Abstract
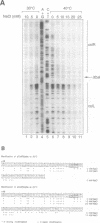
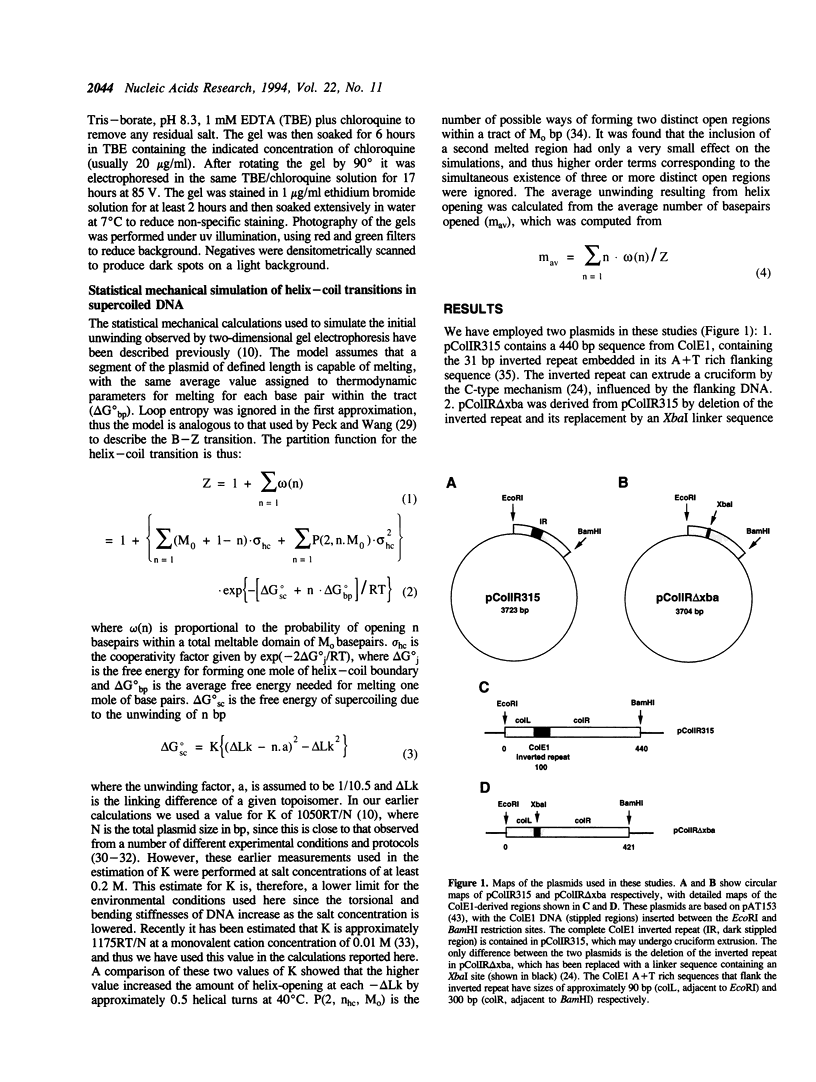
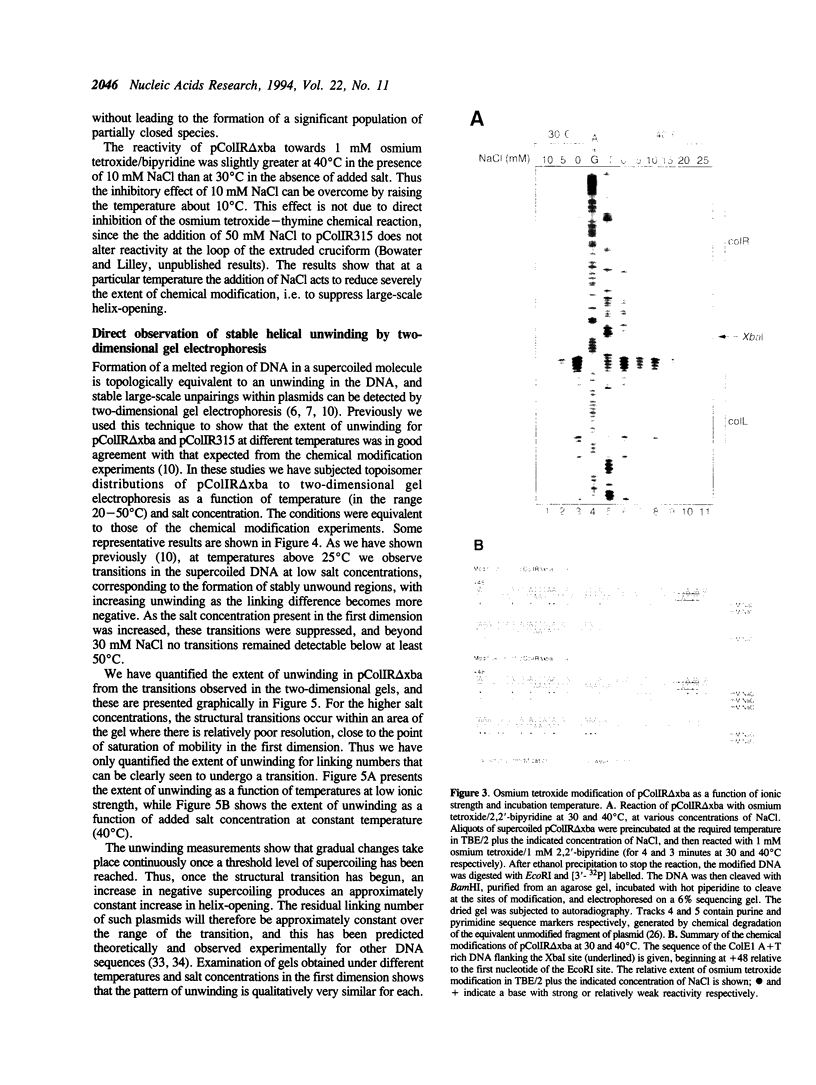
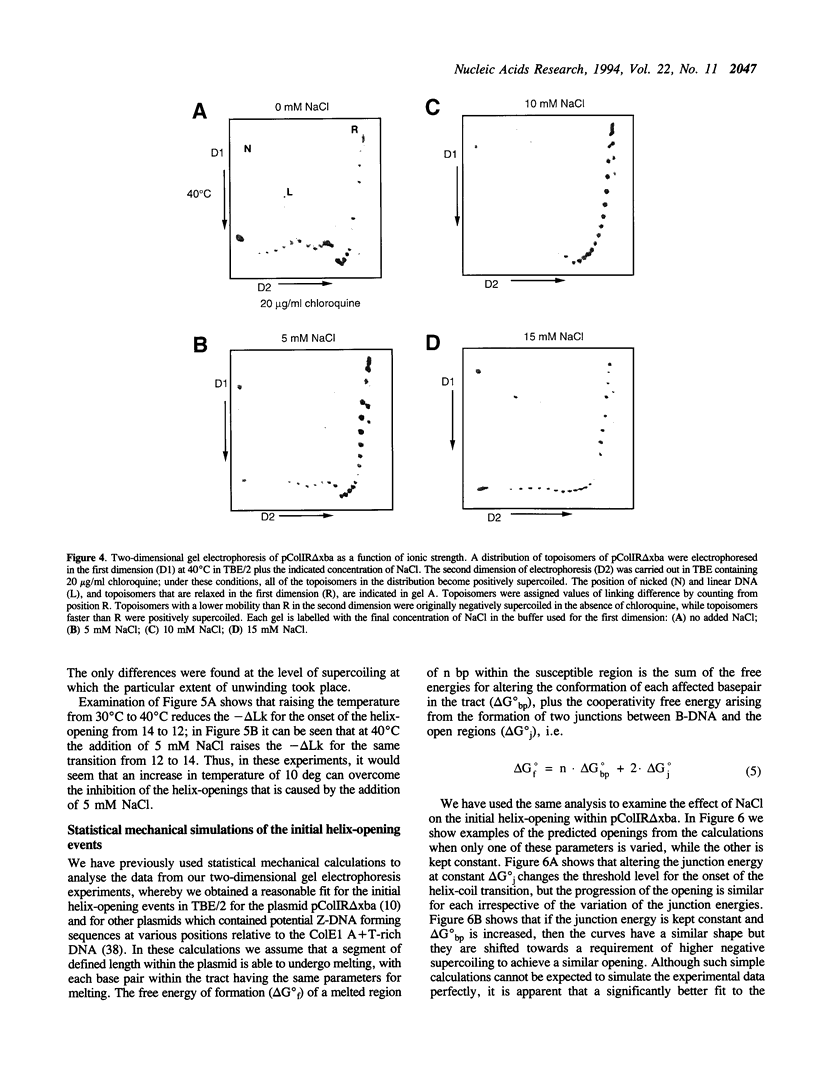
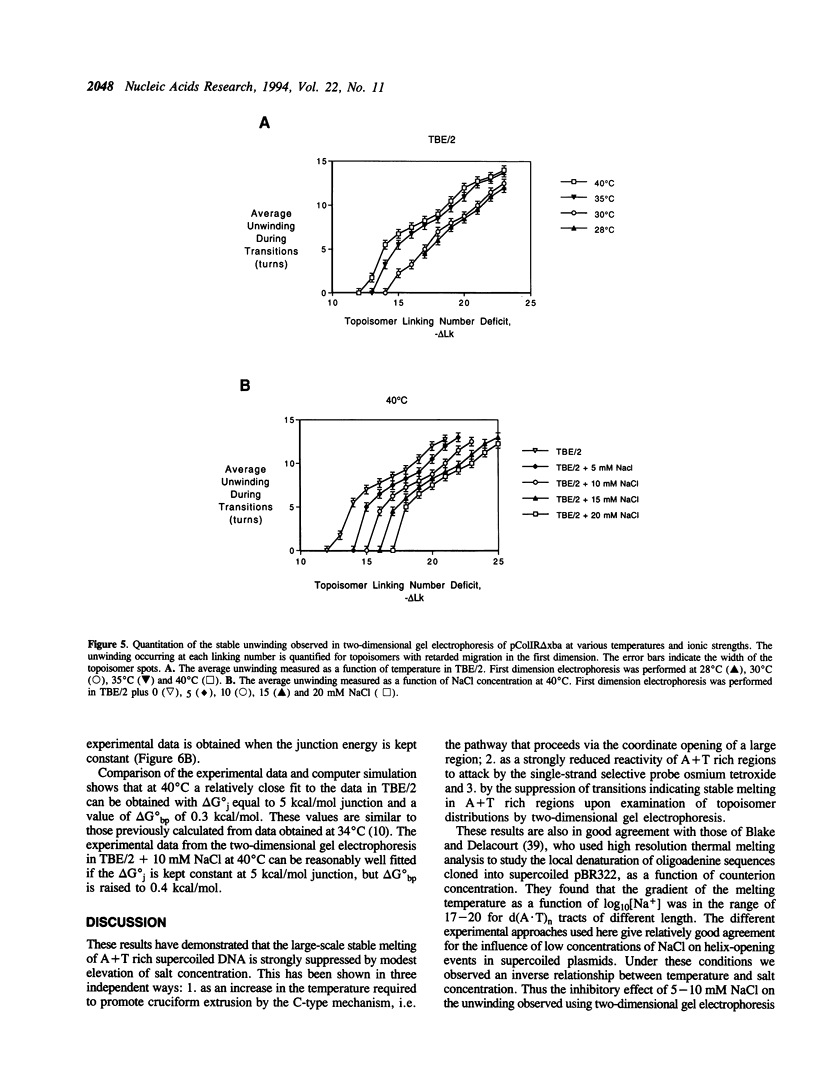
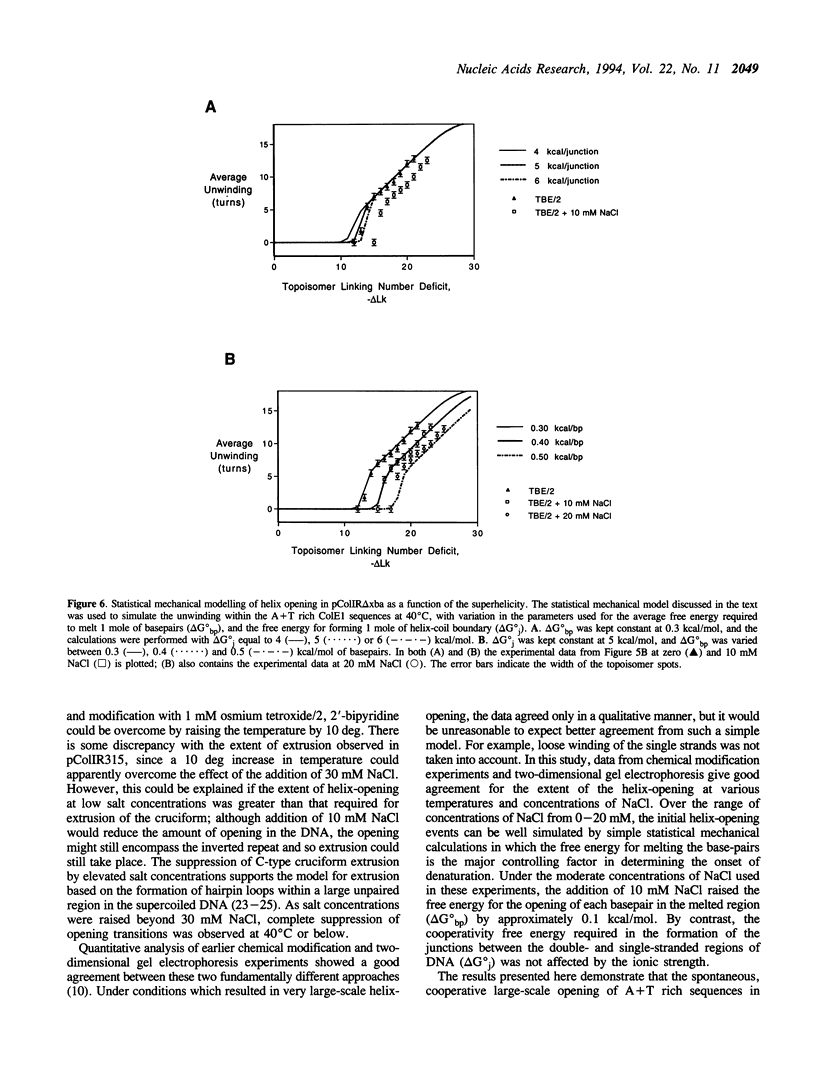
Large-scale cooperative helix opening has been previously observed in A + T rich sequences contained in supercoiled DNA molecules at elevated temperatures. Since it is well known that helix melting of linear DNA is suppressed by addition of salt, we have investigated the effects of added salts on opening transitions in negatively supercoiled DNA circles. We have found that localised large-scale stable melting in supercoiled DNA is strongly suppressed by modest elevation of salt concentration, in the range 10 to 30 mM sodium. This has been shown in a number of independent ways: 1. The temperature required to promote cruciform extrusion by the pathway that proceeds via the coordinate large-scale opening of an A + T rich region surrounding the inverted repeat (the C-type pathway, first observed in the extrusion of the ColE1 inverted repeat) is elevated by addition of salt. The temperature required for extrusion was increased by about 4 deg for an addition of 10 mM NaCl. 2. A + T rich regions in supercoiled DNA exhibit hyperreactivity towards osmium tetroxide as the temperature is raised; this reactivity is strongly suppressed by the addition of salt. At low salt concentrations of added NaCl (10 mM) we observe that there is an approximate equivalence between reducing the salt concentration, and the elevation of temperature. Above 30 mM NaCl the reactivity of the ColE1 sequences is completely supressed at normal temperatures. 3. Stable helix opening transitions in A + T rich sequences may be observed with elevated temperature, using two-dimensional gel electrophoresis; these transitions become progressively harder to demonstrate with the addition of salt. With the addition of low concentrations of salt, the onset of opening transitions shifts to higher superhelix density, and by 30 mM NaCl or more, no transitions are visible up to a temperature of 50 degrees C. Statistical mechanical simulation of the data indicate that the cooperativity free energy for the transition is unaltered by addition of salt, but that the free energy cost for opening each basepair is increased. These results demonstrate that addition of even relatively low concentrations of salt strongly suppress the large-scale helix opening of A + T rich regions, even at high levels of negative supercoiling. While the opening at low salt concentrations may reveal a propensity for such transitions, spontaneous opening is very unlikely under physiological conditions of salt, temperature and superhelicity, and we conclude that proteins will therefore be required to facilitate opening transitions in cellular DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Bowater R. P., Lilley D. M. Competing B-Z and helix-coil conformational transitions in supercoiled plasmid DNA. J Biol Chem. 1992 Jan 25;267(3):1776–1785. [PubMed] [Google Scholar]
- Bauer W. R., Benham C. J. The free energy, enthalpy and entropy of native and of partially denatured closed circular DNA. J Mol Biol. 1993 Dec 20;234(4):1184–1196. doi: 10.1006/jmbi.1993.1669. [DOI] [PubMed] [Google Scholar]
- Benham C. J. Energetics of the strand separation transition in superhelical DNA. J Mol Biol. 1992 Jun 5;225(3):835–847. doi: 10.1016/0022-2836(92)90404-8. [DOI] [PubMed] [Google Scholar]
- Benham C. J. Sites of predicted stress-induced DNA duplex destabilization occur preferentially at regulatory loci. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2999–3003. doi: 10.1073/pnas.90.7.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake R. D., Delcourt S. G. Electrostatic forces at helix-coil boundaries in DNA. Biopolymers. 1990 Feb 5;29(2):393–405. doi: 10.1002/bip.360290211. [DOI] [PubMed] [Google Scholar]
- Bode J., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992 Jan 10;255(5041):195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Bowater R., Aboul-Ela F., Lilley D. M. Two-dimensional gel electrophoresis of circular DNA topoisomers. Methods Enzymol. 1992;212:105–120. doi: 10.1016/0076-6879(92)12007-d. [DOI] [PubMed] [Google Scholar]
- Bowater R., Aboul-ela F., Lilley D. M. Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A + T)-rich regions that promote low-salt cruciform extrusion. Biochemistry. 1991 Dec 10;30(49):11495–11506. doi: 10.1021/bi00113a003. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Burd J. F., Wartell R. M., Dodgson J. B., Wells R. D. Transmission of stability (telestability) in deoxyribonucleic acid. Physical and enzymatic studies on the duplex block polymer d(C15A15) - d(T15G15). J Biol Chem. 1975 Jul 10;250(13):5109–5113. [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. Complete enzymatic replication of plasmids containing the origin of the Escherichia coli chromosome. J Biol Chem. 1986 Apr 25;261(12):5616–5624. [PubMed] [Google Scholar]
- Furlong J. C., Sullivan K. M., Murchie A. I., Gough G. W., Lilley D. M. Localized chemical hyperreactivity in supercoiled DNA: evidence for base unpairing in sequences that induce low-salt cruciform extrusion. Biochemistry. 1989 Mar 7;28(5):2009–2017. doi: 10.1021/bi00431a008. [DOI] [PubMed] [Google Scholar]
- Gille H., Messer W. Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 1991 Jun;10(6):1579–1584. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenwedel D. W., Hsu C. H., Lu D. S. The effects of aqueous neutral-salt solutions on the melting temperatures of deoxyribonucleic acids. Biopolymers. 1971;10(1):47–68. doi: 10.1002/bip.360100106. [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry. 1990 Oct 16;29(41):9551–9560. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Eddy M. J. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989 Dec 20;8(13):4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Natale D. A., Eddy M. J. Stable DNA unwinding, not "breathing," accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F. S., Bauer W. R. Temperature dependence of the gel electrophoretic mobility of superhelical DNA. Nucleic Acids Res. 1985 Mar 11;13(5):1665–1682. doi: 10.1093/nar/13.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The kinetic properties of cruciform extrusion are determined by DNA base-sequence. Nucleic Acids Res. 1985 Mar 11;13(5):1443–1465. doi: 10.1093/nar/13.5.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Bowater R., Aboul-ela F., Lilley D. M. Helix opening transitions in supercoiled DNA. Biochim Biophys Acta. 1992 May 7;1131(1):1–15. doi: 10.1016/0167-4781(92)90091-d. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989 Feb 24;56(4):521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer F., Yeramian E., Lilley D. M. Long-range structural effects in supercoiled DNA: statistical thermodynamics reveals a correlation between calculated cooperative melting and contextual influence on cruciform extrusion. Biopolymers. 1989 Aug;28(8):1449–1473. doi: 10.1002/bip.360280810. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. A dominant influence of flanking sequences on a local structural transition in DNA. Cell. 1986 Dec 5;47(5):817–827. doi: 10.1016/0092-8674(86)90524-6. [DOI] [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. Helix stability and the mechanism of cruciform extrusion in supercoiled DNA molecules. Nucleic Acids Res. 1988 Feb 11;16(3):1079–1093. doi: 10.1093/nar/16.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2486–2490. doi: 10.1073/pnas.87.7.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]