Abstract
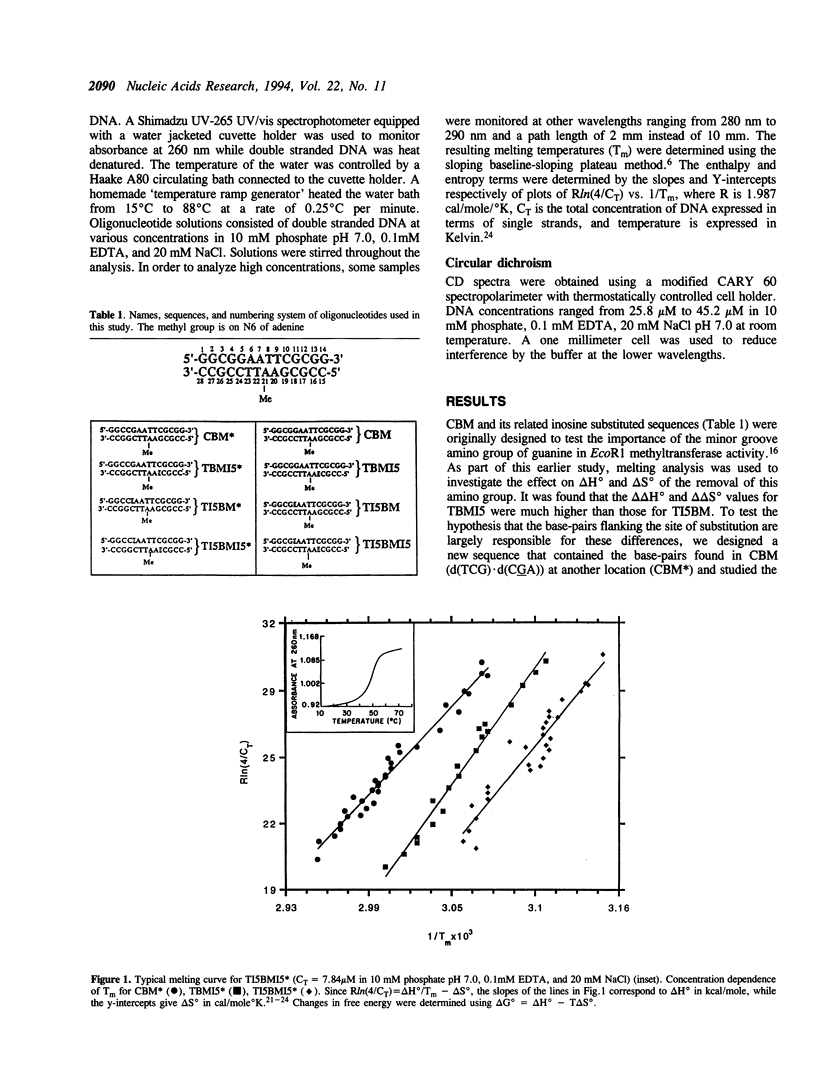
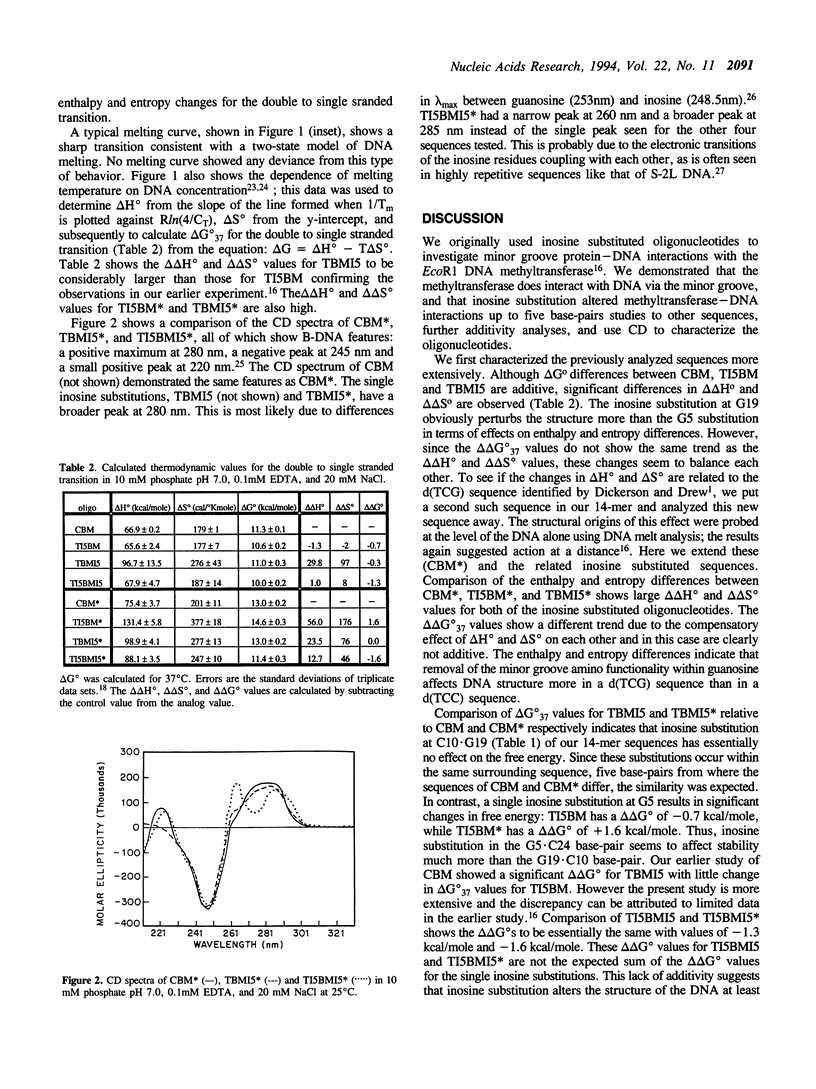
The effect of replacing a G.C base-pair with an I.C base-pair on DNA stability was investigated for a related set of 14-mers. DNA melting analysis of the 14-mers was used to determine delta Hzero, delta Szero and delta G(zero)37 of the double to single stranded transition. All 14mers were shown to have B-DNA character by circular dichroism analysis. 14mers substituted with a single inosine in place of guanosine at different positions showed that consequences on DNA stability are sequence-dependent. Large changes in delta Hzero and delta Szero result when inosine is substituted within the trinucleotide sequence d(TCG).d(CGA) while substitution within d(TCC).d(GGA) causes minor changes in enthalpy and entropy. Moreover, some 14-mers with two inosine substitutions five base-pairs apart showed non-additive free energy changes for the double to single stranded transition. These results clearly indicate that the structural consequences of replacing a single guanosine with an inosine are transmitted over a significant distance.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the cleavage by the EcoRI restriction endonuclease of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7270–7278. [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Corfield P. W., Hunter W. N., Brown T., Robinson P., Kennard O. Inosine.adenine base pairs in a B-DNA duplex. Nucleic Acids Res. 1987 Oct 12;15(19):7935–7949. doi: 10.1093/nar/15.19.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosstick R., Li X., Tuli D. K., Williams D. M., Connolly B. A., Newman P. C. Molecular recognition in the minor groove of the DNA helix. Studies on the synthesis of oligonucleotides and polynucleotides containing 3-deaza-2'-deoxyadenosine. Interaction of the oligonucleotides with the restriction endonuclease EcoRV. Nucleic Acids Res. 1990 Aug 25;18(16):4771–4778. [PMC free article] [PubMed] [Google Scholar]
- Cruse W. B., Aymani J., Kennard O., Brown T., Jack A. G., Leonard G. A. Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs. Nucleic Acids Res. 1989 Jan 11;17(1):55–72. doi: 10.1093/nar/17.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Diekmann S., McLaughlin L. W. DNA curvature in native and modified EcoRI recognition sites and possible influence upon the endonuclease cleavage reaction. J Mol Biol. 1988 Aug 20;202(4):823–834. doi: 10.1016/0022-2836(88)90561-x. [DOI] [PubMed] [Google Scholar]
- Falsafi S., Reich N. O. Molecular dynamics simulations of B-DNA: an analysis of the role of initial molecular configuration, randomly assigned velocity distribution, long integration times, and nonconstrained termini. Biopolymers. 1993 Mar;33(3):459–473. doi: 10.1002/bip.360330312. [DOI] [PubMed] [Google Scholar]
- Fliess A., Wolfes H., Rosenthal A., Schwellnus K., Blöcker H., Frank R., Pingoud A. Role of thymidine residues in DNA recognition by the EcoRI and EcoRV restriction endonucleases. Nucleic Acids Res. 1986 Apr 25;14(8):3463–3474. doi: 10.1093/nar/14.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran T. E., Joachimiak A., Sigler P. B. The DNA target of the trp repressor. EMBO J. 1992 Aug;11(8):3021–3030. doi: 10.1002/j.1460-2075.1992.tb05372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashibara K. C., Verdine G. L. Template-directed interference footprinting of cytosine contacts in a protein-DNA complex: potent interference by 5-aza-2'-deoxycytidine. Biochemistry. 1992 Nov 24;31(46):11265–11273. doi: 10.1021/bi00161a002. [DOI] [PubMed] [Google Scholar]
- Johnson B. B., Dahl K. S., Tinoco I., Jr, Ivanov V. I., Zhurkin V. B. Correlations between deoxyribonucleic acid structural parameters and calculated circular dichroism spectra. Biochemistry. 1981 Jan 6;20(1):73–78. doi: 10.1021/bi00504a013. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Jr Circular dichroism and its empirical application to biopolymers. Methods Biochem Anal. 1985;31:61–163. doi: 10.1002/9780470110522.ch2. [DOI] [PubMed] [Google Scholar]
- Karslake C., Botuyan M. V., Gorenstein D. G. 31P NMR spectra of oligodeoxyribonucleotide duplex lac operator-repressor headpiece complexes: importance of phosphate ester backbone flexibility in protein-DNA recognition. Biochemistry. 1992 Feb 18;31(6):1849–1858. doi: 10.1021/bi00121a038. [DOI] [PubMed] [Google Scholar]
- Lesser D. R., Kurpiewski M. R., Waters T., Connolly B. A., Jen-Jacobson L. Facilitated distortion of the DNA site enhances EcoRI endonuclease-DNA recognition. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7548–7552. doi: 10.1073/pnas.90.16.7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipanov A., Kopka M. L., Kaczor-Grzeskowiak M., Quintana J., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA. Biochemistry. 1993 Feb 9;32(5):1373–1389. doi: 10.1021/bi00056a024. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987 Sep;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- Modrich P., Rubin R. A. Role of the 2-amino group of deoxyguanosine in sequence recognition by EcoRI restriction and modification enzymes. J Biol Chem. 1977 Oct 25;252(20):7273–7278. [PubMed] [Google Scholar]
- Newman P. C., Williams D. M., Cosstick R., Seela F., Connolly B. A. Interaction of the EcoRV restriction endonuclease with the deoxyadenosine and thymidine bases in its recognition hexamer d(GATATC). Biochemistry. 1990 Oct 23;29(42):9902–9910. doi: 10.1021/bi00494a021. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Poncin M., Hartmann B., Lavery R. Conformational sub-states in B-DNA. J Mol Biol. 1992 Aug 5;226(3):775–794. doi: 10.1016/0022-2836(92)90632-t. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Reich N. O., Danzitz M. J., Jr Non-additivity of sequence-specific enzyme-DNA interactions in the EcoRI DNA methyltransferase. Nucleic Acids Res. 1991 Dec 11;19(23):6587–6594. doi: 10.1093/nar/19.23.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992 Apr 10;256(5054):217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- Schroeder S. A., Roongta V., Fu J. M., Jones C. R., Gorenstein D. G. Sequence-dependent variations in the 31P NMR spectra and backbone torsional angles of wild-type and mutant Lac operator fragments. Biochemistry. 1989 Oct 17;28(21):8292–8303. doi: 10.1021/bi00447a006. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Vorlicková M., Khudyakov IYa, Hejtmánková I., Kypr J. Circular dichroism studies of salt- and alcohol- induced conformational changes in cyanophage S-2L DNA which contains amino 2 adenine instead of adenine. J Biomol Struct Dyn. 1991 Aug;9(1):81–85. doi: 10.1080/07391102.1991.10507894. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]



