Abstract
Increased expression of the invasion- and metastasis-associated protein S100A4 is found in many types of cancer, but the regulation of S100A4 expression is poorly understood. The microenvironment surrounding tumors has a significant effect on tumor progression, and in the present study, we investigated the role of the microenvironment in the expression of S100A4. Tumors of three different human carcinoma cell lines were established in the tongue or skin of mice, and S100A4 expression was assessed by quantitative RT-PCR, Western blotting, and immunohistochemical analysis in tumors and stromal tissue and in cancer cells grown in vitro. Tongue tumors of the oral squamous cell carcinoma cell line HSC-4 showed a pronounced increase in S100A4 expression during tumor growth, whereas only a minor increase was detected in skin tumors of the same cell line. The S100A4 expression correlated with the methylation status of cytosine-guanine sites in the first intron of the gene. For all cell lines, S100A4 expression in the tumor stroma was related to the presence of inflammatory cells rather than to the level of S100A4 in the tumor cells.
In a tumor, extensive cross talk occurs between the malignant cancer cells and the “nonmalignant” stroma. Numerous studies have shown that tumor-associated stromal cells are important suppliers of cytokines, growth factors, and proteases that can facilitate tumor progression, invasion, and metastasis.1,2
The metastasis-promoting protein S100A4 belongs to the S100 family of calcium-binding proteins. Many studies have verified S100A4 as an important player in the metastatic process, and increased expression of the protein has been associated with poor prognosis in various human cancer types.3,4 The mechanism of the metastasis-promoting function of S100A4 is, however, not well-defined. The protein seems to have multiple intracellular and extracellular functions that may contribute to its prometastatic effects (reviewed in several papers5,6). S100A4 has been shown to be expressed in cancer cells7,8 and in several types of stromal cells, eg, fibroblasts, lymphocytes, and macrophages.9,10 The protein is also secreted from cell lines in vitro8,10,11 and can be detected in the tumor interstitial fluid,10,11 suggesting a role for S100A4 in the tumor-stroma interplay.
Squamous cell carcinomas (SCCs) can arise at several locations in the body. In the oral cavity, SCCs are characterized by an aggressive behavior, with frequent lymph node metastasis and poor prognosis.12 Skin SCCs usually have a much more benign course; they seldom metastasize, and very few patients die of the disease.13,14 This difference may be due to inherent properties of the cancer cells, but it may also be influenced by local factors in the oral cavity and the skin. To study the stromal effect on cancer growth and progression, we established a xenograft model where we inject human cancer cells into the tongue or skin of BALB/c nude mice. We previously found that the various cell lines formed larger tumors and behaved more aggressively when they grew in tongue compared with in skin, irrespective of organ of origin of the cancer cells.15 Tongue tumors showed, in contrast to skin tumors, an infiltrative growth pattern accompanied by increased proteolytic activity toward the invasive front.16 Lymph node metastases were detected from some of the tongue tumors but not from any of the skin tumors.15 Because S100A4 is reported to affect expression and activation of matrix metalloproteinases17 and to promote invasion and metastasis of cancer cells, we hypothesized that this protein could be differentially expressed in tongue and skin tumors in our xenograft model. In the present study, we investigated the role of the tumor microenvironment for expression of S100A4 in tumor cells and the surrounding stroma.
Materials and Methods
Cell Lines and Xenograft Tumors
The SCC9 cell line was purchased from American Type Culture Collection (Rockville, MD). Invitrogen's Flp-In Technology (Invitrogen, Carlsbad, CA) was used to establish SCC9 cells stably overexpressing S100A4 (SCC9/A4) or enhanced green fluorescent protein (SCC9/EGFP). HSC-4 was a kind gift from Prof. M. Yanagishita, Tokyo Medical and Dental University, Tokyo, Japan. The Ishikawa cell line was purchased from Sigma-Aldrich (St. Louis, MO), and UT-SCC-12A was generously provided by Prof. Reidar Grénman (University of Turku, Turku, Finland) and established as described.18 Cell lines and culture media are listed in Supplemental Table S1 (available at http://ajp.amjpathol.org). Xenograft tumors of the various cell lines were established in the tongue or skin of BALB/c nude mice as previously described.15 The mice were sacrificed after 5 to 28 days of tumor growth, and tumor tissue was either submerged in RNAlater (Sigma-Aldrich) or fixed in Zn-based fixative.19 Tissue/tumor samples stored in RNAlater were dissected under a dissecting microscope and were divided into three different fractions: i) tumor, ii) stroma close to tumor (<0.5 mm from the tumor), and iii) stroma further away from the tumor (>0.5 mm from the tumor).
Immunohistochemical Analysis and Immunoblotting
Immunohistochemical analysis was performed as previously described19 on xenograft tumors and cell lines grown in vitro using two different rabbit polyclonal antibodies against S100A4 (see Supplemental Table S2 at http://ajp.amjpathol.org). Immunohistochemical staining for S100A4 in the tumors was scored according to the method of Kobel et al,20 with staining intensity designated as nonexistent (0), weak (1), moderate (2), or strong (3). The number of positive cells was scored as no cells stained (0), <10% (1), 10% to 50% (2), 51% to 80% (3), or >80% (4). The final score was calculated by multiplication of these two variables. Stromal S100A4 staining was scored as negative (0), weak (1), moderate (2), or strong (3). For immunoblotting, all cell lysates were prepared using NuPAGE LDS Sample Buffer (Invitrogen) with 75 mmol/L dithiothreitol and was run on NuPAGE Novex 4% to 12% or 10% Bis-Tris gels (Invitrogen). Samples were blotted onto a Hybond ECL nitrocellulose membrane (Amersham Biosciences, Buckinghamshire, England), blocked in 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween 20, and probed with the antibodies listed in Supplemental Table S2 (available at http://ajp.amjpathol.org). Western Blotting Luminol Reagent (Santa Cruz Biotechnology Inc., Santa Cruz, CA) was used for detection.
RNA Extraction and RT-PCR
Tumor and stroma samples were homogenized in a TissueLyzer homogenizer (QIAGEN, Hilden, Germany) with 5-mm steel beads, and total RNA was extracted using the RNeasy Fibrous Tissue Mini Kit (QIAGEN) according to the manufacturer's instructions. RNA was also extracted from the cell lines grown in vitro using an RNeasy Mini Kit (QIAGEN) according to the protocol. On-column DNase treatment of the RNA samples was performed using QIAGEN's RNase-Free DNase according to the product manual. Quantity and purity of the extracted RNA was determined using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE), and RNA integrity was assessed using the Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA).
The mRNA expression levels in cell lines and tissue samples were quantified by quantitative RT-PCR performed using a Stratagene Mx3000P instrument (Stratagene, La Jolla, CA). Reverse transcription of total RNA was performed using a Reverse Transcriptase Core Kit (Eurogentec SA, Searing, Belgium) with random nonamer primers according to the manufacturer's recommendations using 100 ng of RNA per 10 μL of cDNA reaction.
For the analysis of S100A4, hydrolysis probe–based assays were used. Primer pairs and FAM/Dark quencher–labeled hydrolysis probes were designed using Primer Express v.2.0 (Applied Biosystems, Foster City, CA) and were synthesized by Eurogentec (Eurogentec SA). All the sequences are listed in Table 1. cDNA corresponding to 10 ng of RNA was amplified for 40 cycles in a 25-μL PCR mix (qPCR Mastermix Plus Low ROX; Eurogentec SA) containing a final concentration of 5 mmol/L MgCl2, 100 nmol/L probe, and 400 nmol/L of each primer. Cycling conditions: 95°C for 10 minutes, 40 cycles at 95°C for 30 seconds, and 60°C for 1 minute.
Table 1.
Sequences of Primers and Probes Synthesized by Eurogentec
| Gene symbol | Forward primer Reverse primer | Probe | Amplicon length (bp) | RefSeq accession no. |
|---|---|---|---|---|
| hS100A4 |
|
5′-CTGACCCGGGAGCTGCCCAG-3′ | 79 | NM_002961.2NM_019554.2 |
| hRPLP0 |
|
5′-TGGGCTCCAAGCAGATGCAGCAGAT-3′ | 149 | NM_001002.3NM_053275.3 |
| mS100A4 |
|
5′-TCAAGGAGCTACTGACCAGGGAGCTGC-3′ | 76 | NM_011311.2 |
| mPpia |
|
5′-CATGTGCCAGGGTGGTGACTTTACACG-3′ | 77 | NM_008907.1 |
| mNono |
|
5′-TAAGGATAAAGGCTTTGGCTTTATTCGCTTGG-3′ | 80 | NM_023144.2 |
h, human; m, mouse.
A SYBR Green (Applied Biosystems)–based method was used for analysis of human DNMT3b [DNA (cytosine-5)methyltransferase-3b]. Primers were purchased from QIAGEN (assay name: Hs_DNMT3B_1_SG; catalog number QT00032067). cDNA corresponding to 10 ng of RNA was amplified for 40 cycles in a 25-μL PCR mix (RT2 Real-Time SYBR Green/ROX PCR Master Mix (SA Biosciences, Frederick, MD). Cycling conditions: 95°C for 10 minutes, 40 cycles at 95°C for 30 seconds, 60°C for 1 minute, and 72°C for 1 minute.
Duplicate reverse transcription reactions were performed for each RNA sample, and duplicate PCR analyses were performed on each cDNA sample. Primer specificities and absence of primer dimers were determined by SYBR Green melting curve analysis and agarose gel electrophoresis. The absence of genomic DNA was confirmed by performing a no–reverse transcription control, and absence of contaminations was assessed by including a no-template control in every run.
The expression of 12 human and mouse candidate reference genes was tested using human and mouse housekeeping genes RT2 Profiler PCR Array (SA Biosciences), respectively. From this, hRPLP0 was chosen as the reference gene for the samples derived from human cell lines, and mPpia and mNono were chosen for the stromal samples. The ΔΔCq method21 was used to determine the relative amount of target mRNA in the different samples.
Methylation Analysis
To study the effect of DNA methylation on S100A4 expression in vitro, HSC-4, Ishikawa, and UT-SCC-12A cells were treated for 4 days with 1 μmol/L of the DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidine (Sigma-Aldrich). Fresh medium was added daily. For sequencing studies, genomic DNA was isolated from HSC-4 cells grown in vitro and from HSC-4 skin and tongue tumor tissue using the QIAmp DNA Mini Kit (QIAGEN) according to the protocol. DNA was bisulfite modified using the EpiTect Bisulfite Kit from QIAGEN following the manufacturer's recommendations. A set of previously published primers (S100A4-F: 5′-TGTTTTTGAGATGTGGGTTTG-3′ and S100A4-R: 5′-CACAATTACCTTCTACCTTTC-3′)22 was used to amplify a human-specific region in the first intron of the S100A4 gene, which encompasses three cytosine-guanine (CpG) sites whose methylation status has been found to be associated with S100A4 expression.22,23 PCR products were purified using a QIAquick PCR Purification Kit (QIAGEN) according to the instructions, and purified products were sequenced in both directions using the BigDye v.3.1 Terminator Cycle Sequencing Kit and the Applied Biosystems 3130xl Genetic Analyzer (both from Applied Biosystems, Warrington, England).
Statistical Analysis
Two-tailed t-tests were used to evaluate differences in S100A4 expression. A P < 0.05 was accepted as statistically significant.
Results
S100A4 Expression Was Up-Regulated in Tongue Tumors
To study the effect of the tumor microenvironment on S100A4 expression in tumor cells, we analyzed three different cell lines when grown in vitro and as xenograft tumors in tongue and skin. The cell lines originate from patients with tongue SCC (HSC-4), endometrial adenocarcinoma (Ishikawa), or skin SCC (UT-SCC-12A), thus enabling growth of orthotopic and heterotopic tumors.
Immunohistochemical staining of the HSC-4, Ishikawa, and UT-SCC-12A cell lines grown in vitro were found to be negative for S100A4 (Figure 1, A–C), whereas immunoblotting showed that all cell lines expressed low to moderate levels of S100A4 (Figure 1J).
Figure 1.
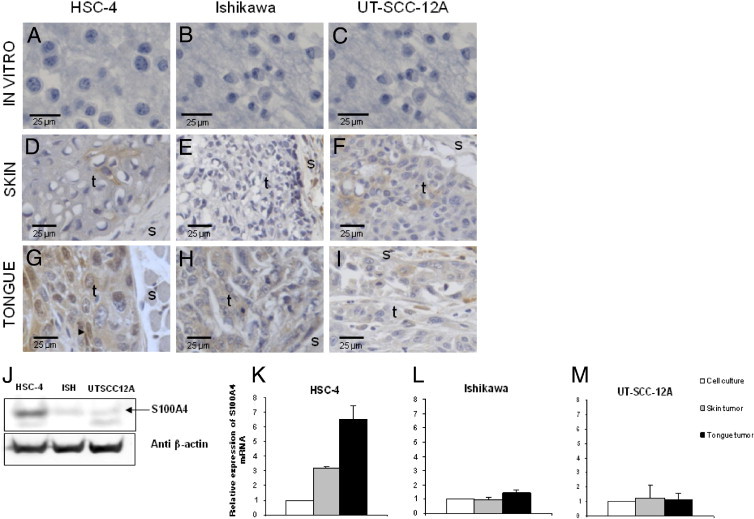
Increased S100A4 protein and mRNA expression levels in tumors compared with cells grown in vitro. Immunohistochemical S100A4 staining of the paraffin-embedded HSC-4 (A), Ishikawa (B), and UT-SCC-12A (C) cell lines grown in vitro and of skin (D, E, and F) and tongue (G, H, and I) tumors of the same cell lines. Representative areas of four to six parallel tumors are presented. S100A4 staining is shown in brown, and nuclei are stained blue. t, tumor area; and s, stromal area. Nuclear localization of S100A4 in an HSC-4 tongue tumor is indicated with an arrowhead in G. Tumors of the HSC-4 and Ishikawa cell lines were analyzed after 28 days of tumor growth, and tumors of the UT-SCC-12A cell line were analyzed after 15 days. J: Immunoblots of HSC-4, Ishikawa (ISH), and UT-SCC12A cells grown in vitro probed with anti-S100A4 and anti–β-actin. S100A4 mRNA levels in HSC-4 (K), Ishikawa (L), and UT-SCC-12A (M) analyzed by RT-PCR. Expression levels in tumors from skin (gray bars = skin tumor; n = 3) and tongue (black bars = tongue tumor; n = 3) were quantified relative to the expression levels in cells grown in vitro (white bars = cell culture; n = 1) Error bars represent SEM. All expression levels were normalized to expression of the reference gene RPLP0. RNA was isolated from tumors harvested after 15 days of tumor growth.
Skin and tongue tumors established from the three cell lines were also subjected to immunohistochemical staining for S100A4. The staining pattern and intensity were similar for the two S100A4 antibodies used, and Table 2 provides the immunohistochemical scores of the tumors. The skin tumors of the HSC-4 cell line showed focal weak S100A4 staining (Figure 1D). In contrast, tongue tumors of the same cell line had moderate to strong cytoplasmic staining in almost all tumor cells; in addition, nuclear staining was seen in approximately 20% of tumor cells (Figure 1G). Skin tumors of the Ishikawa cell line were negative for S100A4 (Figure 1E), whereas expression was induced in the tongue tumors, where cytoplasmic staining was seen toward the invasive front of the tumors (Figure 1H). In contrast to the two former cell lines, no significant difference in S100A4 expression was detected between tongue and skin tumors of the UT-SCC-12A cell line (Figure 1, F and I).
Table 2.
Quantification of S100A4 Expression Based on Immunohistochemical Staining of HSC-4 and Ishikawa (28 Days) and UT-SCC-12A (15 Days) Tongue and Skin Tumors
| Tumor cells (mean ± SEM) |
Stroma (mean ± SEM) |
|||||
|---|---|---|---|---|---|---|
| Cell line | Tongue | Skin | P value | Tongue | Skin | P value |
| HSC-4 (n = 4) | 6.0 ± 0.0 | 1.0 ± 0.6 | 0.003 | 1.0 ± 0.4 | 0.0 ± 0.0 | 0.050 |
| Ishikawa (n = 6) | 1.8 ± 0.5 | 0.0 ± 0.0 | 0.012 | 1.0 ± 0.4 | 0.7 ± 0.2 | 0.448 |
| UT-SCC-12A (n = 5) | 1.8 ± 1.1 | 1.4 ± 0.2 | 0.724 | 1.8 ± 0.4 | 1.2 ± 0.2 | 0.195 |
| Pooled tongue and skin tumors (n = 15) | 2.9 ± 0.6 | 0.7 ± 0.2 | 0.004 | 1.3 ± 0.2 | 0.7 ± 0.2 | 0.041 |
The relative amount of S100A4 mRNA in tongue and skin tumors was analyzed using RT-PCR. For the HSC-4 cell line, S100A4 mRNA levels in tongue tumors were up-regulated approximately twofold compared with those in skin tumors and approximately sixfold compared with those in cells grown in vitro (Figure 1K). For the Ishikawa cell line, mRNA levels were only slightly up-regulated in tongue compared with in vitro levels (Figure 1L), whereas tumors of the UT-SCC-12A cells had no increase in S100A4 mRNA levels compared with cells grown in vitro (Figure 1M).
S100A4 Expression Was Gradually Increased During Tumor Growth
Tongue tumors of the HSC-4 cell line obtained after 5, 10, 15, and 28 days of tumor growth were immunohistochemically stained for S100A4 to study the onset and progression of S100A4 protein expression. After 5 days in vivo, all the tumor cells were negative (Figure 2A), and some cells showed weak positive staining after 10 days (Figure 2B). The number of positive cells gradually increased, and at day 15, more than half of the tumor cells showed weak S100A4 staining (Figure 2C). After 28 days, most tumor cells revealed moderate to strong positive staining (Figure 2D). The staining of the tumors was scored (Figure 2E), and the increase in staining from day 5 to days 10, 15, and 28 was significant (P = 0.038, P = 0.004 and P = 0.003, respectively).
Figure 2.
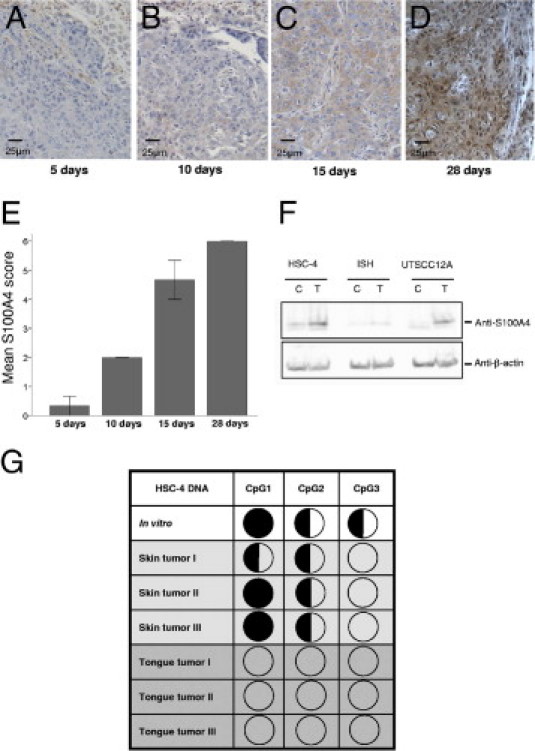
S100A4 expression is gradually increased during tumor growth and is dependent on hypomethylation of the gene. Immunohistochemical S100A4 staining of HSC-4 tongue tumors after 5 days (A), 10 days (B), 15 days (C), and 28 days (D) of tumor growth. Representative areas of three parallel tumors are presented. S100A4 staining is shown in brown, and nuclei are stained blue. E: Graph shows quantification of S100A4 staining at 5, 10, 15, and 28 days of tumor growth. n = 3 for all time points. Error bars represent SEM. F: Immunoblots of HSC-4, Ishikawa (ISH), and UT-SCC-12A cells cultured in the absence (control; C) or presence (treated; T) of 1 μmol/L 5-Aza-2′deoxycytidine, probed with anti-S100A4 and anti–β-actin. G: Methylation status of three CpG residues in the first intron of S100A4 in HSC-4 skin (n = 3) and tongue (n = 3) tumors and in cells grown in vitro determined by bisulphite sequencing and estimation of relative peak heights. Filled circles, >75% methylation; half-filled circles, 25% to 75% methylation; and empty circles, <25% methylation.
Hypomethylation of CpG Sites in the S100A4 Gene Increased Expression
S100A4 expression has previously been shown to be dependent on the methylation status of CpG sites in the first intron of the gene.22,24–26 To determine whether methylation status affected S100A4 expression in the cells used in the present study, cells were cultured in vitro in the presence of the DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidine. RT-PCR analyses showed that the relative amount of S100A4 mRNA increased approximately twofold in treated samples compared with in control samples of all three cell lines (data not shown). Immunoblotting revealed increased S100A4 protein expression in treated cells compared with control samples (Figure 2F), demonstrating the relevance of CpG methylation for S100A4 expression in the HSC-4, Ishikawa, and UT-SCC-12A cell lines.
To study whether hypomethylation was involved in the observed induction of S100A4 expression during tumor growth, the methylation status of three CpG sites in the first intron of the S100A4 gene was investigated in HSC-4 tongue and skin tumors and in HSC-4 cells grown in vitro. Bisulphite sequencing showed that methylation status correlated well with the observed expression patterns of S100A4; the highest methylation level was found in cells grown in vitro, and all three CpG sites were unmethylated in HSC-4 tongue tumors (Figure 2G).
The methylation of DNA is catalyzed by DNA methyltransferases (DNMTs), and DNMT3B is reported to be one of the main enzymes responsible for the establishment of new methylation patterns of DNA.27,28 To determine whether the expression level of this DNMT could explain the observed differences in methylation status between HSC-4 tongue and skin tumors, quantitative RT-PCR was used to quantify the relative amount of DNMT3B mRNA in the tumors. No significant difference in expression was found between tongue and skin tumors (data not shown), suggesting that other DNMTs or other mechanisms were involved in modification of the methylation pattern in the tumors.
S100A4 Expression in Tumor Stroma Was Not Correlated with S100A4 Levels in Tumor Cells but Rather with the Degree of Inflammation
Because stromal S100A4 also has been reported to affect the invasive and metastatic potential of cancer cells,11 stromal S100A4 expression was assessed in the xenograft model (Table 2). Prominent immunohistochemical S100A4 staining was seen in specific cells and in the extracellular matrix of the subepithelial connective tissue in normal tongue (Figure 3A) and skin (Figure 3B) of BALB/c nude mice. In general, staining was stronger in normal skin tissue compared with in normal tongue tissue.
Figure 3.
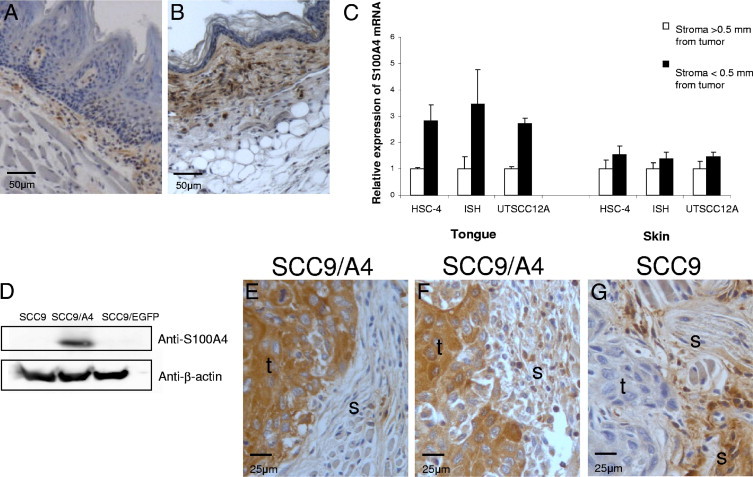
S100A4 expression is increased in tumor-associated stroma but is not correlated with S100A4 levels in the tumor cells. Sections of normal tongue (A) and skin (B) tissue from BALB/C nude mice were immunohistochemically stained for S100A4. Representative areas of the samples are presented. S100A4 staining is shown in brown, and nuclei are stained blue. C: S100A4 mRNA levels in tumor-associated stroma in tongue and skin were analyzed by RT-PCR. Expression levels in stroma close to the tumors of HSC-4, Ishikawa (ISH), and UT-SCC-12A (black bars; n = 3) were quantified relative to the expression levels in stroma further away (>0.5 mm) from the tumors (white bars; n = 3). All expression levels were normalized to the expression of the reference genes Ppia and Nono. Error bars represent SEM. D: Immunoblots of cell lysates from SCC9, SCC9/A4, and SCC9/EGFP cells probed with anti-S100A4 and anti–β-actin. Immunohistochemical S100A4 staining of tongue tumors of the SCC9/A4 cell line with sparse stromal staining (E) and more pronounced stromal staining (F) and of the SCC9 cell line with strong stromal staining (G). Representative areas of three parallel tumors are presented; S100A4 staining is shown in brown, and nuclei are stained blue. t, tumor area; and s, stromal area.
RT-PCR was used to compare the S100A4 mRNA expression levels in the stromal tissue close to and further away (>0.5 mm) from the tumors. S100A4 expression was approximately twice as high in skin as in tongue further away from the tumors (data not shown); however, the stromal cells close to the tumors showed a larger increase in S100A4 mRNA expression in tongue than in skin (Figure 3C). Similarly, immunohistochemical S100A4 staining was stronger in stroma close to tongue and skin tumors of all the cell lines compared with that seen in normal tongue and skin tissues. The increase was slightly higher in stroma surrounding tongue tumors compared with stroma surrounding skin tumors; see the immunohistochemical scores in Table 2. The intensity of stromal staining varied considerably between the parallel tumors of each cell line and did not correlate with the staining intensity of the cancer cells but rather with the level of inflammation. In a previous study using the same xenograft tumors, we found that the inflammatory infiltrate was more pronounced in tongue tumors compared with in the corresponding skin tumors for all three cell lines. Neutrophils were the predominant cell type in the inflammatory infiltrate,15 and in the present study, stromal S100A4 staining was mainly seen in and surrounding cells with morphologic features similar to those of neutrophils and macrophages (data not shown).
Previous studies have shown that some cell lines release S100A4 into the growth medium in vitro. The protein can also be detected in tumor interstitial fluid,10,29 but the source of extracellular S100A4 in the tumor is unknown. From the tongue SCC cell line SCC9, we successfully established a cell line stably overexpressing S100A4 (SCC9/A4) (Figure 3D). To study whether S100A4 in tumor cells affected stromal S100A4 levels, by either secreting the protein or inducing S100A4 expression in stromal cells, SCC9/A4 cells were used to establish tongue tumors in the xenograft model. Cells overexpressing EGFP (SCC9/EGFP) were used as a control. Very faint S100A4 bands were detected on immunoblots of conditioned media from the S100A4-overexpressing cells, indicating some release from the SCC9/A4 cell line in vitro (data not shown). Tumors of the SCC9/A4 cell line showed intense S100A4 staining in the cancer cells (Figure 3, E and F). However, only focal stromal staining was seen in areas with sparse inflammation (Figure 3E), whereas much more staining was seen in areas with a more pronounced inflammatory reaction (Figure 3F). Tumors of the parental SCC9 cell line showed weak S100A4 staining in the cancer cells, and the stromal staining was strong in areas with a pronounced inflammatory reaction (Figure 3G). Tumors of the control cell line SCC9/EGFP showed the same staining pattern as the SCC9 tumors (data not shown). Thus, S100A4 expression in the tumor-associated stromal cells was unaffected by S100A4 expression levels in the cancer cells but rather was correlated with the degree of inflammation.
Discussion
Tumor-stroma interactions are known to be important for cancer progression. Using a xenograft model where the same cancer cell lines were used to establish tumors in tongue and skin, we previously found that the cells show remarkably different growth patterns in the two environments.15,16 In the present study, we investigated the role of the tumor microenvironment in regulation of the metastasis-associated protein S100A4. We found increased expression of S100A4 in tongue tumors of the oral SCC cell line HSC-4 and the endometrial adenocarcinoma cell line Ishikawa compared with the corresponding skin tumors. In contrast, tumors of the skin SCC cell line UT-SCC-12A showed similar S100A4 expression in tongue and skin.
The signaling events regulating S100A4 expression are not well characterized, but several studies have shown that S100A4 expression is regulated epigenetically by methylation of CpG dinucleotides in the first intron of the gene.23,25,30 Hypomethylation of these cytosine residues is thought to be involved in cancer-associated up-regulation of S100A4 expression.22,23,25,30 In the present study, we found increased expression of S100A4 in the cell lines after treatment with a DNA methyltransferase inhibitor in vitro. When examining the intronic sequence in DNA from HSC-4 tongue and skin tumors and HSC-4 cells grown in vitro, we also found a correlation between methylation status and S100A4 expression, showing that hypomethylation was involved in the observed induction of S100A4 expression in the tumors. Several studies have demonstrated DNA hypermethylation of cancer cells grown in vitro,31,32 and oral SCC cell lines have shown a higher propensity to become hypermethylated than have cell lines isolated from other cancer types.32 This may explain the observation that the oral SCC cell line HSC-4 showed the strongest induction of S100A4 in vivo. S100A4 was recently shown to be down-regulated in epidermal cancers compared with expression in normal epidermal tissue.33 DNA sequencing of cancer cells isolated from epidermal cancers revealed methylation of the regulatory CpG dinucleotides in the first intron of the S100A4 gene. This epigenetic silencing of S100A4 expression in skin cancers may contribute to their generally low metastatic potential. The report is in accordance with our observation that xenograft skin tumors expressed low levels of S100A4, although normal skin tissue of the mice showed stronger S100A4 expression than did normal tongue tissue. The results indicate that factors in the tumor microenvironment affect the DNA methylation status of the cancer cells. These findings also have clinical implications because DNA methyltransferase inhibitors have been recognized as promising candidate anticancer drugs because of their ability to reactivate expression of tumor suppressor genes silenced by aberrant methylation.28,34 The present findings demonstrate that inhibition of methyltransferases can activate prometastatic genes, raising concerns that the treatment could actually promote metastasis.
S100A4 expression has also been shown to be induced by some extracellular growth factors, eg, epidermal growth factor, basic fibroblast growth factor, and transforming growth factor-β.35,36 Proteolytic enzymes, including plasmin and matrix metalloproteinases, can release and activate growth factors stored in the extracellular matrix, such as latent transforming growth factor-β and basic fibroblast growth factor.37,38 In a previous study using the same xenograft tumors, we found increased proteolytic activity in tongue tumors compared with skin tumors.16 This may yield higher levels of biologically active transforming growth factor-β and basic fibroblast growth factor with a subsequent induction of S100A4 expression in the tongue tumors. Because S100A4 is known to induce increased expression and activation of several matrix metalloproteinases,17 a positive feedback loop may exist.
S100A4 expression in tumor stroma is also shown to contribute to increased invasion and metastasis of cancer cells. This was demonstrated in a study where tumors of highly metastatic carcinoma cells did not metastasize in S100A4 knockout mice, but the metastatic phenotype was partly restored by co-injecting the cancer cells with S100A4-expressing stromal cells.11 In the xenograft model, tongue tumors of the UT-SCC-12A cell line were the most aggressive, with lymph node metastases detected in half of the mice.15 Still, S100A4 staining of these tumors was not prominent and did not differ significantly from staining of the noninvasive skin tumors of the same cell line. However, UT-SCC-12A tongue tumors provoked a pronounced inflammatory reaction, and this was accompanied by marked stromal S100A4 expression. This indicates that S100A4 expression in the tumor stroma may be as important for metastases as S100A4 expression in the tumor cells.
In the present study, the staining intensity of S100A4 in tumor and stroma did not correlate. Instead, stromal S100A4 staining reflected the degree of inflammation and was seen within and outside tumor-associated inflammatory cells. Although immunodeficient mice (Charles River Laboratories, Wilmington, MA) were used in the present experiments, they do have B cells, natural killer cells, and all the cells of the innate immune system. High numbers of tumor-associated inflammatory cells, such as macrophages and neutrophils, are associated with poor prognosis in several cancer types. Because many inflammatory cells are reported to express S100A4,10,39 a role as suppliers of stromal S100A4 may contribute to their cancer-promoting effect.
The present findings show that the microenvironment can affect S100A4 expression in cancer cells by regulating the methylation of CpG units in the gene. The ability of tongue stroma to increase S100A4 expression in cancer cells, and the increased number of tumor-associated, S100A4-expressing inflammatory cells, may contribute to the aggressive behavior of oral SCCs. This study, therefore, merits further investigation of the significance of S100A4 expression in patients with oral cancer.
Acknowledgments
We thank Dr. Kjetil Boye, Ph.D., for critical reading and Dr. Peter McCourt, Ph.D., for critical reading and linguistic revision of the manuscript.
Footnotes
Supported by grants from The Norwegian Cancer Society, The North Norwegian Regional Health Authorities, The Erna and Olav Aakre Foundation for Cancer Research, and The University of Tromsø.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.01.022.
Contributor Information
Hilde Ljones Wetting, Email: hilde.ljones@uit.no.
Elin Hadler-Olsen, Email: elin.hadler-olsen@uit.no.
Supplemental data
References
- 1.Mueller M.M., Fusenig N.E. Friends or foes: bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R., Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 3.Rudland P.S., Platt-Higgins A., Renshaw C., West C.R., Winstanley J.H., Robertson L., Barraclough R. Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res. 2000;60:1595–1603. [PubMed] [Google Scholar]
- 4.Gongoll S., Peters G., Mengel M., Piso P., Klempnauer J., Kreipe H., von Wasielewski R. Prognostic significance of calcium-binding protein S100A4 in colorectal cancer. Gastroenterology. 2002;123:1478–1484. doi: 10.1053/gast.2002.36606. [DOI] [PubMed] [Google Scholar]
- 5.Garrett S.C., Varney K.M., Weber D.J., Bresnick A.R. S100A4, a mediator of metastasis. J Biol Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- 6.Sherbet G.V. Metastasis promoter S100A4 is a potentially valuable molecular target for cancer therapy. Cancer Lett. 2009;280:15–30. doi: 10.1016/j.canlet.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 7.Ebralidze A., Tulchinsky E., Grigorian M., Afanasyeva A., Senin V., Revazova E., Lukanidin E. Isolation and characterization of a gene specifically expressed in different metastatic cells and whose deduced gene product has a high degree of homology to a Ca2+-binding protein family. Genes Dev. 1989;3:1086–1093. doi: 10.1101/gad.3.7.1086. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi N., Horiuchi A., Osada R., Imai T., Wang C., Chen X., Konishi I. Nuclear expression of S100A4 is associated with aggressive behavior of epithelial ovarian carcinoma: an important autocrine/paracrine factor in tumor progression. Cancer Sci. 2006;97:1061–1069. doi: 10.1111/j.1349-7006.2006.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt-Hansen B., Klingelhofer J., Grum-Schwensen B., Christensen A., Andresen S., Kruse C., Hansen T., Ambartsumian N., Lukanidin E., Grigorian M. Functional significance of metastasis-inducing S100A4(Mts1) in tumor-stroma interplay. J Biol Chem. 2004;279:24498–24504. doi: 10.1074/jbc.M400441200. [DOI] [PubMed] [Google Scholar]
- 10.Cabezon T., Celis J.E., Skibshoj I., Klingelhofer J., Grigorian M., Gromov P., Rank F., Myklebust J.H., Maelandsmo G.M., Lukanidin E., Ambartsumian N. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int J Cancer. 2007;121:1433–1444. doi: 10.1002/ijc.22850. [DOI] [PubMed] [Google Scholar]
- 11.Grum-Schwensen B., Klingelhofer J., Berg C.H., El-Naaman C., Grigorian M., Lukanidin E., Ambartsumian N. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 2005;65:3772–3780. doi: 10.1158/0008-5472.CAN-04-4510. [DOI] [PubMed] [Google Scholar]
- 12.Funk G.F., Karnell L.H., Robinson R.A., Zhen W.K., Trask D.K., Hoffman H.T. Presentation, treatment, and outcome of oral cavity cancer: a National Cancer Data Base report. Head Neck. 2002;24:165–180. doi: 10.1002/hed.10004. [DOI] [PubMed] [Google Scholar]
- 13.Miller D.L., Weinstock M.A. Nonmelanoma skin cancer in the United States: incidence. J Am Acad Dermatol. 1994;30:774–778. doi: 10.1016/s0190-9622(08)81509-5. [DOI] [PubMed] [Google Scholar]
- 14.McGuire J.F., Ge N.N., Dyson S. Nonmelanoma skin cancer of the head and neck, I: histopathology and clinical behavior. Am J Otolaryngol. 2009;30:121–133. doi: 10.1016/j.amjoto.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Hadler-Olsen E., Wetting H.L., Rikardsen O., Steigen S.E., Kanapathippillai P., Grenman R., Winberg J.O., Svineng G., Uhlin-Hansen L. Stromal impact on tumor growth and lymphangiogenesis in human carcinoma xenografts. Virchows Arch. 2010;457:677–692. doi: 10.1007/s00428-010-0980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadler-Olsen E., Wetting H.L., Ravuri C., Omair A., Rikardsen O., Svineng G., Kanapathippillai P., Winberg J.O., Uhlin-Hansen L. Organ specific regulation of tumour invasiveness and gelatinolytic activity at the invasive front. Eur J Cancer. 2011;47:305–315. doi: 10.1016/j.ejca.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Elenjord R., Ljones H., Sundkvist E., Loennechen T., Winberg J.O. Dysregulation of matrix metalloproteinases and their tissue inhibitors by S100A4. Connect Tissue Res. 2008;49:185–188. doi: 10.1080/03008200802143125. [DOI] [PubMed] [Google Scholar]
- 18.Grenman R., Pekkola-Heino K., Joensuu H., Aitasalo K., Klemi P., Lakkala T. UT-MUC-1, a new mucoepidermoid carcinoma cell line, and its radiosensitivity. Arch Otolaryngol Head Neck Surg. 1992;118:542–547. doi: 10.1001/archotol.1992.01880050096023. [DOI] [PubMed] [Google Scholar]
- 19.Hadler-Olsen E., Kanapathippillai P., Berg E., Svineng G., Winberg J.O., Uhlin-Hansen L. Gelatin in situ zymography on fixed, paraffin-embedded tissue: zinc and ethanol fixation preserve enzyme activity. J Histochem Cytochem. 2010;58:29–39. doi: 10.1369/jhc.2009.954354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobel M., Weichert W., Cruwell K., Schmitt W.D., Lautenschlager C., Hauptmann S. Epithelial hyaluronic acid and CD44v6 are mutually involved in invasion of colorectal adenocarcinomas and linked to patient prognosis. Virchows Arch. 2004;445:456–464. doi: 10.1007/s00428-004-1095-0. [DOI] [PubMed] [Google Scholar]
- 21.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Rosty C., Ueki T., Argani P., Jansen M., Yeo C.J., Cameron J.L., Hruban R.H., Goggins M. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tulchinsky E., Grigorian M., Tkatch T., Georgiev G., Lukanidin E. Transcriptional regulation of the mts1 gene in human lymphoma cells: the role of DNA-methylation. Biochim Biophys Acta. 1995;1261:243–248. doi: 10.1016/0167-4781(95)00013-7. [DOI] [PubMed] [Google Scholar]
- 24.Rehman I., Goodarzi A., Cross S.S., Leiblich A., Catto J.W., Phillips J.T., Hamdy F.C. DNA methylation and immunohistochemical analysis of the S100A4 calcium binding protein in human prostate cancer. Prostate. 2007;67:341–347. doi: 10.1002/pros.20401. [DOI] [PubMed] [Google Scholar]
- 25.Xie R., Loose D.S., Shipley G.L., Xie S., Bassett R.L., Jr., Broaddus R.R. Hypomethylation-induced expression of S100A4 in endometrial carcinoma. Mod Pathol. 2007;20:1045–1054. doi: 10.1038/modpathol.3800940. [DOI] [PubMed] [Google Scholar]
- 26.Dokun O.Y., Florl A.R., Seifert H.H., Wolff I., Schulz W.A. Relationship of SNCG, S100A4, S100A9 and LCN2 gene expression and DNA methylation in bladder cancer. Int J Cancer. 2008;123:2798–2807. doi: 10.1002/ijc.23893. [DOI] [PubMed] [Google Scholar]
- 27.Luczak M.W., Jagodzinski P.P. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 28.Kelly T.K., De Carvalho D.D., Jones P.A. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen K.B., Andersen K., Fodstad O., Maelandsmo G.M. Sensitization of interferon-γ induced apoptosis in human osteosarcoma cells by extracellular S100A4. BMC Cancer. 2004;4:52. doi: 10.1186/1471-2407-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsey J.C., Lusher M.E., Anderton J.A., Gilbertson R.J., Ellison D.W., Clifford S.C. Epigenetic deregulation of multiple S100 gene family members by differential hypomethylation and hypermethylation events in medulloblastoma. Br J Cancer. 2007;97:267–274. doi: 10.1038/sj.bjc.6603852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 32.Smiraglia D.J., Rush L.J., Fruhwald M.C., Dai Z., Held W.A., Costello J.F., Lang J.C., Eng C., Li B., Wright F.A., Caligiuri M.A., Plass C. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10:1413–1419. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Liu Z.L., Zhang K.L., Chen X.Y., Kong Q.Y., Wu M.L., Sun Y., Liu J., Li H. Methylation-associated silencing of S100A4 expression in human epidermal cancers. Exp Dermatol. 2009;18:842–848. doi: 10.1111/j.1600-0625.2009.00922.x. [DOI] [PubMed] [Google Scholar]
- 34.Issa J.P., Kantarjian H.M. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada H., Danoff T.M., Kalluri R., Neilson E.G. Early role of Fsp1 in epithelial-mesenchymal transformation. Am J Physiol. 1997;273:F563–F574. doi: 10.1152/ajprenal.1997.273.4.F563. [DOI] [PubMed] [Google Scholar]
- 36.Strutz F., Zeisberg M., Ziyadeh F.N., Yang C.Q., Kalluri R., Muller G.A., Neilson E.G. Role of basic fibroblast growth factor-2 in epithelial-mesenchymal transformation. Kidney Int. 2002;61:1714–1728. doi: 10.1046/j.1523-1755.2002.00333.x. [DOI] [PubMed] [Google Scholar]
- 37.Yu Q., Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- 38.Folgueras A.R., Pendas A.M., Sanchez L.M., Lopez-Otin C. Matrix metalloproteinases in cancer: from new functions to improved inhibition strategies. Int J Dev Biol. 2004;48:411–424. doi: 10.1387/ijdb.041811af. [DOI] [PubMed] [Google Scholar]
- 39.Takenaga K., Nakamura Y., Sakiyama S. Expression of a calcium binding protein pEL98 (mts1) during differentiation of human promyelocytic leukemia HL-60 cells. Biochem Biophys Res Commun. 1994;202:94–101. doi: 10.1006/bbrc.1994.1898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


