Abstract
Many plant viral RNAs lack the 5′-cap structure that is required on all host mRNAs for interacting with essential translation initiation factors. Instead, uncapped viral RNAs take over the host translation machinery by harbouring sequences that functionally replace the 5′-cap. Recent reports reveal at least eight different classes of CITE (cap-independent translation element) located in the 3′-UTRs (untranslated regions) of various viruses. We describe how the structure and behaviour of each class of element differs from the other classes, suggesting that they recruit translation factors and, ultimately, the ribosome by diverse mechanisms. These results greatly expand our understanding of ways in which mRNAs can recruit ribosomes, and they provide insight into the regulation of virus gene expression.
Keywords: internal ribosome entry site, kissing stem–loop, long-distance base-pairing, Luteoviridae, Tombusviridae, translation initiation factor
Introduction
All RNA viruses are parasites of their host's translation machinery [1]. Hence, plant viral RNAs are among the most efficient mRNAs in eukaryotes. Understanding how viral RNAs usurp the cell's translational apparatus sheds light on mechanisms of translation in plants and provides important knowledge of regulation of viral gene expression.
All plant mRNAs have a m7GpppG [7-methylguanosine-(5′)triphospho(5′)guanosine] 5′-cap structure and a poly(A) tail at the 3′-end. These modifications are essential for efficient binding of translation initiation factors that recruit the ribosome to the mRNA [2]. The cap structure is bound by the eIF (eukaryotic initiation factor) 4E subunit of eIF4F, which is a heterodimer of eIF4E and eIF4G. eIF4G also binds poly(A) tail-bound PABP [poly(A)-binding protein], thus circularizing the mRNA, and other factors. eIF4G then recruits the 40S ribosomal subunit via interaction with eIF3 [3]. Plants contain a second form of eIF4F, called eIFiso4F which consists of eIFiso4E and eIFiso4G [4].
In contrast with the mRNAs of their hosts, the mRNAs of a large portion of all plant viruses, including those in the Potyviridae, Comoviridae, Tombusviridae and Luteoviridae families and the Sobemovirus and Umbravirus genera, lack a m7GpppG 5′ cap structure. Yet all of these viral RNAs are very efficient messages. Many uncapped animal viral RNAs harbour an IRES (internal ribosome entry site) that allows efficient cap-independent translation initiation [5,6]. Animal virus IRESs facilitate entry of the ribosome at a site distant from the 5′-end of the mRNA to a position located closely upstream of, or at, the initiation codon. IRESs have been reported in several plant virus families as well [7-9].
A new type of CITE (cap-independent translation element) (3′-CITE), known only in plant viral RNAs, has emerged in an ever-increasing number of plant virus genera. The 3′-CITE is located in the 3′-UTR (untranslated region), yet confers very efficient initiation at the 5′-proximal AUG codon on the mRNA [10]. These CITEs do not act as IRESs, although some co-operate with an IRES. To date, 3′-CITEs fall into at least eight structural classes. These 3′-CITE classes show no apparent sequence or structural similarity to each other, although almost all contain a stem–loop in which the loop is known or predicted to base-pair to the 5′-UTR.
TED (translation enhancer domain) of STNV (satellite tobacco necrosis virus)
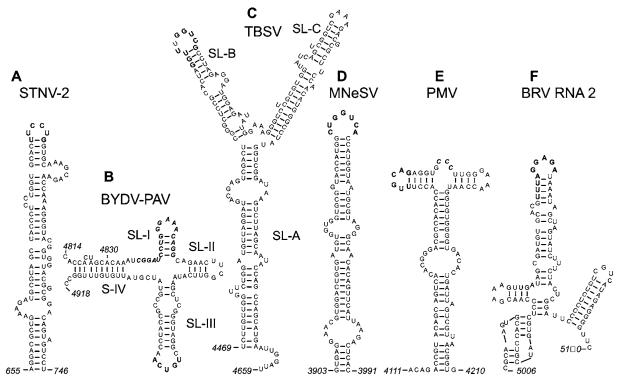
The first discovered 3′-CITE was the TED of STNV [11,12]. The TED consists of a rather loosely structured, ~100 nt, bulged stem–loop (Figure 1A), located at the 5′-end of the 620 nt 3′-UTR of STNV RNA [13]. Although the terminal loop is predicted to base-pair to a loop in the short 5′-UTR, mutagenesis experiments showed only a slight reduction in translation activity when this base-pairing was disrupted [14]. The STNV TED confers cap-independent translation when moved to the 5′-UTR or even within a coding region [13]. It bears no resemblance to the CITE in the 3′-end of its helper virus, TNV (tobacco necrosis virus) on which it depends for replication (see below).
Figure 1. Sequences and secondary structures of representative members of six classes of plant viral 3′-CITE.
Bold bases are known to or predicted to base-pair to the 5′-UTR. Bold italics indicate conserved sequences. (A) STNV TED, known only in STNV-1 [37] and STNV-2 [42] RNAs. (B) The BTE, found in all members of the Luteovirus [10], Dianthovirus [19], and Necrovirus [20,21] genera, and in two umbraviruses. (C) The TBSV CITE, found in nine members of genus Tombusvirus [23]. (D) The predicted MNeSV CITE secondary structure is [25] also predicted for MNSV. (E) The PTE, also present in PEMV RNA 2. (F) Predicted structure of the BRV RNA2 CITE [32]. Shaded text indicates the stop codon of the RNA 2 ORF.
BTEs [BYDV (barley yellow dwarf virus)-like cap-independent translation elements]
The BTEs vary in length, but all contain a 17 nt conserved sequence (bold italic text, Figure 1B) [15] and a nearby stable stem–loop (SL-III) that base-pairs to the 5′-end of the mRNA via a kissing stem–loop interaction [16]. Deletion or mutagenesis of the approx. 100 nt BTE of BYDV reduces translation efficiency 8–20-fold in wheat germ extract and 50–100-fold in protoplasts [15]. Addition of a 5′-cap restores translation. Like the STNV TED, the BTE functions equally well in the 3′-and 5′-UTRs [17]. There is little structural requirement in the 5′-UTR other than an available sequence complementary to the BTE loop. The BTE conferred cap-independent translation even when added to the 3′-end of a reporter RNA containing DV (dengue virus) UTRs [18]. As in other flaviviruses, the 5′-end and 3′-UTR of DV RNA base-pair to each other. Thus the 3′-BTE need not base-pair to the 5′-UTR as long as sequences in part of the 3′-UTR can base-pair with the 5′-end.
Compared with the BYDV BTE, the BTE in RNA 1 of RCNMV (red clover necrotic mosaic dianthovirus) contains two extra helices protruding from the central hub [19]. Like the STNV TED, but unlike many other 3′-CITEs (see below), mutagenesis of the loop predicted to base-pair to the 5′-UTR did not disrupt translation. However, unpredicted base-pairing to the 5′-UTR may have occurred. Given the strong phylogenetic conservation of potential kissing stem–loop interactions with the 5′-UTR in all BTEs, a biological function is likely.
The BTEs in TNV [20,21] and other necrovirus RNAs [22] lack SL (stem–loop)-II (Figure 1B). Disruption of the predicted kissing of TNV SL-III to the 5′-UTR greatly reduced translation [20,21]. The relative translation efficiencies of the TNV BTE and the unrelated TED of STNV RNA may be important to maintain the parasitic relationship between STNV and its helper virus. However, they have yet to be compared directly.
Two umbraviruses, ground nut rosette and tobacco bushy top, harbour apparent BTEs. They contain the 17 nt consensus sequence and a stable SL-III homologue with complementarity to the very short (10–20 nt) 5′-UTR (Z. Wang and W.A. Miller, unpublished work). Interestingly, two other umbraviruses, including PEMV (pea enation mosaic virus) RNA 2 (below), lack BTE motifs.
TBSV (tomato bushy stunt virus)
Probably all nine genera of the Tombusviridae have a 3′-CITE, but the structures differ greatly among genera. For example, the necro- and diantho-viruses appear to be the only Tombusviridae genera that contain a BTE. In contrast, cap-independent translation of TBSV (genus Tombusvirus) relies on a 3′-UTR region called R3.5 (along with flanking bases) that does not resemble a BTE. Instead, R3.5 forms a Y-shaped structure (Figure 1C) that is highly conserved among all nine viruses in the genus [23]. Long distance base-pairing between nine nucleotides in the loop and stem of SL-B (bold text, Figure 1C) and a T-shaped replication element in the 5′-UTR is essential for translation [23,24]. Interestingly, unlike the BTE–5′-UTR interaction, the long-distance kissing loops of TBSV tolerate many base changes, as long as complementarity is preserved [23].
MNeSV (maize necrotic streak virus)-like elements
The 3′-CITE of MNeSV (Tombusviridae, no genus assigned) is predicted to form a long bulged stem–loop (Figure 1D) [25]. It superficially resembles STNV TED, but the MNeSV CITE has no sequence or structural motifs in common with the STNV TED, suggesting that it comprises a separate class of 3′-CITE. Its predicted kissing loop to the 5′-UTR does resemble that of TBSV at five bases (bold text, Figures 1C and 1D). Nieto et al. [26] have suggested that MNSV (melon necrotic spot carmovirus) (Tombusviridae), may contain a 3′-CITE. We found a potential MNeSV-like structure in the MNSV 3′-UTR (Z. Wang, unpublished work), suggesting that it fits in the MNeSV class of 3′-CITE.
PTEs [PMV (panicum mosaic virus)-like translation elements]
PMV (Tombusviridae) RNA contains a 3′-CITE with a predicted T-shaped structure [27]. One loop (bold text, Figure 1E) has the potential to base-pair to the 5′ UTR. As with the STNV TED and the BTE, the PTE can function in various regions of the genome [27]. PEMV RNA 2 (genus Umbravirus, no family assigned) harbours a CITE very similar to that of PMV (Z. Wang, unpublished work). It has a predicted T-shaped structure with stem–loops of the same lengths as those in the PMV CITE. The only notable sequence similarity between the two CITEs is the CCC tract (bold italic text, Figure 1E) in the four-base bulge that separates the two branches.
TCV (turnip crinkle virus)
TCV (Tombusviridae) has a 3′-CITE in the 5′ 155 nt of the 3′-UTR [28]. Using the CLC RNA Workbench folding program, we predict that this CITE forms a Y-shaped structure (different from that of TBSV). Two laboratory groups failed to find potential base-pairing between this 3′-CITE and the 5′-UTR [23,28]. However, we predict that a loop (nucleotides 3836–3841) at the end of one branch of the putative Y-shaped structure forms a kissing interaction with a prominent loop in the first ORF (open reading frame) (nucleotides 545–550) in TCV RNA (GenBank® accession number AY312063). Although this is far from the 5′-end in linear sequence, the secondary structure of the 5′-end is predicted to bring this kissing loop close to the 5′-terminus (K. Treder, unpublished work), as is the case for other 3′-CITEs. Surprisingly, the 3′ CITE may be host-specific, because it is necessary for TCV accumulation in Hibiscus protoplasts, but deletion of the entire 3′-CITE did not affect virus accumulation in Arabidopsis protoplasts [29].
HCRSV (hibiscus chlorotic ringspot virus)
A totally different interaction is predicted for HCRSV (Tombusviridae). It has an ill-defined 3′-UTR CITE of at most 180 nt [30] that is reported to co-operate with a 100 nt IRES located in the middle of the genomic RNA, 123 nt upstream of the coat protein ORF [31]. In wheat germ extract, the 3′-CITE and IRES synergistically enhance translation of the 5′-distal ORFs that they flank by a proposed mechanism in which portions of each element base pair to different portions of 18S rRNA [31]. This is the first report of an IRES in the Tombusviridae.
BRV (blackcurrant reversion virus)
Base-pairing between a sequence in the 3′-UTR of BRV RNA 2 and a sequence in the 5′-UTR was shown by co-variation mutagenesis to be required for cap-independent translation in vitro and in vivo [32]. The predicted structure of the 3′-element (Figure 1F) has not been determined experimentally, neither have the ends been defined. However, kissing stem–loop interactions between the 3′- and 5′-UTRs, usually featuring AU- and GU-rich stem regions, are conserved in both RNAs of all 14 nepovirus genomes examined. The presence of a 3′-CITE in a virus, with a VPg (viral genome-linked protein) and a poly(A) tail (both lacking in all viruses above), which is totally unrelated to Tombusviridae, greatly expands the range of viruses that can harbour a 3′-CITE. The 5′-UTR of BRV RNA 2 serves as an IRES [33]. In bicistronic constructs containing the BRV RNA 2 5′-UTR sequence in the intergenic region, the downstream ORF was translated efficiently, independently of the translatability of the upstream ORF in vitro and in vivo. Neither the 5′-IRES nor the 3′-CITE was shown to function in the absence of the other, so together they may be function as a single element split between the 5′-and 3′-UTRs.
Why a 3′-CITE?
Location of the CITE in the 3′-UTR provides two advantages. For the Tombusviridae-like viruses (all except for BRV), it allows the single copy of the element in genomic RNA to be present in the 3′-UTRs of the viral sgRNAs (subgenomic RNAs), all of which have the same 3′-end as genomic RNA. This helps minimize viral genome length. In TCV [28], MNeSV [25] and BYDV [34], the sgRNAs are translated more efficiently than genomic RNA in competitive conditions due to their different 5′-UTRs. In BYDV infections, a second sgRNA that consists of the viral 3′-UTR including the 3′-CITE, acts in trans to favour translation of the coat protein sgRNA over the genomic RNA [34]. Preferential translation of sgRNA facilitates a switch from early (mainly genomic RNA) to late (mainly sgRNA) translation. A second reason for the 3′ location of the CITE is that it can allow the viral replicase to shut off translation of the genomic RNA that it is copying before the replicase encounters ribosomes moving in the opposite direction on the same RNA [10,35]. This would free the genome of ribosomes that are known to block replicases on the same RNA template [36] before the replicase reached that portion of the genome.
Mechanisms
Investigation of the mechanisms by which 3′-CITEs recruit translation machinery is in its infancy. Specific binding of the STNV TED to eIF4F or eIFiso4F is required for translation [37]. Both the eIF4E and eIF4G subunits of eIF4F participate in this interaction. In contrast, the BYDV BTE relies on eIF4F [15], but binds primarily to the eIF4G subunit and has little need for the eIF4E subunit or eIFiso4F [38].
Genetic evidence supports roles for eIF4F and eIFiso4F components in infection by many viruses with uncapped RNAs [39]. Mutations in eIF4G and eIF4E confer recessive resistance to TCV [40] and MNSV [26] respectively. The mutation in eIF4G (cum2 resistance gene) prevented TCV translation [40], whereas mutations in eIFiso4E that conferred resistance to potyviruses did not affect TCV infection [41]. The dependence of TCV, MNSV, STNV and BYDV RNAs on different initiation factors reveal that the diverse 3′-CITEs use diverse mechanisms of recruiting the ribosome.
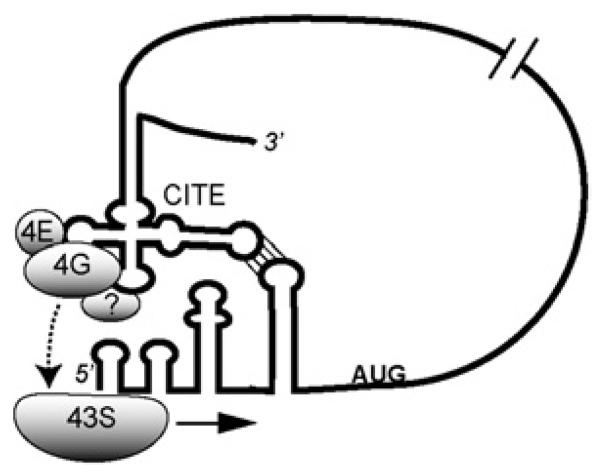
We propose that the diverse 3′-CITEs have evolved like RNA aptamers to bind different domains on various translation initiation factors. These factors are then delivered to the 5′-end by long-distance base-pairing (Figure 2). They then recruit the 40S subunit which scans to the first AUG. Obviously, much work remains to test this and other mechanisms for the elements described here.
Figure 2. Model for 3′-CITE recruitment of translational machinery to viral mRNA.
Using the BYDV BTE as an example, eIF4F and possibly other factors bind the 3′-CITE. Long-distance base-pairing juxtaposes the CITE-bound factors near the 5′-end. This delivers eIF4F and possibly other factors to the 5′-end, where they interact with the ribosome to facilitate scanning.
Whatever the mechanisms, ultimately they achieve the same end result, because different 3′-CITEs are interchangeable: the distribution of 3′-CITE classes does not correlate with virus taxonomy. For example, the four genera known to harbour BTEs are in at least two virus families, whereas viruses in the same genus (Umbravirus) or family (Tombusviridae) can have entirely different classes of 3′-CITE. Thus recombination between viruses to exchange 3′-CITEs occurs relatively frequently.
Acknowledgments
This work was funded by NIH (National Institutes of Health) grant number GM067104.
Abbreviations used
- BRV
blackcurrant reversion virus
- BYDV
barley yellow dwarf virus
- BTE
BYDV-like cap-independent translation element
- CITE
cap-independent translation element
- DV
dengue virus
- eIF
eukaryotic initiation factor
- HCRSV
hibiscus chlorotic ringspot virus
- IRES
internal ribosome entry site
- m7GpppG
7-methylguanosine(5 )triphospho(5 )guanosine
- MNeSV
maize necrotic streak virus
- MNSV
melon necrotic spot virus
- ORF
open reading frame
- PEMV
pea enation mosaic virus
- PMV
panicum mosaic virus
- PTE
PMV-like translation element
- sgRNA
subgenomic RNA
- SL
stem–loop
- STNV
satellite tobacco necrosis virus
- TBSV
tomato bushy stunt virus
- TCV
turnip crinkle virus
- TED
translation enhancer domain
- TNV
tobacco necrosis virus
- UTR
untranslated region
References
- 1.Flint SJ, Enquist LW, Racaniello VR, Skalka AM. Principles of Virology. ASM Press; Washington, DC: 2004. [Google Scholar]
- 2.Gallie DR. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey J, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2007. pp. 747–774. [Google Scholar]
- 3.Pestova TV, Lorsch JR, Hellen CU. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey J, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2007. pp. 87–128. [Google Scholar]
- 4.Browning KS. Biochem. Soc. Trans. 2004;32:589–591. doi: 10.1042/BST0320589. [DOI] [PubMed] [Google Scholar]
- 5.Jan E. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Jackson RJ. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 7.Niepel M, Gallie DR. J. Virol. 1999;73:9080–9088. doi: 10.1128/jvi.73.11.9080-9088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kneller EL, Rakotondrafara AM, Miller WA. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorokhov YL, Skulachev MV, Ivanov PA, Zvereva SD, Tjulkina LG, Merits A, Gleba YY, Hohn TJ, Atabekov JG. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller WA, White KA. Annu. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danthinne X, Seurinck J, Meulewaeter F, Van Montagu M, Cornelissen M. Mol. Cell. Biol. 1993;13:3340–3349. doi: 10.1128/mcb.13.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmer RT, Benkowski LA, Schodin D, Lax SR, Metz AM, Ravel JM, Browning KS. J. Biol. Chem. 1993;268:9504–9510. [PubMed] [Google Scholar]
- 13.Meulewaeter F, Van Montagu M, Cornelissen M. RNA. 1998;4:1347–1356. doi: 10.1017/s135583829898092x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meulewaeter F, Danthinne X, Van Montagu M, Cornelissen M. Plant J. 1998;14:169–176. doi: 10.1046/j.1365-313x.1998.00104.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Browning KS, Miller WA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L, Allen E, Miller WA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- 17.Guo L, Allen E, Miller WA. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakotondrafara AM, Polacek C, Harris E, Miller WA. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T. J. Virol. 2003;77:12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meulewaeter F, van Lipzig R, Gultyaev AP, Pleij CW, Van Damme D, Cornelissen M, van Eldik G. Nucleic Acids Res. 2004;32:1721–1730. doi: 10.1093/nar/gkh338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen R, Miller WA. J. Virol. 2004;78:4655–4664. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen R, Miller WA. Virology. 2007;358:448–458. doi: 10.1016/j.virol.2006.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabian MR, White KA. J. Biol. Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- 24.Fabian MR, White KA. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheets K, Redinbaugh MG. Virology. 2006;350:171–183. doi: 10.1016/j.virol.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Nieto C, Morales M, Orjeda G, Clepet C, Monfort A, Sturbois B, Puigdomenech P, Pitrat M, Caboche M, Dogimont C, et al. Plant J. 2006;48:452–462. doi: 10.1111/j.1365-313X.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 27.Batten JS, Desvoyes B, Yamamura Y, Scholthof KB. FEBS Lett. 2006;580:2591–2597. doi: 10.1016/j.febslet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Qu F, Morris TJ. J. Virol. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Wong SM. J. Gen. Virol. 2007;88:680–687. doi: 10.1099/vir.0.82536-0. [DOI] [PubMed] [Google Scholar]
- 30.Koh DC-Y, Liu DX, Wong S-M. J. Virol. 2002;76:1144–1153. doi: 10.1128/JVI.76.3.1144-1153.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh DC, Wong SM, Liu DX. J. Biol. Chem. 2003;278:20565–20573. doi: 10.1074/jbc.M210212200. [DOI] [PubMed] [Google Scholar]
- 32.Karetnikov A, Keranen M, Lehto K. Virology. 2006;354:178–191. doi: 10.1016/j.virol.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 33.Karetnikov A, Lehto K. J. Gen. Virol. 2007;88:286–297. doi: 10.1099/vir.0.82307-0. [DOI] [PubMed] [Google Scholar]
- 34.Shen R, Rakotondrafara AM, Miller WA. J. Virol. 2006;80:10045–10054. doi: 10.1128/JVI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barry JK, Miller WA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamarnik AV, Andino R. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gazo BM, Murphy P, Gatchel JR, Browning KS. J. Biol. Chem. 2004;279:13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- 38.Treder K, Pettit Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. RNA. 2008 doi: 10.1261/rna.777308. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robaglia C, Caranta C. Trends Plant Sci. 2006;11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. J. Virol. 2004;78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lellis AD, Kasschau KD, Whitham SA, Carrington JC. Curr. Biol. 2002;12:1046–1051. doi: 10.1016/s0960-9822(02)00898-9. [DOI] [PubMed] [Google Scholar]
- 42.van Lipzig R, Gultyaev AP, Pleij CW, van Montagu M, Cornelissen M, Meulewaeter F. RNA. 2002;8:229–236. doi: 10.1017/s1355838202018071. [DOI] [PMC free article] [PubMed] [Google Scholar]