegans at a significantly (P<0.05) faster rate than the wild-type Alg− PAO1 (Fig. 4). The lowest survival was seen at the second hour, 19 % with Alg− PAOampR, as compared to 34 % with Alg− PAO1 (Fig. 4). In addition, Alg+ PDOampR also showed a higher virulence than Alg+ PDO300 with 85 and 98 % survival at 4 h post-incubation, respectively (Fig. 4). The increase in virulence in Alg− PAOampR and Alg+ PDOampR could be restored using pAmpR (Fig. 4), suggesting that AmpR acts as a negative regulator of P. aeruginosa virulence.
Fig. 4.
Kinetics of the paralysis of C. elegans by P. aeruginosa Alg− PAO1 wild-type strain (—), Alg− PAOampR (– . . –), complemented Alg− PAOampR(pAmpR) (– . –), Alg+ PDO300 (-.-), Alg+ PDOampR (--) and complemented Alg+ PDOampR(pAmpR) (…). L4 stage larval hermaphrodite Bristol N2 C. elegans were placed on each brain heart infusion agar plate containing a bacterial lawn and scored for dead worms by microscopic examination. E. coli OP50 (— —) was used as a negative control. Values are the mean±sd of triplicate analyses. Results were statistically significant (P<0.05 for PAO1 vs PAOΔampR at 2 h, and PDO300 and PDOampR at 4 h).
DISCUSSION
AmpR is the master transcriptional regulator involved in β-lactam antibiotic resistance. We have demonstrated previously that in addition to regulating AmpC and PoxB β-lactamases, P. aeruginosa AmpR plays a role in controlling the expression of some virulence factors (Kong et al., 2005b). In this study, we have shown that there is a complex regulatory network between β-lactam resistance, alginate production, and QS and virulence gene expression, factors determining the establishment of both acute and chronic P. aeruginosa infection.
ampR autoregulation requires AlgT/U
Previous studies in Enterobacteriaceae spp. have demonstrated that the transcription of ampR is autoregulatory (Lindquist et al., 1989); but, we reported otherwise for the non-mucoid strain of P. aeruginosa (Kong et al., 2005b). Data presented here show that the autoregulatory mechanism of ampR could be seen in the presence of inducers in an alginate overproducing strain, Alg+ PDO300, but was lost in the absence of ampR (Table 2). This suggests that the regulation of ampR transcription requires AlgT/U, and is AmpR-dependent in Alg+ PDO300. The requirement of these factors for autoregulation explains the inconsistency seen with the Enterobacteriaceae models: in an in vivo system using the heterologous host E. coli, the PampR-lacZ activity was repressed threefold in the presence of Citrobacter freundii AmpR (Lindquist et al., 1989). However, this mode of regulation was lost in a minicell system with Enterobacter cloacae AmpR (Lindberg & Normark, 1987).
The intriguing autoregulatory mechanism seen in the alginate-overproducing PAO1 derivative may have important clinical implications: the data with Alg− PAO1 suggest that this early colonizer is able to induce the production of β-lactamases upon β-lactam chemotherapy. However, this non-mucoid strain is unable to autoregulate ampR, indicating that the production of β-lactamases is induced only upon contact with β-lactam antibiotics. This phenomenon may be disadvantageous during antibiotic selections. Persistence of the organism in the lungs of patients with CF will ultimately result in the selection of mucoid strains that hyperproduce alginate (Høiby, 1975). This phenotypic alteration is accompanied by resistance to antibiotics and immune clearance (Giwercman et al., 1991). Data from Alg+ PDO300 suggest that the selected mucoid P. aeruginosa strains are primed to β-lactam resistance by the increased production of AmpR, and hence β-lactamases, upon contact with β-lactam antibiotics. This observation should be further verified using clinical strains with commonly used β-lactams.
AmpR is a negative regulator of algT/U
The simultaneous presence of β-lactam resistance and alginate-overproduction suggests a possible co-regulation of these phenomena. We have shown here that autoregulation of ampR is AlgT/U-dependent. Loss of the ampR gene in Alg− PAO1 resulted in a significant increase in the promoter activity of algT/U operon (Fig. 2). However, this did not phenotypically alter Alg− PAO1 to an Alg+ phenotype due to the post-transcriptional control of AlgT/U by the anti-sigma factor, MucA, expressed downstream of algT/U (Hughes & Mathee, 1998). In Alg+ PDO300, like in Alg− PAO1, there is an increase of algT/U expression upon loss of ampR. Data from these two strain backgrounds suggest that AmpR suppresses the expression of algT/U.
The possible mechanistic interaction between the alg and amp regulons has been reported in Azotobacter vinelandii, where a mutation in ampDE, encoding negative regulators of β-lactamases, resulted in elevated expression of alginate biosynthetic genes (Núñez et al., 2000). In addition, microarray data also have demonstrated that alginate production is induced upon antibiotic challenge (Bagge et al., 2004), and a later study identified AlgT, AlgW and Prc proteases as being involved in this process (Wood et al., 2006). Our results are further supportive of their findings in which β-lactam resistance and alginate production of P. aeruginosa are co-regulated. This co-regulation is likely mediated by AmpR-AlgT/U interaction. Future studies will address this potential interaction.
AlgT/U and AmpR are regulators of virulence factors
Multiple QS-dependent phenotypes, including LasA and pyocyanin production, are differentially regulated in an ampR mutant, and are probably an indirect effect of AmpR on the QS system. We have previously shown that deletion of ampR gene increased the production of LasA protease in an Alg− strain, suggesting that lasA expression is suppressed by AmpR (Kong et al., 2005b). We postulated that a similar observation should be obtained in an Alg+ strain. As expected, the absence of ampR in the presence of functional AlgT/U elevated the promoter expression of lasA and the production of LasA protease (Table 3). This alteration in LasA synthesis suggests that both AlgT/U and AmpR negatively impact transcription of the lasA gene. Although the inverse correlation between alginate and protease production has been repeatedly reported, our results establish that this correlation is mediated through the downregulation of QS in Alg+ strains. Comparing two isogenic strains, Alg− PAO1 and Alg+ PDO300, the see-saw effect is brought upon by the ECF sigma factor, AlgT/U. Since sigma factor, an essential component of RNA polymerase, is unlikely to be involved in the repression of gene expression, AlgT/U-mediated downregulation of QS genes is probably indirect.
To determine whether the in vitro alterations in virulence factor expression could be translated into significant in vivo killing, the C. elegans–P. aeruginosa interaction model was employed. As predicted, loss of ampR strongly correlated with an increase in virulence with both Alg− PAOampR and Alg+ PDOampR showing higher rates of C. elegans paralysis as compared to their parent strains (Alg− PAO1 and Alg+ PDO300, respectively). The significantly higher amounts of pigmentation produced by ampR mutants compared to the isogenic wild-type strain explains the higher killing rate, which is in agreement with other studies (Tan et al., 1999).
Concluding remarks
The data presented here reveal a complex co-regulatory network between β-lactam resistance, alginate production, QS and virulence gene expression. We have previously shown that AmpR regulates AmpC and PoxB β-lactamases and QS-dependent proteases (Kong et al., 2005a, b). In this paper, that observation is further extended to include the alginate master regulator, AlgT/U. Importantly, we show that the positive autoregulation of ampR requires AlgT/U, whereas AmpR negatively regulates algT/U expression (Fig. 2) serving as a negative feedback loop to limit the AlgT/U expression. We propose that this intimate crosstalk between these two global regulators provides a potential molecular framework for the simultaneous occurrence of β-lactam resistance and alginate-overproducing strains in chronic CF lung infections. Further studies on clinical isolates are warranted to understand the complex regulatory network linking all these critical factors in establishing infections. Delineating the interplaying factors and regulatory network is of fundamental significance to understanding the pathogenesis of P. aeruginosa.
Acknowledgments
This work was partially supported by the NIH (MBRS SCORE grant S06 GM08205 to K. M.), NIGMS (RISE grant R25 GM61347 to R. T. S.), Florida International University teaching assistantships to D. B., K.-F. K., S. R. J. and R. T. S.), Biomedical Research Initiative student research award (R25 GM61347 to R. T. S) and Cystic Fibrosis Foundation student traineeship (KONG05HO to K.-F. K.; LEAL05HO to S. M. L.). We thank Barbara Iglewski for generously sharing all the promoter-fusion plasmids.
Abbreviations
CF, cystic fibrosis
ECF, extracytoplasmic function
qPCR, quantitative real-time PCR
QS, quorum sensing
References
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K.(editors) (1999). Short Protocols in Molecular Biology. New York. : Wiley.
- Bagge, N., Schuster, M., Hentzer, M., Ciofu, O., Givskov, M., Greenberg, E. P. & Høiby, N. (2004). Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob Agents Chemother 48, 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher, A. & Schweizer, H. P. (2000). Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29, 948–952. [DOI] [PubMed] [Google Scholar]
- Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L. & Lory, S. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73, 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries, C. A. & Ohman, D. E. (1994). Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol 176, 6677–6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett, R. G. (1969). Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl Microbiol 18, 936–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, L. R. & Linker, A. (1973). Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol 116, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha, M. A. & Kropinski, A. M. (1990). Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol 172, 3496–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fick, R. B., Jr, Sonoda, F. & Hornick, D. B. (1992). Emergence and persistence of Pseudomonas aeruginosa in the cystic fibrosis airway. Semin Respir Infect 7, 168–178. [PubMed] [Google Scholar]
- Figurski, D. H. & Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved, A. M. & Deretic, V. (2003). Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol 185, 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved, A. M., Boucher, J. C. & Deretic, V. (2002). Global genomic analysis of AlgU (σE)-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol 184, 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, L. A. & Manoil, C. (2001). Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol 183, 6207–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlich, D., Naas, T. & Nordmann, P. (2004). Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob Agents Chemother 48, 2043–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giwercman, B., Jensen, E. T., Høiby, N., Kharazmi, A. & Costerton, J. W. (1991). Induction of β-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother 35, 1008–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan, J. R. & Harris, G. S. (1986). Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci 3, 302–308. [PubMed] [Google Scholar]
- Greenberg, E. P. (2000). Bacterial genomics. Pump up the versatility. Nature 406, 947–948. [DOI] [PubMed] [Google Scholar]
- Griffith, K. L. & Wolf, R. E., Jr (2002). Measuring β-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun 290, 397–402. [DOI] [PubMed] [Google Scholar]
- Heeb, S., Itoh, Y., Nishijyo, T., Schnider, U., Keel, C., Wade, J., Walsh, U., O'Gara, F. & Haas, D. (2000). Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact 13, 232–237. [DOI] [PubMed] [Google Scholar]
- Hentzer, M., Wu, H., Andersen, J. B., Riedel, K., Rasmussen, T. B., Bagge, N., Kumar, N., Schembri, M. A., Song, Z. & other authors (2003). Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22, 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger, C. D., Ye, R. W., Parsek, M. R., Xie, Z.-D. & Chakrabarty, A. M. (1995). The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigmaE). Proc Natl Acad Sci U S A 92, 7941–7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby, N. (1975). Prevalence of mucoid strains of Pseudomonas aeruginosa in bacteriological specimens from patients with cystic fibrosis and patients with other diseases. Acta Pathol Microbiol Scand Suppl 83, 549–552. [PubMed] [Google Scholar]
- Holloway, B. W. & Morgan, A. F. (1986). Genome organization in Pseudomonas. Annu Rev Microbiol 40, 79–105. [DOI] [PubMed] [Google Scholar]
- Hughes, K. T. & Mathee, K. (1998). The anti-sigma factors. Annu Rev Microbiol 52, 231–286. [DOI] [PubMed] [Google Scholar]
- Kessler, E., Safrin, M., Olson, J. C. & Ohman, D. E. (1993). Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem 268, 7503–7508. [PubMed] [Google Scholar]
- Kong, K. F., Jayawardena, S. R., Del Puerto, A., Wiehlmann, L., Laabs, U., Tummler, B. & Mathee, K. (2005a). Characterization of poxB, a chromosomal-encoded Pseudomonas aeruginosa oxacillinase. Gene 358, 82–92. [DOI] [PubMed] [Google Scholar]
- Kong, K. F., Jayawardena, S. R., Indulkar, S. D., Del Puerto, A., Koh, C. L., Høiby, N. & Mathee, K. (2005b). Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother 49, 4567–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, K. F., Schneper, L. & Mathee, K. (2010). β-Lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS 118, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, F. & Normark, S. (1987). Common mechanism of ampC β-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacae P99 β-lactamase gene. J Bacteriol 169, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist, S., Lindberg, F. & Normark, S. (1989). Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J Bacteriol 171, 3746–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge, J. M., Minchin, S. D., Piddock, L. J. & Busby, J. W. (1990). Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem J 272, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak, J. B., Cannon, C. L. & Pier, G. B. (2002). Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15, 194–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra, S., Silo-Suh, L. A., Mathee, K. & Ohman, D. E. (2000). Proteome analysis of the effect of mucoid conversion on global protein expression in Pseudomonas aeruginosa strain PAO1 shows induction of the disulfide bond isomerase, dsbA. J Bacteriol 182, 6999–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. W., Schurr, M. J., Mudd, M. H., Govan, J. R., Holloway, B. W. & Deretic, V. (1993). Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A 90, 8377–8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee, K., McPherson, C. J. & Ohman, D. E. (1997). Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179, 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee, K., Ciofu, O., Sternberg, C., Lindum, P. W., Campbell, J. I., Jensen, P., Johnsen, A. H., Givskov, M., Ohman, D. E. & other authors (1999). Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145, 1349–1357. [DOI] [PubMed] [Google Scholar]
- Mohr, C. D., Rust, L., Albus, A. M., Iglewski, B. H. & Deretic, V. (1990). Expression patterns of genes encoding elastase and controlling mucoidy: co-ordinate regulation of two virulence factors in Pseudomonas aeruginosa isolates from cystic fibrosis. Mol Microbiol 4, 2103–2110. [DOI] [PubMed] [Google Scholar]
- Ng, W.-L. & Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu Rev Genet 43, 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez, C., Moreno, S., Cárdenas, L., Soberón-Chávez, G. & Espín, G. (2000). Inactivation of the ampDE operon increases transcription of algD and affects morphology and encystment of Azotobacter vinelandii. J Bacteriol 182, 4829–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman, D. E. & Chakrabarty, A. M. (1982). Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun 37, 662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, S. S. (1992). Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl 28, 1–79. [PubMed] [Google Scholar]
- Pesci, E. C., Pearson, J. P., Seed, P. S. & Iglewski, B. H. (1997). Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol 179, 3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, M. J., Seed, P. C., Toder, D. S., Iglewski, B. H., Ohman, D. E., Gustin, J. K., Goldberg, J. B. & Pier, G. B. (1997). Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect Immun 65, 3086–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G. & Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. [DOI] [PubMed] [Google Scholar]
- Ramos, D., Heydorn, A., Koh, C.-L. & Mathee, K. (2003). Loss of alginate production in mucoid Pseudomonas aeruginosa occurs via deregulation of the alternative sigma factor AlgT. In Proceedings of the National Conference on Undergraduate Research (NCUR), pp. 1–9. University of Utah, Salt Lake City, Utah.
- Rolinson, G. N. (1998). Forty years of β-lactam research. J Antimicrob Chemother 41, 589–603. [DOI] [PubMed] [Google Scholar]
- Schuster, M. & Greenberg, E. P. (2006). A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296, 73–81. [DOI] [PubMed] [Google Scholar]
- Schweizer, H. P. (1993). Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15, 831–834. [PubMed] [Google Scholar]
- Schweizer, H. P. & Hoang, T. T. (1995). An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158, 15–22. [DOI] [PubMed] [Google Scholar]
- Sifri, C. D., Begun, J. & Ausubel, F. M. (2005). The worm has turned – microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol 13, 119–127. [DOI] [PubMed] [Google Scholar]
- Tan, M.-W., Mahajan-Miklos, S. & Ausubel, F. M. (1999). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Delden, C. & Iglewski, B. H. (1998). Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis 4, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, V. E., Gillis, R. J. & Iglewski, B. H. (2004). Transcriptome analysis of quorum-sensing regulation and virulence factor expression in Pseudomonas aeruginosa. Vaccine 22, S15–S20. [DOI] [PubMed] [Google Scholar]
- Wood, L. F., Leech, A. J. & Ohman, D. E. (2006). Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol 62, 412–426. [DOI] [PubMed] [Google Scholar]